-
干旱是限制植物生长、发育以及作物产量的最主要非生物胁迫之一[1]。水分亏缺不仅会导致细胞膨压下降,还会引发植物体的一系列形态和生理改变,包括气孔关闭、活性氧(ROS)积累以及光合作用受抑等[2]。为应对干旱胁迫,植物形成了多层次的适应策略来维持体内水分平衡和氧化还原稳态[3−4]。
黄酮类化合物(flavonoids)是一类广泛存在于植物中的次生代谢产物,主要包括黄酮类(flavones),黄酮醇类(flavonols)和花青素(anthocyanidins)等[5]。黄酮类化合物具有显著的抗氧化活性和ROS清除能力,被认为是非酶类抗氧化系统的重要组成部分[6]。已有研究表明:黄酮类化合物通过其显著的抗氧化活性,在植物应对干旱胁迫的生理响应中扮演着至关重要的角色。如干旱可诱导银杏Ginkgo biloba和沙棘Hippophae rhamnoide体内黄酮类化合物含量升高,并伴随着黄酮类合成相关结构基因表达的上调[7−8];干旱胁迫下,玉米Zea mays耐旱突变体doi57保卫细胞中积累的黄酮醇较野生型更多,而过氧化氢(H2O2)更少[9];在拟南芥Arabidopsis thalian中,异源过表达类黄酮合成酶编码基因或转录因子(AmDEL和PgF3H)显著促进黄酮类化合物的积累,并增强植株的干旱耐受性[10−11]。黄酮类化合物生物合成的转录调控机制已在多种植物中得到阐释。该过程受到多类别转录因子构成的复杂网络精密调控,其中MYB、bHLH、WD40等家族成员通过形成MYB-bHLH-WD40(MBW)三元复合体,协同激活或抑制下游结构基因的表达,从而精确调控黄酮类物质的合成时序、组织分布及积累水平[12]。其中,R2R3-MYB转录因子在不同类黄酮分支途径中发挥核心调控作用,如拟南芥 AtMYB12/AtPFG1和AtMYB75/AtPAP1的单独和同时过表达均可提升黄酮醇含量并增强抗旱性[13],而pfg3和pfg3-d1突变体则表现出黄酮类化合物(尤其是黄酮醇类)的积累缺陷,因而对干旱和渗透胁迫更为敏感[14]。除MYB家族外,bHLH和WD40蛋白作为MBW复合体的关键组件,不仅能够增强MYB转录因子的DNA结合特异性与稳定性,还广泛参与光信号、激素(如茉莉酸甲酯)及非生物胁迫(如干旱、低温)应答,从而将内外源信号整合至黄酮类合成途径中[15],如光照可诱导梨Pyrus pyrifolia愈伤组织黄酮类物质积累[16]。此外,其他转录因子如WRKY、NAC、ERF等也被报道通过直接或间接方式参与该调控网络,进一步丰富了类黄酮生物合成的转录调控维度,如葡萄Vitis vinifera中过表达VvNAC17可以诱导黄酮类化合物积累[17]。可见,黄酮类化合物的生物合成受到多层次、多因子协同的转录调控,其中R2R3-MYB转录因子(尤其是SG7亚家族)通过靶向关键酶基因(如FLS)启动子,在精确调控黄酮醇合成中扮演核心角色,为作物品质改良及合成生物学应用提供了重要的靶点基因与理论依据[18]。
太行菊Opisthopappus taihangensis为菊科Asteraceae太行菊属Opisthopappus多年生草本植物。已有研究表明:在干旱胁迫下,蒸腾速率、气孔导度、光合速率等多种指标显示太行菊的抗旱能力明显强于其他几种广义菊属Chrysanthemum植物,是优良的抗旱种质[19],菊花脑C. nankingense在形态上较太行菊更快萎蔫[20]。野菊C. indicum过表达CiFNSII可以抑制黄酮醇合成并提高黄酮合成,从而增强植株抗旱性[21]。而太行菊的抗旱能力是否与黄酮类化合物的积累有关尚不明确,其干旱响应基因也有待深入挖掘。本研究通过测定太行菊和菊花脑在模拟干旱胁迫下黄酮类化合物含量及种类的变化,探究类黄酮在太行菊抗旱中的作用;并结合系统发育分析、实时荧光定量PCR(RT-qPCR)和亚细胞定位实验,对黄酮类合成相关转录因子OtMYB12的表达进行鉴定和检测,以解析它们在太行菊抗旱中的功能。本研究不仅可揭示黄酮类化合物在太行菊干旱胁迫响应过程中的作用,还可为耐干旱菊花新品种的创制提供可利用的基因资源。
-
选用抗旱种质太行菊和干旱敏感种质菊花脑为研究材料,均栽培于温室。2种材料的扦插苗先在体积比为1∶1的蛭石与珍珠岩基质中生根,随后移栽至体积比为6∶3∶1的草炭、珍珠岩与松针土基质中培养45 d。培养条件为光周期12 h光照/12 h黑暗、温度25 ℃、相对湿度60%。
-
选取生长状态良好且一致的植株,将扦插苗的根部用去离子水清洗干净后转入霍格兰德(Hoagland)营养液培养,缓苗7 d后进行干旱胁迫模拟。干旱模拟采用200 g·L−1聚乙二醇6000 (PEG 6000),并设置复水处理。共设7个处理:0、3、6、9、12、24 h干旱胁迫和胁迫后复水24 h。每个处理分别对叶片和根系进行取样,部分样品经锡纸包裹和液氮速冻后,保存于−80 ℃超低温冰箱,用于RT-qPCR检测;部分样品经阴干、粉碎、过40目筛后,保存于干燥器,用于黄酮类化合物测定。每个处理设置3个生物学重复。
-
芦丁标准曲线建立参考刘海芳等[22]并稍做改动,具体步骤如下:精准称取芦丁对照品20.00 mg,溶解于体积分数为1%的盐酸-甲醇溶液,制备成0.2 mg·mL−1的储备液,于4 ℃保存备用。采用硝酸铝络合-紫外分光光度法建立标准曲线,将储备液稀释为不同质量浓度(0、0.008、0.016、0.024、0.032、0.040、0.048 mg·mL−1)的标准品,在紫外可见分光光度计中于波长510 nm处分别检测吸光度。用最小二乘法进行线性拟合,得质量浓度(y)与吸光度(x)的线性回归方程为y=13.576x+0.003 6 (R2=0.999 8)。
-
精准称取5种类黄酮类标准品(芦丁、木犀草素、槲皮素、山奈酚和芹菜素)各10.00 mg,分别溶解于甲醇制备成的0.2 mg·mL−1储备液。随后,将储备液稀释成20 ng·mL−1的混标工作液,用于高效液相色谱(HPLC)分析。检测条件如下:色谱柱为ZORBAX Eclipse XDB-C18柱,柱温为30 ℃,流速为1.0 mL·min−1,进样量为10 μL,检测波长为254 nm。流动相A相为甲醇,B相为体积分数为0.01%的磷酸水溶液,梯度洗脱程序如下:0~40 min,体积分数为25%的A;40~41 min,62.5%~100%A;41~56 min,100%A;56~57 min,100%~25%A;57~67 min,25%A,以实现5种标准品的良好分离。为确保测定准确性,在全部样品分析完成后,根据各化合物的峰面积选定其标准曲线的线性范围,并建立线性回归方程用于定量分析(表1)。
表 1 5种类黄酮类物质的线性回归方程
Table 1. Linear regression equations of 5 flavonoids
化合物 回归方程 R2 线性范围/(mg·L−1) 芦丁 y=27.124x+48.556 0.999 8 1.10~550.00 槲皮素 y=53.12x−13.131 0.999 6 0.05~25.00 木犀草素 y=25.646x+59.855 0.999 7 0.40~200.00 山奈酚 y= 3.8849 x−4.2011 0.999 3 0.02~10.00 芹菜素 y=29.03x− 3.7862 0.999 6 0.032~16.000 说明:x为经HPLC分析获得的峰面积,y为黄酮类物质的质量浓度。 供试色谱仪:Agilent1100高效液相色谱仪。供试试剂:芦丁(153-18-4,HPLC≥98%)、木犀草素(491-70-3,HPLC≥98%)、槲皮素(117-39-5,HPLC≥98%)、山奈酚(520-18-3,HPLC≥98%)、芹菜素(520-36-5,HPLC≥98%),均购自上海源叶公司。色谱纯甲醇(67-56-1)、磷酸(7664-38-2)购自艾览公司;超纯水由超纯水机CASCADA制得。
-
分别精准称取0.20 g太行菊及菊花脑样品,加入10 mL 体积分数为1%的盐酸-甲醇溶液4 ℃静置4 h,离心过滤后用体积分数为1%的盐酸-甲醇溶液定容至25 mL容量瓶中,为样品溶液。平行制备3份样品溶液,将样品溶液浓度稀释至40%,按1.3.1方法测定吸光度,根据芦丁线性回归方程计算。
-
黄酮类物质提取及检测参考孙翊等[23]并稍做改动,具体步骤如下:分别精准称取0.20 g太行菊及菊花脑样品,加入1.5 mL 体积分数为1%的盐酸-甲醇溶液4 ℃避光提取24 h,每8 h震荡1次。离心后0.22 μm微孔滤膜过滤,为样品溶液。平行制备3份样品溶液用于HPLC检测,HPLC检测条件同1.3.2。据5种物质的线性回归方程计算样品溶液中黄酮类物质的质量浓度。
-
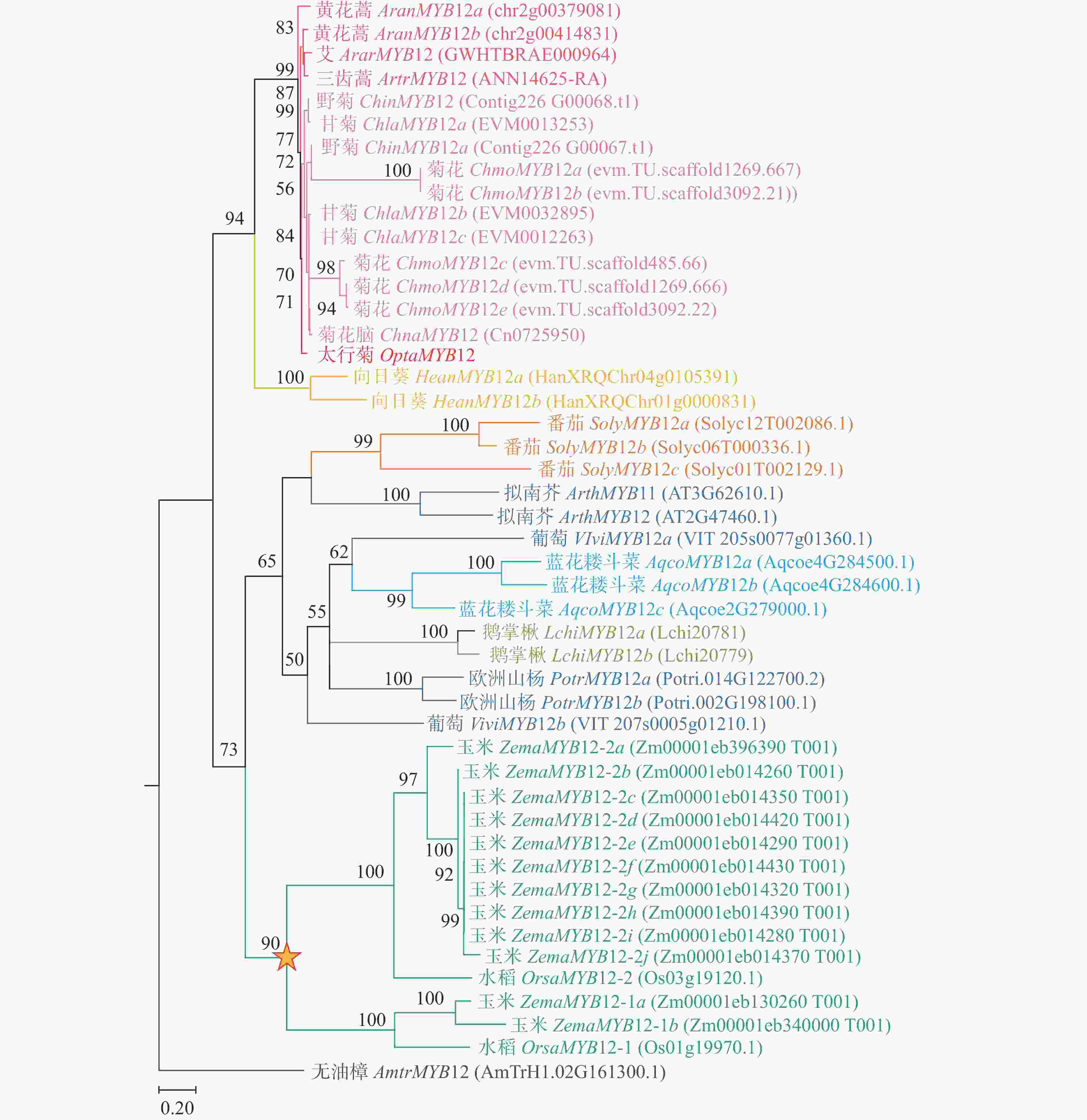
以拟南芥AtMYB12基因的蛋白质序列为查询序列[24],使用BLASTP v2.5.0+在太行菊转录组及其他18个被子植物的基因组中搜索对应的同源基因,E-value 阈值设为1.0×10−5[25]。为进行系统发育分析,各基因的氨基酸序列使用MAFFT v7.505进行比对[26],并通过PAL2NAL v14生成对应的基于密码子的核苷酸比对[27]。随后使用IQ-TREE 2.1.4-beta构建最大似然系统发育树(maximum likelihood tree),参数设置为“-bb
1000 -nt AUTO”[28],并利用MEGA11进行可视化[29]。本研究用到的转录组和基因组数据均下载于已发表论文或者公共数据库(表2)。表 2 系统发育分析所用的基因组或转录组数据
Table 2. Genomic and transcriptomic data used for phylogenetic analyses
科名 种名 缩写 文献 科名 种名 缩写 文献 菊科 Asteraceae 太行菊Opisthopappus taihangensis Opta [30] 菊科 Asteraceae 三齿蒿Artemisia tridentata Artr [40] 野菊Chrysanthemum indicum Chin [31] 茄科 Solanaceae 番茄Solanum lycopersicum Soly [41] 菊花C. × morifolium Chmo [32] 十字花科 Brassicaceae 拟南芥Arabidopsis thaliana Arth [42] 菊花脑C. nankingense Chna [33] 杨柳科 Salicaceae 欧洲山杨Populus trichocarpa Potr [43] 甘菊C. lavandulifolium Chla [34] 葡萄科 Vitaceae 葡萄Vitis vinifera Vivi [44] 生菜Lactuca sativa Lasa [35] 毛茛科 Ranunculaceae 蓝花耧斗菜Aquilegia coerulea Aqco [45] 向日葵Helianthus annuus Hean [36] 禾本科 Poaceae 玉米Zea mays Zema [46] 牛蒡Arctium lappa Alap [37] 水稻Oryza sativa Orsa [47] 艾Artemisia argyi Arar [38] 木兰科Magnoliaceae 鹅掌楸Liriodendron chinense Lich [48] 黄花蒿Artemisia annua Aran [39] 无油樟科Amborellaceae 无油樟Amborella trichopoda Amtr [49] -
取太行菊胁迫各阶段根系及叶片于液氮中研磨,使用OMEGA试剂盒提取太行菊RNA。根据Takara试剂盒说明书,采用两步法进行反转录实验。RT-qPCR引物由Integrated DNA Technologies在线设计,基因的表达水平以OtActin基因为内参。
-
以太行菊cDNA为模板进行PCR扩增,扩增产物经电泳分离回收后进行连接并转化大肠埃希菌Escherichia coli,挑取单克隆菌落测序验证。选择Kpn I作为酶切位点进行无缝克隆,构建pSuper1300::OtMYB12::GFP重组载体,并采用唯地公司冻融法将载体转化入农杆菌Agrobacterium tumefaciens GV3101。配置侵染液后,将重组质粒注射入烟草Nicotiana benthamiana叶片,暗培养3 d。随后将注射叶片切块制片,并在激光共聚焦显微镜下进行观察。
-
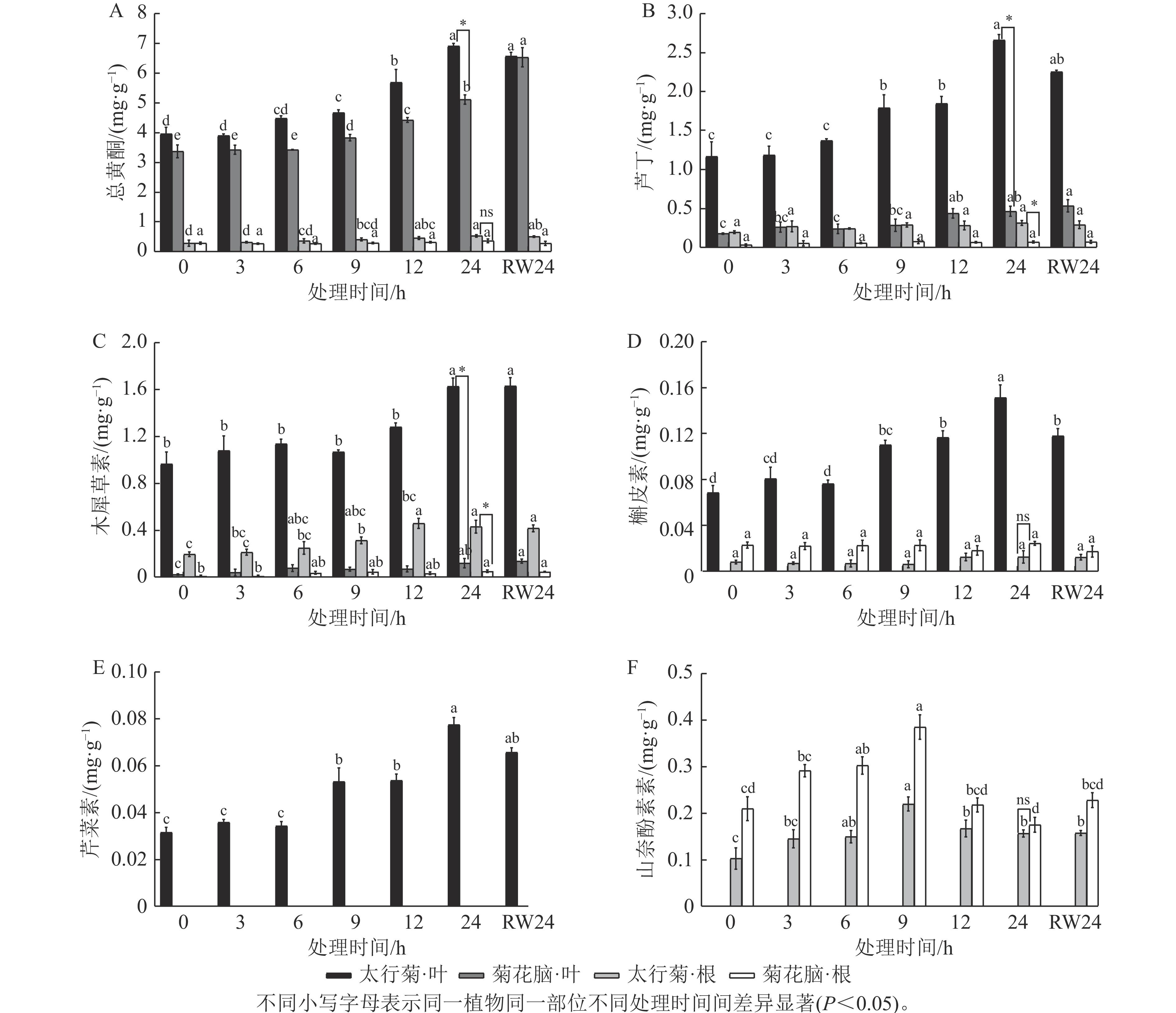
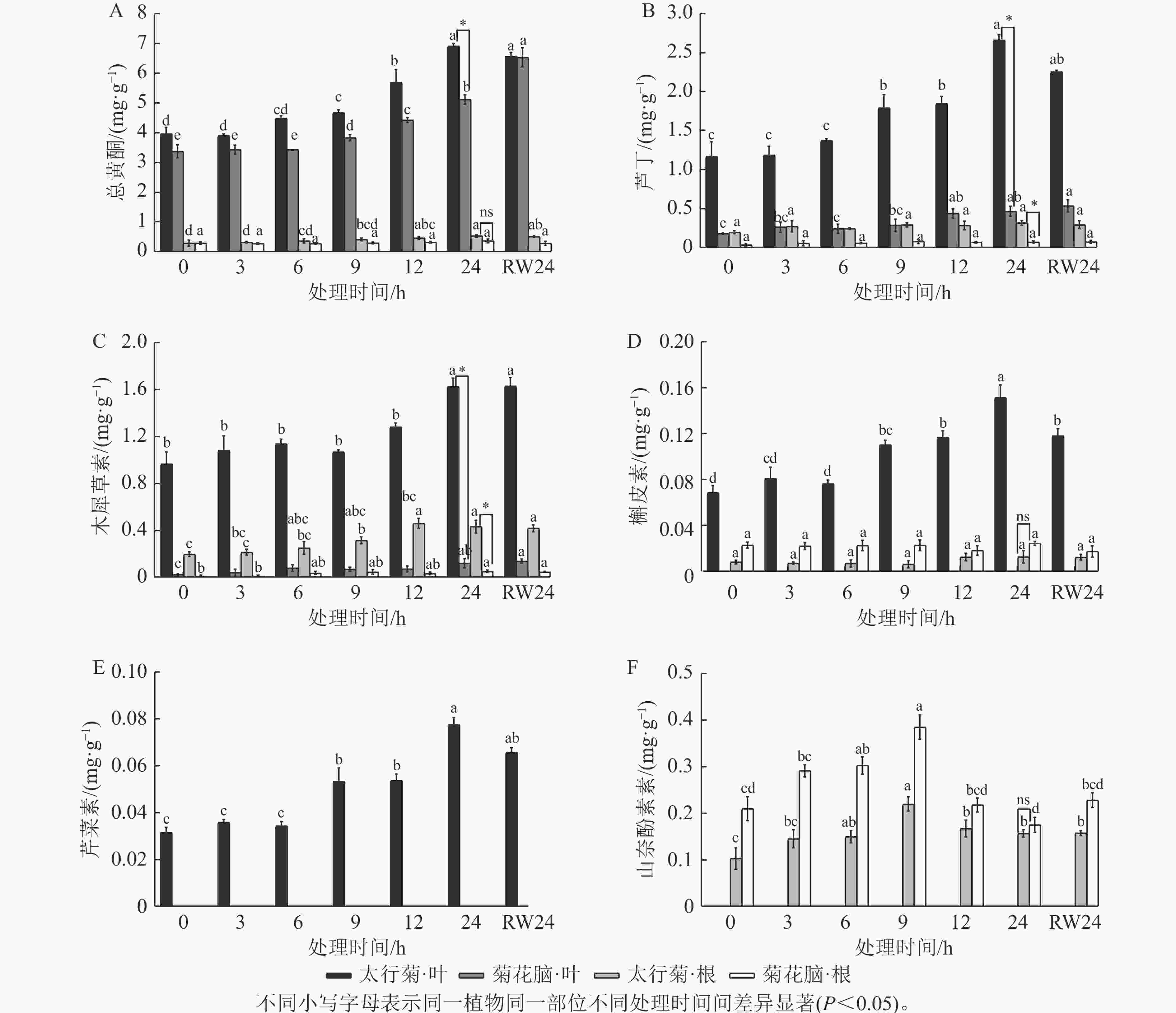
由图1A可见:在太行菊中,无论是根还是叶片,总黄酮质量分数总体均随干旱胁迫时间持续上升,在胁迫24 h时达最大,分别为0.561和 6.929 mg·g−1;复水24 h后,总黄酮质量分数下降,但与胁迫24 h差异不显著。菊花脑中干旱胁迫及复水后总黄酮变化趋势类似于太行菊,但复水处理后,菊花脑叶片总黄酮质量分数仍略有上升,与胁迫24 h时差异不显著。太行菊根和叶片中的总黄酮质量分数均高于菊花脑。上述结果表明:虽然干旱胁迫均可诱导黄酮类物质积累,但太行菊在黄酮合成与储存方面表现出更高的能力,可为其优异抗旱性提供生理基础。
-
在干旱胁迫响应过程中,不同黄酮类物质的积累式样可能存在差异。由图1B~E可见:在太行菊叶片中,芦丁、木犀草素、槲皮素和芹菜素质量分数总体在干旱处理后随胁迫时间延长均上升,均在胁迫24 h达峰值,显著高于其他处理(P<0.05),复水后下降,其中,木犀草素和芹菜素质量分数在复水24 h后与胁迫24 h时差异不显著,而槲皮素差异显著(P<0.05)。在太行菊的根中,木犀草素质量分数虽变化幅度不大,但整体趋势仍表现为随干旱胁迫延长而上升,在胁迫12 h时达峰值,复水后下降,但复水24 h与胁迫24 h时差异不显著;芦丁和槲皮素质量分数虽表现为干旱胁迫下上升、复水后下降,但上升与下降趋势均不显著。上述几类物质,在菊花脑叶片中仅检测到2种(芦丁和木犀草素),且二者在复水后仍呈上升趋势,但与胁迫24 h时差异不显著;在菊花脑的根中积累非常少,未呈现明显积累规律。如图1F可知:山奈酚的积累模式最为特殊,且在2个物种中较为保守:随干旱延长而上升,于胁迫9 h时达峰值,随后显著下降(P<0.05),复水后变化不显著。
在干旱胁迫响应过程中,不同黄酮类物质的总量也可能存在差异。在太行菊叶片中,4种黄酮类物质质量分数均在胁迫24 h时达到峰值,其中,芦丁质量分数最高,达2.939 mg·g−1;其次为木犀草素、槲皮素和芹菜素,分别为1.635、0.142 和0.078 mg·g−1;叶片中未检测到山奈酚。在太行菊的根中,芦丁质量分数最高,为0.648 mg·g−1;木犀草素和山奈酚相对较低,分别为0.474和0.172 mg·g−1;而槲皮素积累极为微量,且未检测到芹菜素。胁迫24 h时,太行菊根与叶中的芦丁和木犀草素质量分数均显著高于菊花脑(P<0.05);太行菊根中槲皮素和山奈酚质量分数低于菊花脑,但并无显著差异。
上述结果表明:干旱胁迫均可诱导2种植物黄酮类化合物积累,但不同化合物在不同器官中的积累优势存在差异:太行菊叶片以芦丁为主,但根部差异较小,这说明不同成分可能在不同器官中发挥特异性作用。同时,太行菊在多数黄酮类化合物的合成与储存能力上均明显优于菊花脑,进一步凸显其在干旱适应方面的优势。
-
为鉴定太行菊中参与黄酮类物质合成、响应干旱胁迫的关键基因,进行了同源搜索和系统发育分析。由图2可见:在太行菊转录组中筛选到一个胁迫后显著上调的基因MYB12,命名为OtMYB12。进化分析显示:该基因在太行菊的进化过程中未经历基因重复或丢失事件,始终保持单拷贝状态。上述结果表明:OtMYB12进化较为保守,其功能可能与其他物种中的MYB12一致,主要参与调控黄酮类化合物的合成。
-
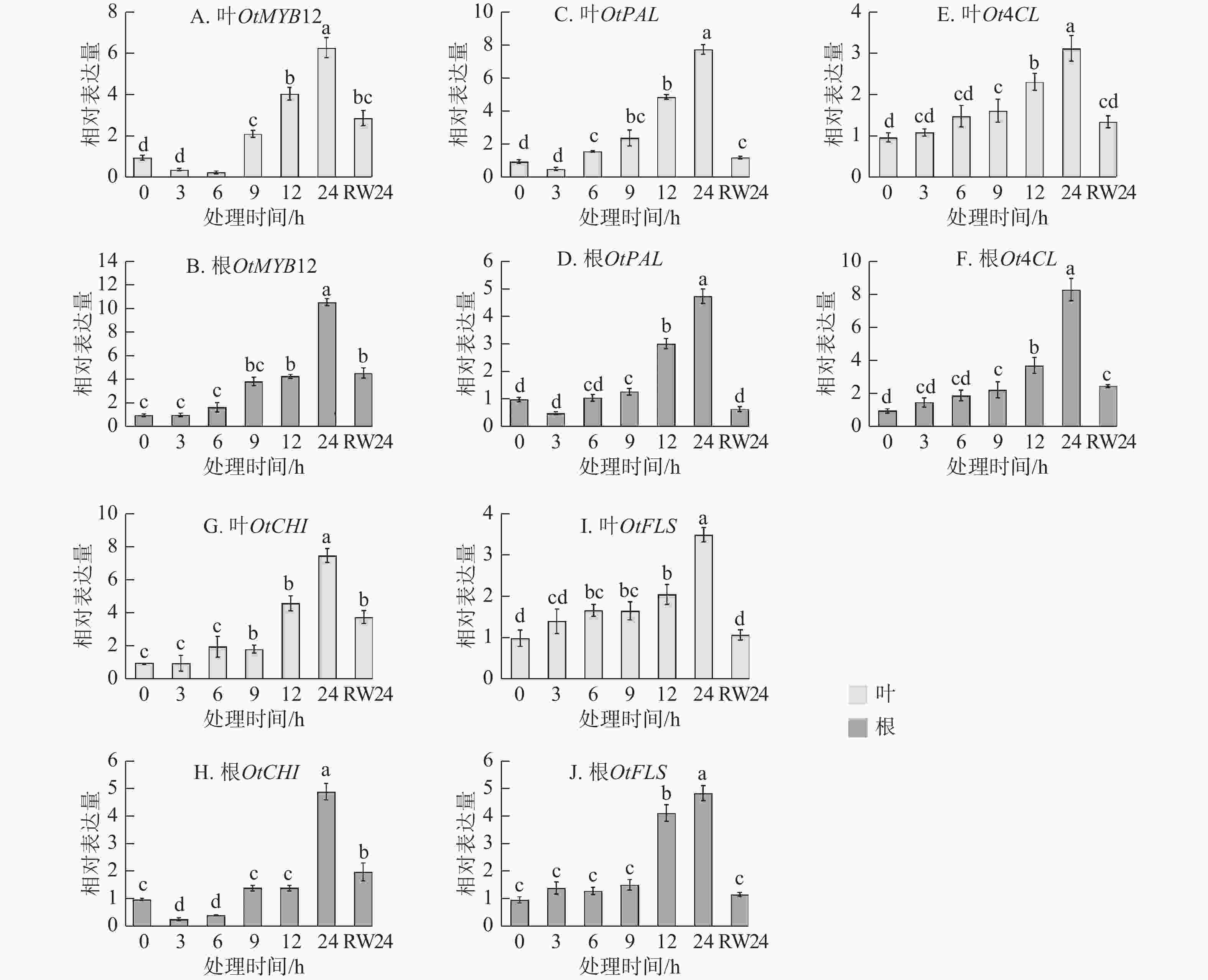
为明确OtMYB12在干旱响应过程中的作用,对干旱胁迫及复水处理后其表达动态进行了检测。由图3A~B可见:无论在叶片还是根部,OtMYB12的表达量均随胁迫时间延长升高,在胁迫24 h时达到峰值,复水24 h后显著下调(P<0.05),其表达趋势与干旱及复水条件下黄酮类化合物质量分数的变化高度一致。对部分黄酮合成结构基因进行了检测,由图3C~J可见:OtMYB12与黄酮合成结构基因表达趋势基本一致。上述结果表明:OtMYB12可能参与调控太行菊黄酮类化合物的生物合成,并在干旱胁迫响应中发挥重要作用。
-
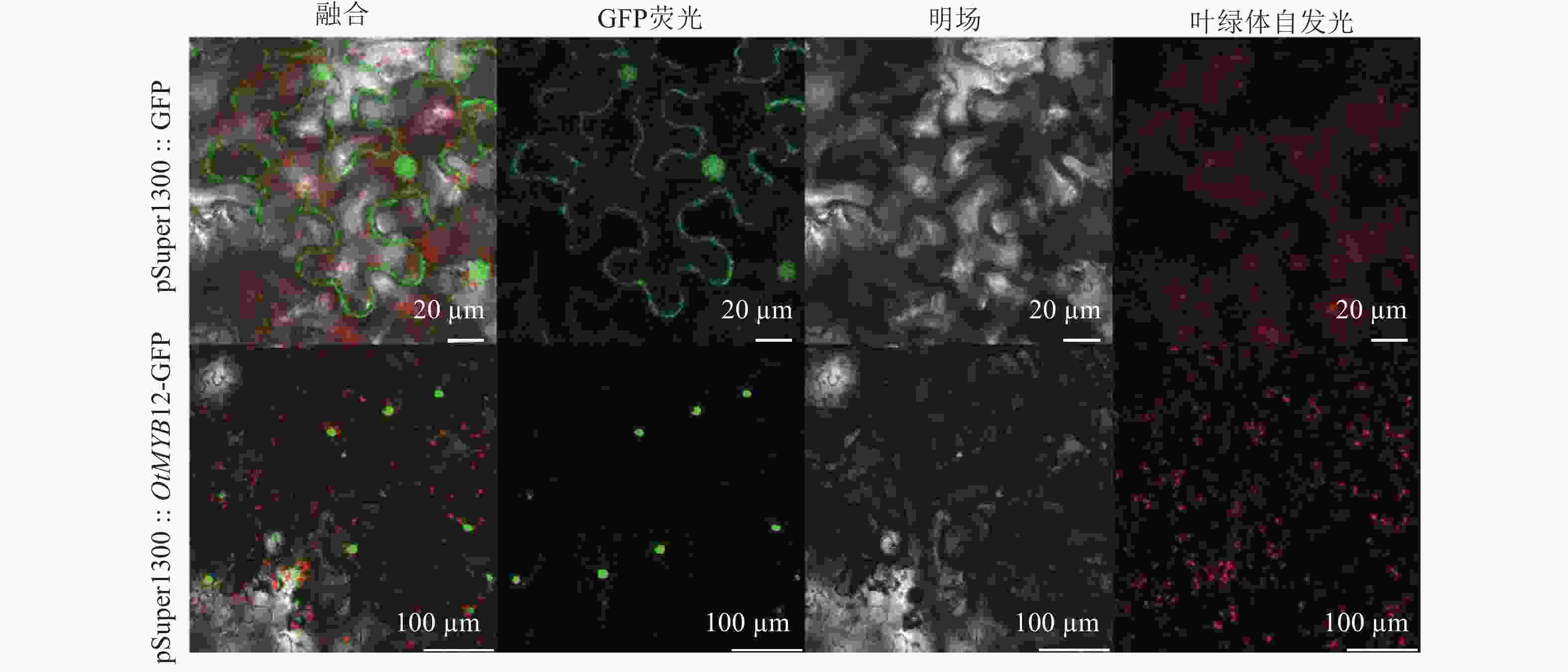
由图4可见:绿色荧光蛋白(GFP)空载荧光信号定位于细胞核和细胞膜,而太行菊OtMYB12荧光信号定位于细胞核中,表明OtMYB12于细胞核中发挥调控作用,符合转录因子的特征。
-
植物生长发育过程中,黄酮类物质在多种胁迫中发挥着关键调控作用,如干旱胁迫下类黄酮含量增加可降低白车轴草Trifolium repens 的ROS含量[50];拟南芥中黄酮类化合物可能通过激活抗氧化系统间接提高渗透调节能力,从而提高耐旱性[51]。本研究结果表明:干旱胁迫可同时诱导太行菊和菊花脑体内黄酮类物质积累,与前人研究结果相似。但太行菊在黄酮合成与调节方面表现出了更高的能力,无论是根还是叶片,总黄酮量均显著高于菊花脑,可能是其优异抗旱性的生理基础。5种黄酮类物质质量分数变化进一步揭示了黄酮类物质在太行菊干旱胁迫中的作用。在太行菊叶中,芦丁的质量分数最高,而根部各种黄酮类物质的积累差异较小。这种特异性积累模式暗示了不同化合物可能在不同部位行使特异性的功能,如刺桐Erythrina variegata根中类黄酮可能吸引土壤微生物,从而调节胁迫下的水分状况[52];干旱胁迫下黄酮类化合物(橙皮苷、异泽兰黄素等)在蓝莓Vaccinium uliginosum叶中显著积累,赋予蓝莓更强的耐旱性[53]。
分子水平上,黄酮类合成途径关键调控转录因子OtMYB12在干旱胁迫下表现出显著差异表达。系统发育分析表明:该基因在太行菊的进化过程始终保持单拷贝状态,这暗示着OtMYB12进化较为保守,可能与其他物种中MYB12一样参与黄酮类生物合成途径,为太行菊的优异抗旱性提供了分子基础。拟南芥中AtMYB11、AtMYB12、AtMYB111通过控制局部黄酮醇积累特异性介导气孔运动,从而维持细胞内的活性氧稳态[54];AtMYB12通过调控乙烯和生长素介导的黄酮合成通路,控制拟南芥黄酮类化合物合成[55];梨中PbMYB12通过靶向PbCHS、PbFLS正向调控黄酮醇合成[56]。本研究所选择的OtMYB12如何深入参与黄酮类生物合成途径,有待进一步研究。
-
干旱胁迫下太行菊、菊花脑的总黄酮质量分数均显著升高,但太行菊根和叶片中总黄酮质量分数均高于菊花脑,且在多数黄酮类化合物的合成与调节能力上均明显优于菊花脑。此外,通过荧光定量和系统发育分析,发现OtMYB12定位于细胞核中,表达动态与黄酮类化合物及其结构基因的变化趋势高度一致,且它的进化较为保守,可能同样参与调控太行菊黄酮类生物合成途径。本研究不仅揭示了黄酮类化合物在太行菊干旱胁迫响应中的作用,也为抗旱菊花新品种培育提供了可利用的基因资源。
Role of flavonoids and mining of drought-tolerant genes in the drought response of Opisthopappus taihangensis
-
摘要:
目的 探究黄酮类化合物在太行菊Opisthopappus taihangensis响应干旱胁迫中的作用,并为抗旱菊花Chysanthemum × morifolium新品种培育提供可利用的基因资源。 方法 以耐旱种质太行菊和干旱敏感种质菊花脑C. nankingense为材料,在模拟干旱及复水条件下,测定太行菊的总黄酮及5种黄酮类化合物的质量分数,同时鉴定可能调控黄酮类化合物合成的关键候选基因OtMYB12,并分析其表达动态及亚细胞定位。 结果 ①在干旱胁迫下,太行菊叶片和根部黄酮类物质质量分数随胁迫时间延长持续升高,于24 h达峰,复水后显著下降,且始终高于菊花脑对应器官中的质量分数。②在5种黄酮类化合物中,芦丁含量在太行菊叶片和根中都最高,各类物质均随干旱胁迫时间持续积累,复水后下降。③OtMYB12进化过程中未经历重复和丢失事件,定位于细胞核中,其表达水平变化与黄酮类化合物及其结构基因的变化趋势高度相关。 结论 黄酮类物质参与响应太行菊干旱胁迫,OtMYB12可能通过调控类黄酮化合物的合成在此过程中发挥重要作用。图4表2参56 Abstract:Objective This study aims to investigate the role of flavonoids in the drought stress response of Opisthopappus taihangensis and to provide valuable gene resources for the breeding of drought-tolerant chrysanthemums. Method Drought-tolerant O. taihangensis and drought-sensitive Chrysanthemum nankingense were used as experimental materials. Under simulated drought and rehydration conditions, the total flavonoid content and the levels of 5 flavonoids in O. taihangensis were measured. Meanwhile, the key candidate gene OtMYB12, potentially involved in flavonoid biosynthesis, was identified, and its expression dynamics and subcellular localization were analyzed. Result (1) Under drought stress, flavonoid content in O. taihangensis leaves and roots progressively increased with stress duration, peaking at 24 h, and decreased significantly after rehydration, and remained consistently higher than those in the corresponding organs of C. nankingense. (2) Among 5 flavonoids, rutin accumulated most abundantly in leaves and roots. All compounds showed progressive accumulation under drought stress and decreased after rehydration. (3) OtMYB12 remained a single-copy gene during evolution, localized in the nucleus, and its expression dynamics were highly correlated with accumulation patterns of flavonoid and its structural gene. Conclusion Flavonoids participate in the drought stress response of O. taihangensis, and OtMYB12 likely plays a key role in this process by regulating flavonoid biosynthesis. [Ch, 4 fig. 2 tab. 56 ref.] -
Key words:
- Opisthopappus taihangensis /
- drought stress /
- flavonoids /
- MYB 12
-
表 1 5种类黄酮类物质的线性回归方程
Table 1. Linear regression equations of 5 flavonoids
化合物 回归方程 R2 线性范围/(mg·L−1) 芦丁 y=27.124x+48.556 0.999 8 1.10~550.00 槲皮素 y=53.12x−13.131 0.999 6 0.05~25.00 木犀草素 y=25.646x+59.855 0.999 7 0.40~200.00 山奈酚 y= 3.8849 x−4.2011 0.999 3 0.02~10.00 芹菜素 y=29.03x− 3.7862 0.999 6 0.032~16.000 说明:x为经HPLC分析获得的峰面积,y为黄酮类物质的质量浓度。 表 2 系统发育分析所用的基因组或转录组数据
Table 2. Genomic and transcriptomic data used for phylogenetic analyses
科名 种名 缩写 文献 科名 种名 缩写 文献 菊科 Asteraceae 太行菊Opisthopappus taihangensis Opta [30] 菊科 Asteraceae 三齿蒿Artemisia tridentata Artr [40] 野菊Chrysanthemum indicum Chin [31] 茄科 Solanaceae 番茄Solanum lycopersicum Soly [41] 菊花C. × morifolium Chmo [32] 十字花科 Brassicaceae 拟南芥Arabidopsis thaliana Arth [42] 菊花脑C. nankingense Chna [33] 杨柳科 Salicaceae 欧洲山杨Populus trichocarpa Potr [43] 甘菊C. lavandulifolium Chla [34] 葡萄科 Vitaceae 葡萄Vitis vinifera Vivi [44] 生菜Lactuca sativa Lasa [35] 毛茛科 Ranunculaceae 蓝花耧斗菜Aquilegia coerulea Aqco [45] 向日葵Helianthus annuus Hean [36] 禾本科 Poaceae 玉米Zea mays Zema [46] 牛蒡Arctium lappa Alap [37] 水稻Oryza sativa Orsa [47] 艾Artemisia argyi Arar [38] 木兰科Magnoliaceae 鹅掌楸Liriodendron chinense Lich [48] 黄花蒿Artemisia annua Aran [39] 无油樟科Amborellaceae 无油樟Amborella trichopoda Amtr [49] -
[1] GORE N T, SHAIKH S S, MALI A A, et al. Causes, consequences, and responses to drought stress in plants [J]. Life Sciences for Sustainable Development, 2022, 22(1): 127−139. [2] FAROOQ M, WAHID A, ZAHRA N, et al. Advances in plant drought tolerance [J]. Journal of Plant Growth Regulation, 2024, 43: 3337−3369. [3] GUPTA A, RICO-MEDINA A, CAÑO-DELGADO A I. The physiology of plant responses to drought [J]. Science, 2020, 368(6488): 266−269. [4] FAROOQ M, HUSSAIN M, WAHID A, et al. Drought stress in plants: an overview [J]. Plant Responses to Drought Stress, 2015, 1: 1−35. [5] WINKEL-SHIRLEY B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology [J]. Plant Physiology, 2001, 126(2): 485−493. [6] PROCHÁZKOVÁ D, BOUŠOVÁ I, WILHELMOVÁ N. Antioxidant and prooxidant properties of flavonoids [J]. Fitoterapia, 2011, 82(4): 513−523. [7] YU Wanwen, LIU Huimin, LUO Jiaqin, et al. Partial root-zone simulated drought induces greater flavonoid accumulation than full root-zone simulated water deficiency in the leaves of Ginkgo biloba[J/OL]. Environmental and Experimental Botany, 2022, 201: 104998[2025-08-25]. DOI: 10.1016/j.envexpbot.2022.104998. [8] GAO Guori, LÜ Zhongrui, ZHANG Guoyun, et al. An ABA-flavonoid relationship contributes to the differences in drought resistance between different sea buckthorn subspecies [J]. Tree Physiology, 2021, 41(5): 744−755. [9] LI Baozhu, FAN Ruonan, SUN Guiling, et al. Flavonoids improve drought tolerance of maize seedlings by regulating the homeostasis of reactive oxygen species [J]. Plant and Soil, 2021, 461(1): 389−405. [10] WANG Feibing, ZHU Hong, KONG Weili, et al. The Antirrhinum AmDEL gene enhances flavonoids accumulation and salt and drought tolerance in transgenic Arabidopsis [J]. Planta, 2016, 244(1): 59−73. [11] SHIVHARE R, MISHRA P, BADOLA P K, et al. PgF3H gene enhances drought tolerance in transgenic Arabidopsis by regulating flavonoid biosynthesis and stress response[J/OL]. Plant Cell Reports, 2025, 44(7): 150[2025-08-25]. DOI: 10.1007/s00299-025-03524-8. [12] LIU Tianyi, YAN Fan, LIU Yajing, et al. The GmbHLH13-GmCHS7 module positively regulates isoflavones accumulation in soybean (Glycine max. L.)[J/OL]. Plant Physiology and Biochemistry, 2025, 227: 110162[2025-08-25]. DOI: 10.1016/j.plaphy.2025.110162. [13] NAKABAYASHI R, YONEKURA-SAKAKIBARA K, URANO K, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids [J]. The Plant Journal, 2014, 77(3): 367−379. [14] LI Baozhu, FAN Ruonan, FAN Yanting, et al. The flavonoid biosynthesis regulator PFG3 confers drought stress tolerance in plants by promoting flavonoid accumulation[J/OL]. Environmental and Experimental Botany, 2022, 196: 104792[2025-08-25]. DOI: 10.1016/j.envexpbot.2022.104792. [15] BULANOV A N, ANDREEVA E A, TSVETKOVA N V, et al. Regulation of flavonoid biosynthesis by the MYB-bHLH-WDR (MBW) complex in plants and its specific features in cereals[J/OL]. International Journal of Molecular Sciences, 2025, 26(2): 734[2025-08-25]. DOI: 10.3390/ijms26020734. [16] TAO Ruiyan, YU Wenjie, GAO Yuhao, et al. Light-induced basic/helix-loop-Helix64 enhances anthocyanin biosynthesis and undergoes CONSTITUTIVELY PHOTOMORPHOGENIC1-mediated degradation in pear [J]. Plant Physiology, 2020, 184(4): 1684−1701. [17] JIN Zilan, WANG Wanni, NAN Qiong, et al. VvNAC17, a grape NAC transcription factor, regulates plant response to drought-tolerance and anthocyanin synthesis[J/OL]. Plant Physiology and Biochemistry, 2025, 219: 109379[2025-08-25]. DOI: 10.1016/j.plaphy.2024.109379. [18] CAO Yunlin, MEI Yuyang, ZHANG Ruining, et al. Transcriptional regulation of flavonol biosynthesis in plants[J/OL]. Horticulture Research, 2024, 11(4): uhae043[2025-08-25]. DOI: 10.1093/hr/uhae043. [19] 郭彦宏, 张晶星, 杨永娟, 等. 6种野生广义菊属植物对干旱胁迫的生理响应[J]. 浙江农业学报, 2018, 30(8): 1349−1354. GUO Yanhong, ZHANG Jingxing, YANG Yongjuan, et al. Response of six germplasms of Chrysanthemum and related genera to drought stress [J]. Acta Agriculturae Zhejiangensis, 2018, 30(8): 1349−1354. [20] 巴亭亭. 干旱胁迫下太行菊类黄酮含量变化及相关基因的表达分析[D]. 北京: 北京林业大学, 2021. BA Tingting. Changes of Flavonoid Content and Expression Analysis of Related Genes in Opisthopappus taihangensis Under Drought Stress[D]. Beijing: Beijing Forestry University, 2021. [21] LUO Jiayi, LUO Chang, HAN Mingzheng, et al. A natural variation of flavone synthase Ⅱ gene enhances flavone accumulation and confers drought adaptation in Chrysanthemum [J]. New Phytologist, 2025, 247(3): 1445−1459. [22] 刘海芳, 魏东伟, 刘全军, 等. 太行菊不同器官中绿原酸和4种黄酮类物质含量研究[J]. 天然产物研究与开发, 2013, 25(5): 646−651, 671. LIU Haifang, WEI Dongwei, LIU Quanjun, et al. Determination of chlorogenic acid and four flavonoids in different organs of Opisthopappus taihangensis (Ling) Shih [J]. Natural Product Research and Development, 2013, 25(5): 646−651, 671. [23] 孙翊, 李慧, 王亮生, 等. 一种快速有效分析烟草花冠中花青素苷的方法[J]. 植物学报, 2011, 46(2): 189−196. SUN Yi, LI Hui, WANG Liangsheng, et al. Rapid, effective method for anthocyanin analysis in tobacco corolla [J]. Chinese Bulletin of Botany, 2011, 46(2): 189−196. [24] MEHRTENS F, KRANZ H, BEDNAREK P, et al. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis [J]. Plant Physiology, 2005, 138(2): 1083−1096. [25] CAMACHO C, COULOURIS G, AVAGYAN V, et al. BLAST+: architecture and applications[J/OL]. BMC Bioinformatics, 2009, 10: 421[2025-08-25]. DOI: 10.1186/1471-2105-10-421. [26] KATOH K, STANDLEY D M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability [J]. Molecular Biology and Evolution, 2013, 30(4): 772−780. [27] SUYAMA M, TORRENTS D, BORK P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding Codon alignments [J]. Nucleic Acids Research, 2006, 34(suppl 2): W609−W612. [28] MINH B Q, SCHMIDT H A, CHERNOMOR O, et al. Corrigendum to: IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era[J/OL]. Molecular Biology and Evolution, 2020, 37(8): 2461[2025-08-25]. DOI: 10.1093/molbev/msaa131. [29] TAMURA K, STECHER G, KUMAR S. MEGA11: molecular evolutionary genetics analysis version 11 [J]. Molecular Biology and Evolution, 2021, 38(7): 3022−3027. [30] YANG Yongjuan, GUO Yanhong, ZHONG Jian, et al. Root physiological traits and transcriptome analyses reveal that root zone water retention confers drought tolerance to Opisthopappus taihangensis[J/OL]. Scientific Reports, 2020, 10: 2627[2025-08-25]. DOI: 10.1038/s41598-020-59399-0. [31] DENG Yin’ai, YANG Peng, ZHANG Qianle, et al. Genomic insights into the evolution of flavonoid biosynthesis and O-methyltransferase and glucosyltransferase in Chrysanthemum indicum[J/OL]. Cell Reports, 2024, 43(2): 113725[2025-08-25]. DOI: 10.1016/j.celrep.2024.113725. [32] SONG Aiping, SU Jiangshuo, WANG Haibin, et al. Analyses of a chromosome-scale genome assembly reveal the origin and evolution of cultivated Chrysanthemum[J/OL]. Nature Communications, 2023, 14: 2021[2025-08-25]. DOI: 10.1038/s41467-023-37730-3. [33] SONG Chi, LIU Yifei, SONG Aiping, et al. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of Chrysanthemum flowers and medicinal traits [J]. Molecular Plant, 2018, 11(12): 1482−1491. [34] WEN Xiaohui, LI Junzhuo, WANG Lili, et al. The Chrysanthemum lavandulifolium genome and the molecular mechanism underlying diverse Capitulum types[J/OL]. Horticulture Research, 2022, 9: uhab022[2025-08-25]. DOI: 10.1093/hr/uhab022. [35] SHEN Fei, QIN Yajuan, WANG Rui, et al. Comparative genomics reveals a unique nitrogen-carbon balance system in Asteraceae[J/OL]. Nature Communications, 2023, 14: 4334[2025-08-25]. DOI: 10.1038/s41467-023-40002-9. [36] BADOUIN H, GOUZY J, GRASSA C J, et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution [J]. Nature, 2017, 546(7656): 148−152. [37] FAN Wei, WANG Sen, WANG Hengchao, et al. The genomes of chicory, endive, great burdock and yacon provide insights into Asteraceae Palaeo-polyploidization history and plant inulin production [J]. Molecular Ecology Resources, 2022, 22(8): 3124−3140. [38] CHEN Hongyu, GUO Miaoxian, DONG Shuting, et al. A chromosome-scale genome assembly of Artemisia argyi reveals unbiased subgenome evolution and key contributions of gene duplication to volatile terpenoid diversity[J/OL]. Plant Communications, 2023, 4(3): 100516[2025-08-25]. DOI: 10.1016/j.xplc.2023.100516. [39] SHEN Qian, ZHANG Lida, LIAO Zhihua, et al. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis [J]. Molecular Plant, 2018, 11(6): 776−788. [40] MELTON A E, CHILD A W, BEARD R S, et al. A haploid pseudo-chromosome genome assembly for a keystone sagebrush species of western North American rangelands[J/OL]. G3 Genes|Genomes|Genetics, 2022, 12(7): jkac122[2025-08-25]. DOI: 10.1093/g3journal/jkac122. [41] ZHOU Yao, ZHANG Zhiyang, BAO Zhigui, et al. Graph pangenome captures missing heritability and empowers tomato breeding [J]. Nature, 2022, 606(7914): 527−534. [42] LAMESCH P, BERARDINI T Z, LI Donghui, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools [J]. Nucleic Acids Research, 2012, 40(D1): D1202−D1210. [43] TUSKAN G A, DIFAZIO S, JANSSON S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) [J]. Science, 2006, 313(5793): 1596−1604. [44] JAILLON O, AURY J M, NOEL B, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla [J]. Nature, 2007, 449(7161): 463−467. [45] FILIAULT D L, BALLERINI E S, MANDÁKOVÁ T, et al. The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history[J/OL]. eLife, 2018, 7: e36426[2025-08-25]. DOI: 10.7554/eLife.36426. [46] HUFFORD M B, SEETHARAM A S, WOODHOUSE M R, et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes [J]. Science, 2021, 373(6555): 655−662. [47] OUYANG Shu, ZHU Wei, HAMILTON J, et al. The TIGR Rice Genome Annotation Resource: improvements and new features [J]. Nucleic Acids Research, 2007, 35(suppl 1): D883−D887. [48] CHEN Jinhui, HAO Zhaodong, GUANG Xuanmin, et al. Author Correction: Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation[J/OL]. Nature Plants, 2019, 5(3): 328[2025-08-25]. DOI: 10.1038/s41477-019-0368-1. [49] CAREY S B, AKÖZBEK L, LOVELL J T, et al. ZW sex chromosome structure in Amborella trichopoda [J]. Nature Plants, 2024, 10(12): 1944−1954. [50] ZHANG Youzhi, FU Wei, PU Qi, et al. The white clover single-copy nuclear gene TrNAC002 promotes growth and confers drought resistance in plants through flavonoid synthesis[J/OL]. Plants, 2024, 14(1): 31[2025-08-25]. DOI: 10.3390/plants14010031. [51] LI Ningning, WANG Xue, MA Binjie, et al. A leucoanthocyanidin dioxygenase gene (RtLDOX2) from the feral forage plant Reaumuria trigyna promotes the accumulation of flavonoids and improves tolerance to abiotic stresses [J]. Journal of Plant Research, 2021, 134(5): 1121−1138. [52] LIU Chengwu, MURRAY J D. The role of flavonoids in nodulation host-range specificity: an update[J/OL]. Plants, 2016, 5(3): 33[2025-08-25]. DOI: 10.3390/plants5030033. [53] FENG Xinghua, BAI Sining, ZHOU Lianxia, et al. Integrated analysis of transcriptome and metabolome provides insights into flavonoid biosynthesis of blueberry leaves in response to drought stress[J/OL]. International Journal of Molecular Sciences, 2024, 25(20): 11135[2025-08-25]. DOI: 10.3390/ijms252011135. [54] CHANG Yuankai, SHI Mianmian, WANG Xiao, et al. A CRY1-HY5-MYB signaling cascade fine-tunes guard cell reactive oxygen species levels and triggers stomatal opening[J/OL]. The Plant Cell, 2025, 37(4): koaf064[2025-08-25]. DOI: 10.1093/plcell/koaf064. [55] LEWIS D R, RAMIREZ M V, MILLER N D, et al. Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks [J]. Plant Physiology, 2011, 156(1): 144−164. [56] ZHAI Rui, ZHAO Yingxiao, WU Meng, et al. The MYB transcription factor PbMYB12b positively regulates flavonol biosynthesis in pear fruit[J/OL]. BMC Plant Biology, 2019, 19(1): 85[2025-08-25]. DOI: 10.1186/s12870-019-1687-0. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.20250468






 下载:
下载: