-
CONSTANS(CO)基因是植物响应光周期调控的重要基因,位于生物钟的输出途径上,能正调控下游开花基因SOC1和FT,进而调控植物开花。PUTTERILL等[1]首先在拟南芥Arabidopsis thaliana中分离出CO基因,反转录PCR(RT-PCR)检测到CO基因在根和叶中表达。ONOUCHI等[2]对花椰菜Brassica oleracea花叶病毒35S(Cauliflower mosaic virus 35S, CaMV 35S)融合CO(35S::CO)转化拟南芥研究发现,CO蛋白会诱导早花和丧失光周期敏感性。进一步研究发现[3],CO在染色体上的位置介于生物节律钟基因和下游开花基因之间,可将光信号转变为开花信号。对拟南芥CO基因过表达研究[1]发现,CO基因过表达的植株比野生型提前开花,表明CO蛋白的活性决定开花时间;但这种调控在不同成员间并不一致,过表达COL1和COL2对植株开花时间没有影响[4],过表达COL9则导致开花延迟,但COL9缺失突变体在长日照下又表现为早花,说明COL9不但抑制CO基因表达调控开花时间,同时下调FT的表达水平从而延迟成花转变[5]。COL3在拟南芥光形态建成时起正调控作用,促进侧根生长和花色素苷积累,并调节长日照敏感植物的花芽分化[6]。从形态来看,CO基因常以多拷贝的形式存在,如拟南芥的CO家族有17个成员[7],水稻Oryza sativa中有16个成员[8],甘蓝型油菜Brassica napus中也克隆到4个CO同源基因[9]。但各CO家族成员的功能存在明显差异。葡萄Vitis vinifera的VvCOL1主要在芽休眠过程中起作用,表明该基因参与光周期,控制芽休眠的诱导和维持[10]。拟南芥中过表达衣藻Chlamydomonas reinhardtii的CrCO会表现出早花表型,结合衣藻的研究发现:CrCO对淀粉的合成和细胞分裂有调节功能,推测CO在高等植物中可能仍保持调节淀粉合成[11]。大麦Hordeum vulgare的HvCO1和Hd1基因与CO亲缘关系最近,可以通过激活HvFT1诱导大麦开花[8],但在转基因拟南芥中则丢失该功能[12]。拟南芥co突变体过表达牵牛花Ipomoea nil的PnCO基因可促进植物开花[13]。黑麦草Lolium perenne的LpCO可以互补拟南芥co突变体的晚花表型[14],甜菜Beta vulgaris的BvCO1可以修复拟南芥co-2突变体的晚花表型[15]。大豆Glycine max的GmCO9影响根的发育,与种子的成熟密切相关[16]。毛果杨Populus trichocarpa的PtCO促使植株提前开花,也可调控植株的生长和芽的分化[17]。本研究以14个已被测序的物种为试验材料,通过生物信息学手段,从外显子-内含子结构、基因重复、基因差异表达分析等3个方面开展CO家族研究,为探讨不同家族成员的潜在功能提供依据。
-
从植物基因组数据Phytozome()中下载其中13个物种的全基因组序列、蛋白质及对应编码序列(coding sequence, CDS),分别为藻类植物1种,苔藓植物1种,蕨类植物1种,被子植物9种(长日照植物、短日照植物和日中性植物各3种),以无油樟Amborella trichopoda作为被子植物的对照。此外裸子植物1种(挪威云杉Picea abies),其相应序列来源于。
数据查找步骤:①从PFAM蛋白质数据库获得CO结构域的隐马尔可夫模型(PF06203和PF00643)并作为查询序列,得到的数据储存于Windows平台环境下构建的各个物种全基因组氨基酸序列的本地数据库中。②利用HMMER软件包的hmmsearch程序,默认参数条件下在本地数据库进行BLASTP搜索,筛选出符合E-value≤0.01的蛋白质序列作为CO候选同源蛋白。③将备选CO基因的CDS序列通过BLASTN的比对,在全基因组核酸序列中搜索,获得CO在染色体上准确定位信息。④在PFAM蛋白质数据库和SMART蛋白质数据库下对搜索得到的所有同源候选基因蛋白质序列进行鉴定,剔除不含CO(PF06203和PF00643)结构域的氨基酸序列。以此完成各个物种CO家族所有成员的鉴定。
-
利用MUSCLE的默认参数进行蛋白质多序列比对分析;使用MEGA 7.0对完成比对的蛋白质序列构建系统进化树;构建方法选用邻近法(NJ);距离模型采用泊松矫正;空位缺失数据的处理采用两两删除;系统发育统计的可靠性检测采用bootstrap分析,使用1 000次重复。
-
利用在线软件GSDS(Gene Structure Display Server)比较CO家族成员的CDS序列和基因序列,分析CO家族基因的外显子-内含子组成和分布,结合系统发育分析,探究CO在基因结构上的进化规律。
-
利用植物基因组PGDD数据库()搜索染色体上的共线性片段,分析含有的CO基因的共线性区段,研究CO家族不同成员的相互联系和进化过程。
-
搜索14个物种的国际核酸序列数据库(NCBI,https://www.ncbi.nlm.nih.gov/),发现水稻关于繁殖发育的转录组数据最为完整。利用GEO数据库(GSE56463)下载水稻8个不同时期不同部位(花芽、花、开花前的旗叶、开花后的旗叶、开花前的根、开花后的根、未成熟种子、成熟种子)的植物组织转录组数据(RNA-seq)。转录组数据全部采用FPKM标准化后的值。以水稻为例,从转录水平重点分析CO家族不同成员在花发育和种子形成过程中的表达变化,从而探讨它们可能的生物学功能。
-
搜索14个物种蛋白质数据库中的CO家族成员,共鉴定出159个含有CO结构域的蛋白质序列。结果表明:CO常以多拷贝的形式存在于植物中,与以往的研究一致[8]。拷贝数最多的物种是大豆,鉴定出25个家族成员。其次在胡萝卜Daucus carota,小立碗藓Physcomitrella patens,菜豆Phaseolus vulgaris,番茄Solanum lycopersicum,黄瓜Cucumis sativus和蓖麻Ricinus communis中,分别鉴定到18,15,13,13,12和10个拷贝。在小麦,无油樟和卷柏Selaginella moellendorffii中,也发现了5,5和4个CO基因家族成员。挪威云杉和衣藻中拷贝数最少,各存在3个成员。
-
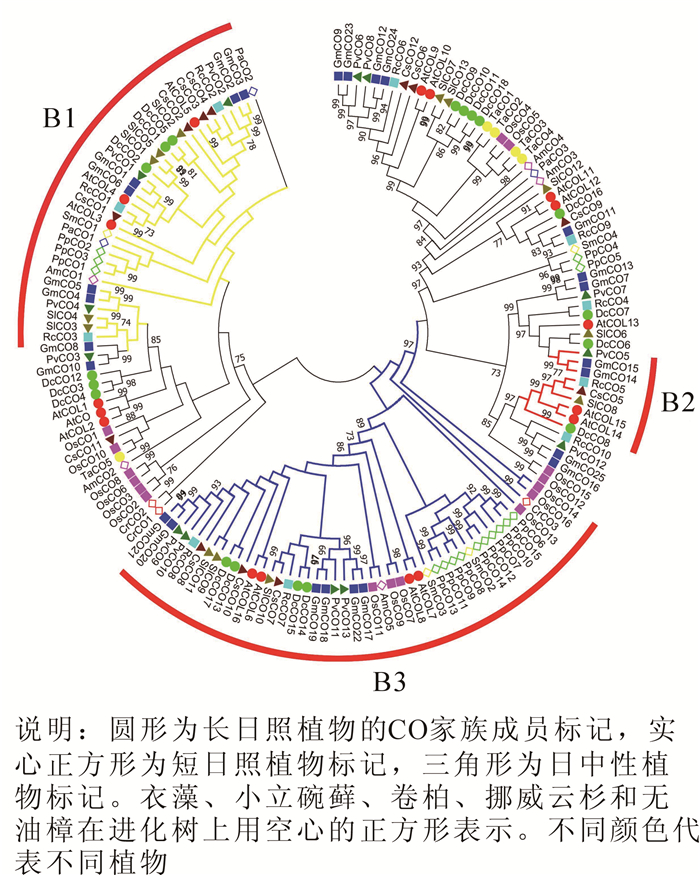
使用邻近法对得到的14个物种159个CO蛋白序列构建系统发育进化树。由生成的无根系统进化树(图 1)可知,植物CO家族在进化中具有多样性,大部分同一谱系的物种基因都能聚类在同一进化枝上。根据结构域特征,选取支持度高且结构稳定的3个亚家族(分别命名为B1,B2和B3)作为后续研究CO基因的基础框架。其中B1亚家族含2个B-box结构域和1个CCT结构域;B2亚家族含1个B-box结构域,1个CCT结构域和1个锌指结构;B3亚家族含1个B-box结构域和1个CCT结构域。
-
CO家族成员的预测结构(图 2)显示,多数物种的CO基因中存在2~4个外显子,同一亚家族内基因的外显子-内含子结构和长度高度相似,说明这些物种的CO基因家族成员之间的亲缘关系也较近,同时也证实了CO基因家族系统进化树的可信度。具体而言,B1亚家族中的大多数基因含2个外显子,1个内含子;B2亚家族中则表现为每个CO基因含4个外显子和3个内含子,且排列相位表现出“0,0,2”规律;B3亚家族相对较为复杂,多数的CO基因含有2个外显子,部分含有4个外显子,但也有例外,如CrCO3基因,不仅长度较大,还发现存在14个外显子和13个内含子,体现在系统进化树中则出现CrCO3分化为独立的进化枝。研究还发现,CO基因包含大量的相位为0的内含子,表明外显子改组可能在CO基因的进化中起着一定的作用,而外显子的插入和内含子的删除可以认为是形成多元化的CO基因以及功能差异的CO蛋白的原因之一。
-
对拟南芥、菜豆、蓖麻3种植物CO家族的基因复制事件的研究可以用来检测CO基因家族中遗传差异间的联系和相应的扩张模式,CO家族的成员可作为锚定基因研究所在染色体区段的分子进化历史。染色体定位分析(图 3,图 4)表明,绝大多数的CO基因在基因组中是随机分布的,仅在少数位点形成串联的基因簇,因此大规模的基因复制事件可能在CO基因家族的进化过程中扮演着重要的角色。共线性分析发现,拟南芥、菜豆和蓖麻的染色体区域间存在强烈且保守的共线性。由图 3和图 4可知:拟南芥CO家族所在的共线性区域最多有22对基因;菜豆和蓖麻内部也存在广泛的共线性情况,例如PvCO5-PvCO7,RcCO5-RcCO10;此外,拟南芥与菜豆和蓖麻之间的共线性情况也非常普遍,例如AtCOL3-RcCO1,AtCOL3-PvCO1,PvCO7-RcCO4。根据这些基因的共线性分析结果可推测,CO家族中重复基因的扩张与基因组重复有着密切的联系。
-
基因表达的差异性反映了基因的功能分化。水稻转录组表达谱显示(图 5),CO基因在花芽、花、根、旗叶和种子中都有表达,以OsCO3,OsCO6,OsCO7,OsCO8,OsCO9,OsCO11,OsCO12和OsCO16这8个基因的表达量较高,尤其是在开花后的根和开花前后的旗叶中的表达量更为明显。具体来看,OsCO3和OsCO6基因在花芽到花的转变过程、开花前后的根和旗叶中的表达量升高,在根中的表达量升高,在乳粒(未成熟的种子)到成熟种子的过程中表达量下降,说明OsCO3和OsCO6基因负向调控花芽到花的转变过程。OsCO8在花芽到花的转变过程、开花前后的根、开花前后的旗叶和乳粒到成熟种子的过程中表达量都呈现上升趋势,说明OsCO8对花的发育以及果实成熟有重要的调控作用。OsCO12在花芽到花的转变过程、开花前后的根和乳粒到成熟种子的过程中表达量上升,在开花前后的旗叶中的表达量下降,说明OsCO12对水稻花的发育起着最为关键的调控作用。OsCO7,OsCO9和OsCO11基因在花芽到花的转变过程、开花前后的旗叶和乳粒到成熟种子的过程中表达量下降,在开花前后的根中的表达量明显上升,说明OsCO7,OsCO9和OsCO11基因可以正向调控花芽到花的转变过程。OsCO16在花芽到花的转变过程中表达量下降,在开花前后的根和旗叶、乳粒到成熟种子的过程中表达量上升,说明OsCO16对花的发育和果实成熟有重要的调控作用。水稻中不同CO基因在不同时期不同组织器官中的表达量不同,表明同一家族不同基因之间存在功能上的差异。
-
CO基因是植物光周期途径中调控开花时间的重要基因。光周期途径中,PHYA,CRY1和CRY2基因相互作用,影响GI等生物节律钟基因,促进CO基因的表达;CO编码转录因子作用于FT[18],使FT从维管束组织转移到茎顶端分生组织,致使花器官发育[19]。通过对14个物种的CO基因的分析,本研究发现:CO基因常以多拷贝的形式存在于植物中,与已有研究结果一致[20];亲缘关系较近的物种,其CO基因的相似性较高;CO基因在裸子植物和被子植物、双子叶植物与单子叶植物、不同科和不同属植物之间都存在明显分化,表明CO基因在植物进化中既相对保守又不断进化,其进化过程与整个物种进化过程相对同步,说明CO基因可能对植物进化起到了重要作用。研究发现:单子叶植物发生过2次基因组重复[21],一半的水稻基因组基因来源于基因组重复[22]。对水稻的基因表达分析发现:CO基因在花、叶、根和茎中都有表达,OsCO3的表达量在花芽到花的转变过程中上升,推测OsCO3负调控花芽到花的转变过程,与KIM等[23]发现的OsCO3通过负调控Hd3a和FT-like(FTL)的表达延迟短日照下水稻开花的结果一致;OsCO7基因在花芽到花的转变过程中表达量下降,说明OsCO7正调控Hd3a的表达,促进短日照下水稻的开花,与XUE等[24]研究结果一致。在短日照条件下,水稻的Hd1抑制Hd3a的转录从而控制开花转型[25],这一结果和拟南芥CO基因在短日照条件下促进FT的表达控制开花转型相反,说明CO基因对于花芽到花的发育起到重要调控作用,进一步证实了水稻不同CO基因在功能上存在差异。本研究结果有助于更加深入地了解CO基因家族成员的潜在功能,为CO基因在光周期途径中调控成花发育过程提供理论依据。
Evolution of the flowering time gene CONSTANS in a photoperiod pathway
-
摘要: CONSTANS(CO)是植物响应光周期调节的重要基因和监测日照长度的重要元件,可将光信号和生物钟信号转变为开花信号,激活下游基因(FT)的表达,从而诱导植物开花。选取14个已被测序的物种,采用生物信息学手段,从外显子-内含子结构、基因重复、基因差异表达等方面开展CO基因家族研究。结果表明:14个物种共鉴定到159个CO家族成员,CO基因常以多拷贝的形式存在,多数含2~4个外显子,在进化过程中表现出多样性。CO家族重复基因的扩张与基因组重复相关。CO在水稻Oryza sativa根、旗叶、花和种子中均有表达,花芽到花的转变过程中OsCO3的表达量上升,而OsCO7下降,说明水稻CO家族成员之间存在功能差异。Abstract: CONSTANS(CO), an essential element for monitoring the duration of a day and an important gene responding to photoperiod regulation, can transform light and biological clock signals into flowering signals, as well as activate the expression of downstream genes (FT), which induces plant flowering. However, their evolutionary history and patterns have not been examined systematically. In this study, a total of 14 species, whose whole-genomes have been sequenced, were selected. Then, via the analyses of bioinformatics, the CO family was studied in regards to exon-intron structure, gene duplication, and differential expression. Results showed 159 members in the CO family. For each plant species, the CO family existed in the form of multiple copies. Almost every copy contained 2 to 4 exons indicating diversity in the evolution process. The synteny analysis showed that expansion of duplicate genes in the CO family was related to genome duplication. In addition, the gene differential expression analysis showed that CO members were expressed in roots, flag leaves, flowers, and seeds of rice (Oryza sativa). Especially, OsCO3 was up-regulated in the process of floral development; whereas, the expression of OsCO7 was down-regulated. This suggested that members of the CO gene family played different roles in rice.
-
Key words:
- botany /
- CO /
- CO family /
- phylogenetic /
- synteny analysis /
- gene duplication
-
-
[1] PUTTERILL J, ROBSON F, LEE K, et al. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors[J]. Cell, 1995, 80(6):847-857. [2] ONOUCHI H, IGEÑO M I, PÉRILLEUX C, et al. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes[J]. Plant Cell, 2000, 12(6):885-900. [3] SUÁREZ-LÓPEZ P, WHEATLEY K, ROBSON F, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis[J]. Nature, 2001, 410(6832):1116-1120. [4] LEDGER S, STRAYER C, ASHTON F, et al. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes[J]. Plant J, 2001, 26(1):15-22. [5] CHENG Xiaofei, WANG Zengyu. Over expression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana[J]. Plant J, 2005, 43(5):758-768. [6] DATTA S, HETTIARACHCHI G H C M, DENG Xingwang, et al. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth[J]. Plant Cell, 2006, 18(1):70-84. [7] ROBSON F, COSTA M M R, HEPWORTH S R, et al. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants[J]. Plant J, 2001, 28(6):619-631. [8] GRIFFITHS S, DUNFORD R P, COUPLAND G, et al. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis[J]. Plant Physiol, 2003, 131(4):1855-1867. [9] ROBERT L S, ROBSON F, SHARPE A, et al. Conserved structure and function of the Arabidopsis flowering time gene CONSTANS in Brassica napus[J]. Plant Mol Biol, 1998, 37(5):763-772. [10] ALMADA R, CABRERA N, CASARETTO J A, et al. VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds[J]. Plant Cell Rep, 2009, 28(8):1193-1203. [11] TENORIO G, OREA A, ROMERO J M, et al. Oscillation of mRNA level and activity of granule-bound starch synthase in Arabidopsis leaves during the day/night cycle[J]. Plant Mol Biol, 2003, 51(6):949-958. [12] CAMPOLI C, DROSSE B, SEARLE I, et al. Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS[J]. Plant J, 2012, 69(5):868-880. [13] HAYAMA R, AGASHE B, LULEY E, et al. A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis[J]. Plant Cell, 2007, 19(10):2988-3000. [14] MARTIN J, STORGAARD M, ANDERSEN C H, et al. Photoperiodic regulation of flowering in perennial ryegrass involving a CONSTANS-like homolog[J]. Plant Mol Biol, 2004, 56(2):159-169. [15] CHIA T Y P, MÜLLER A, JUNG C, et al. Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus[J]. J Exp Bot, 2008, 59(10):2735-2748. [16] HUANG Guowen, MA Jinghua, HAN Yuzhen, et al. Cloning and expression analysis of the soybean CO-Like gene GmCOL9[J]. Plant Mol Biol Rep, 2011, 29:352-359. [17] BÖHLENIUS H, HUANG Tao, CHARBONNEL-CAMPAA L, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees[J]. Science, 2006, 312(5776):1040-1043. [18] PAZHOUHANDEH M, MOLINIER J, BERR A, et al. MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis[J]. Proc Nat Acad Sci USA, 2011, 108(8):3430-3435. [19] AHN J H, MILLER D, WINTER V J, et al. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1[J]. EMBO J, 2006, 25(3):605-614. [20] LAGERCRANTZ U, AXELSSON T. Rapid evolution of the family of CONSTANS LIKE genes in plants[J]. Mol Biol Evol, 2000, 17(10):1499-1507. [21] TANG Haibao, BOWERS J E, WANG Xiyin, et al. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage[J]. Proc Nat Acad Sci USA, 2010, 107(1):472-477. [22] YU Jun, WANG Jun, LIN Wei, et al. The genomes of Oryza sativa:a history of duplications[J]. PLoS Biol, 2005, 3(2):266-281. [23] KIM S K, YUN C H, LEE J H, et al. Os-CO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-LIKE genes under SD conditions in rice[J]. Planta, 2008, 228(2):355-365. [24] XUE Weiya, XING Yongzhong, WENG Xiaoyu, et al. Natural variation in Ghd7 in an important regulator of heading date and yield potential in rice[J]. Nat Genet, 2008, 40(6):761-767. [25] KOJIMA S, TAKAHASHI Y, KOBAYASHI Y, et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions[J]. Plant Cell Physiol, 2002, 43(10):1096-1105. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.2019.01.002







 下载:
下载: