-
开花是高等植物进入生殖生长的标志,是植物完成生活史的必要环节。在拟南芥Arabidopsis thaliana中的研究发现,开花过程受多种内源因素和外源环境因素的影响,由复杂基因网络严格调控[1]。光周期途径中植物开花受季节性昼长变化影响[2];赤霉素(GA)途径是在非诱导光周期途径的条件下,在开花过程中发挥作用[3];春化途径中植物开花要受到低温积累的影响[4];自主途径中通过感知植物自身内部发育状态而调控植物开花[5],多种途径的成花调节信号最终都集中于2个整合因子:开花促进因子FLOERING LOCUS T(FT)和SUPPRESSPR OF OVEREXPRESION OF CONSTANS 1(SOC1)[6-8]。在拟南芥中,过表达FT基因会明显促进SOC1基因的表达量;同时在ft突变体中,SOC1的表达量明显下降[9-11],但是对ft和soc1双突变体与ft单突变体植株的开花时间进行统计,结果显示没明显差异,可知除FT蛋白调控SOC1的表达外,还存在另外的途径影响SOC1基因的表达[10-13]。其他开花控制因子可以通过FT和SOC1参与开花途径的调控,例如开花抑制因子SHORT VEGETATIVE PHASE(SVP)可以结合SOC1和FT的调控序列,对FT和SOC1产生抑制作用[14],进而下调它们的表达量。近年,发现拟南芥中属于J蛋白家族一员的J3蛋白,可以与SVP蛋白相互作用调控SOC1和FT基因的表达量,而实现对成花信号的调节,在调控开花途径中发挥着十分重要的作用[15]。J蛋白(J-domain protein)是广泛存在于动植物中的一个庞大的分子伴侣家族,该类蛋白N末端包含1个约由70个氨基酸构成的保守序列——J结构域而被命名为J蛋白[16-17]。J蛋白的J结构域由4个α-螺旋组成,第2和第3个螺旋之间有一个极为保守的由组氨酸、脯氨酸、天冬氨酸组成的HPD三肽,这是J结构域的重要特征[18];邻近J结构域的是G/F结构域(富含甘氨酸和苯丙氨酸);之后是锌指结构域。根据结构域的不同J蛋白可分为4类[19]:Ⅰ类J蛋白包含所有常见的结构;Ⅱ类J蛋白缺少锌指结构;Ⅲ类J蛋白只有J结构域;Ⅳ类J蛋白也被称作J-like蛋白,这类蛋白的J结构域没有HPD模块[20-21]。目前发现拟南芥中共有120个J蛋白,其中Ⅰ类J蛋白8个,Ⅱ类J蛋白16个,Ⅲ类J蛋白92个,Ⅳ类J蛋白4个[22]。在植物中,有关J蛋白的研究主要集中在拟南芥中,现有的结果表明,J蛋白可以参与多种生物学过程,包括叶绿体的发育[23-25]、植物的抗逆性[26-27]、信号转导[28-29]和成花途径[15]等。在拟南芥开花调控中,J3蛋白通过与SVP等其他开花关键因子相互作用而影响植物成花过程,SVP的突变体svp-41表现为早花现象,J3的突变j3-1表现为晚花现象,同时在j3-1和svp-41的双突变体中,拟南芥表现出早花现象,与svp-41的单突变体的表型相似,这一结果表明J3基因通过与SVP的相互作用实现对开花时间的调控[30-31],进一步研究发现在细胞核中J3基因通过与营养生长短期(SVP)蛋白直接相互作用,衰减SVP蛋白与SOC1和FT调控序列的结合从而上调SOC1和FT基因的表达量[15]。竹类植物开花周期较长并具有时间上的不确定性,而且很多竹子开花后会成片死亡,雷竹Phyllostachys violascens是一种优良的笋用竹种,近年来,雷竹普遍开花,开花后部分雷竹会死亡,导致雷竹笋产量大幅度下降,带来严重的经济损失。有关竹类植物开花的研究已经很多,但是竹子成花机制和花期调控模式至今尚未完全清楚[32]。目前,竹子中还未见有关J3基因生物学功能的研究,本研究以雷竹为研究对象,通过同源克隆技术获得1个J3同源基因,经过亚细胞定位、表达模式和功能分析,以期能为深入研究J3同源基因在竹类植物开花调控机制中的作用提供科学依据。
HTML
-
雷竹材料来源于浙江农林大学智能温室大棚,选取位于同一竹鞭上的笋、花芽、花,开花和不开花雷竹的幼叶、成叶、茎秆,标记分装后放入液氮中速冻,保存在-80 ℃超低温冰箱。
-
用全RNA提取试剂盒(鼎国)分别提取采集的雷竹材料的RNA,以提取的RNA(少于0.5 g·L-1)为模板,用Prime ScriptTM RT Master Mix(Perfect Real Time)试剂盒(Clontech),反转录获得雷竹各组织的cDNA,检测合格后,于-20 ℃保存备用。
-
通过本地Blast,从毛竹Phyllostachys edulis基因组文库中找到一个候选基因序列号(bphylf027h21),根据该候选基因的基因序列设计引物(J3,表 1),雷竹cDNA为模板,进行聚合酶链式反应(PCR)扩增,琼脂糖凝胶电泳检测PCR扩增条带,用胶回收试剂盒(上海生工生物公司)回收纯化电泳条带,将获得的目的片段与pMD19(simple)-T载体置于连接仪上16 ℃连接1.0~3.0 h,之后转化大肠埃希菌Escherichia coli DH5α感受态细胞(热转化法),37 ℃恒温培养箱培养6.0~8.0 h,挑取白色圆形单菌落,菌落PCR鉴定出阳性单克隆,送测序,得到碱基序列。
引物名称 序列(5'→3') 用途 J3-F CTCTCCGTCTCACTCCGCTC 基因开放阅读框(ORF)扩增 J3-R GTCAAATTGCCTTTATAGAA PvJ3-F ATGTTCGGGCGCGCGCCG ORF扩增 PvJ3-R TTACTGTTGTGCGCACTGCAC PvJ3-1-F CCGGAATTCTGGAGAGAGCATTAACG 荧光定量PCR PvJ3-1-R CCGGAATTCCTTCTGCTGGAGGAC PeNTB-F TCTTGTTTGACACCGAAGAGGAG 荧光定量PCR内参引物 PeNTB-R AATAGCTGTCCCTGGAGGAGTTT PvJ3-2-F TCCCCCGGGATGTTCGGGCGCGCG 亚细胞定位 PvJ3-2-R CGCGGATCCCTGTTGTGCGCACTGCACTC PvJ3-3-F tccATCGATATGTTCGGGCGCGCG 过表达载体构建 PvJ3-3-R cgcGGATcCttaCTGTTGTGCGCACTGCACTC Table 1. PCR primers used in this study
用Vector NTI预测基因序列的开放阅读框(ORF)区,据此设计引物(PvJ3,表 1),重复上述实验过程,最终测序得到雷竹PvJ3基因序列。运用TMHMM和ProtScale分析PvJ3蛋白的结构特征和理化性质;Smart在线分析其蛋白的结构域;通过Clustal X2比对雷竹PvJ3与其他物种中的J3同源基因的氨基酸序列的相似性;并用Mega 5.0构建系统发育树,选择邻近法(NJ),bootstrap回复次数为1 000次。
-
用雷竹各组织部位的cDNA为模板,通过实时定量聚合酶链式反应(qRT-PCR),分析雷竹PvJ3基因的表达模式。根据实验要求设计目的基因的定量引物(PvJ3-1,表 1),毛竹的PeNTB基因作为内参基因[33]。各种雷竹组织做3次重复,避光条件下操作,参照SYBR Premix Ex TaqTMⅡ(Takara)试剂盒要求加样。PCR扩增程序(两步法):预变性95 ℃,5 min;95 ℃,30 s;60 ℃,30 s,循环数为40。反应结束后,分析确定数据的可靠性,选择合适的数据,用2-△△CT方法[34]对数据进行分析。
-
根据PvJ3基因ORF区序列(不含终止密码子)和pCaM35S-sGFP载体序列上的酶切位点,在目的基因ORF区的上下游分别引入酶切位点SmaI和BamHI,设计引物(PvJ3-2,表 1),提取pMD19-PvJ3质粒做为模板,进行PCR反应,胶回收纯化得到带有酶切位点的基因片段与pMD19(simple)-T载体连接转化后,阳性单克隆送测序。将测序正确的单克隆进行扩大培养,用质粒提取试剂盒(上海生工生物公司)提取质粒,用限制性内切酶SmaI和BamHI分别对pMD19-PvJ3和pCaM35S-sGFP载体质粒进行双酶切实验,之后用T4 DNA连接酶在16 ℃条件下过夜连接胶回收的酶切产物,连接产物转化大肠埃希菌DH5α感受态细胞,经测序正确,瞬时表达载体PvJ3-GFP构建成功。通过基因枪轰击洋葱Allium cepa表皮,25 ℃下避光暗培养1~2 d后,用荧光显微镜观察绿色荧光蛋白信号,金粉的准备和样品的制备参照文献[35]。
-
根据PvJ3基因ORF区序列和pC1301载体(含有35 S启动子)的序列设计引物(PvJ3-3,表 1),在目的基因ORF区的上下游同样分别引入酶切位点SmaI和BamHI,构建过表达载体pC1301-PvJ3,实验方法参照1.5节。
-
通过热转化法将构建成功的pC1301-PvJ3过表达载体转入农杆菌Agrobacterium tumefaciens GV3101感受态细胞中,提取阳性单克隆的质粒,重新转化大肠埃希菌DH5α感受态细胞,再次测序检验转入重组质粒是否正确。用已正确转入pC1301-PvJ3重组载体的农杆菌配制侵染液,侵染进入花期的野生型拟南芥花序[36],侵染3次,间隔约1周·次-1,侵染后的拟南芥即T0代植株,将消毒处理后的T0代种子均匀撒在加入潮霉素的1/2 MS(Murashige and Skoog)固体培养基上,潮霉素质量浓度为100.00 mg·L-1,按照V(潮霉素溶液):V(1/2MS培养基)=1.50:1 000.00比例加入培养基中,筛选抗性植株,以采用十六烷基三甲基溴化铵(CTAB)法[37]提取的抗性植株的基因组DNA为模板,PCR鉴定出T1代阳性植株,并对植株表型进行初步统计。
1.1. 实验材料
1.2. 核糖核酸(RNA)的提取及互补脱氧核糖核酸(cDNA)的合成
1.3. 目的基因的克隆及序列分析
1.4. 组织特异性表达检测
1.5. 亚细胞定位
1.6. 构建转基因过表达载体
1.7. 拟南芥转基因植株检测
-
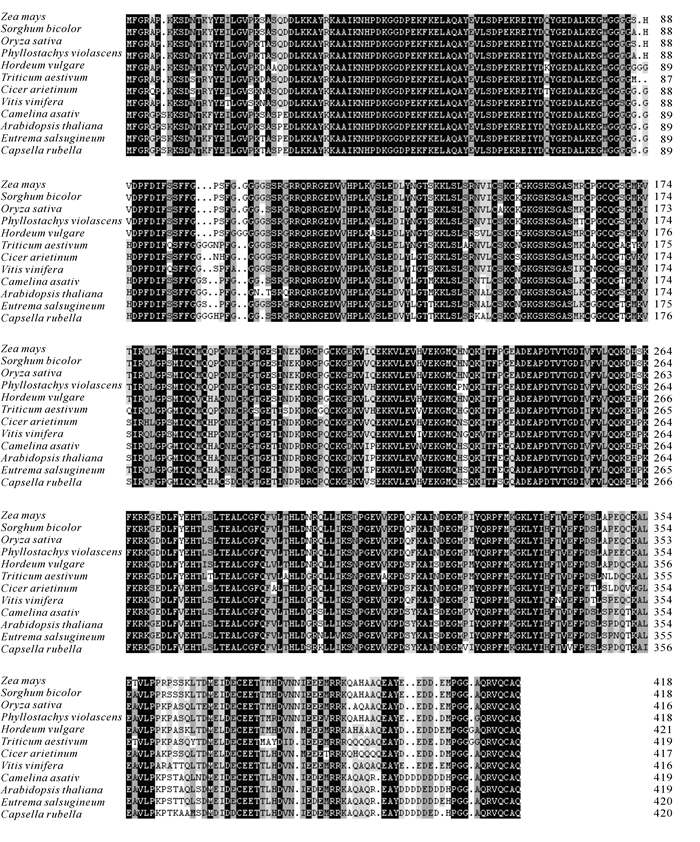
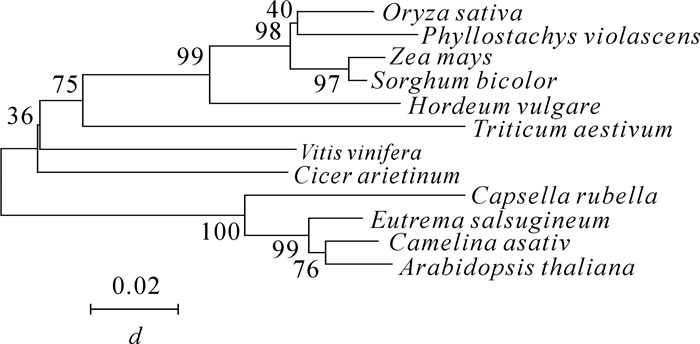
根据在毛竹转录组数据库中挑选的基因设计引物,利用同源克隆的方法,从雷竹中分离得到1个J3同源基因,命名为PvJ3,分析PvJ3基因的碱基序列,可知,其ORF区由1 260 bp个碱基组成,能编码419个氨基酸。经蛋白结构域和氨基酸序列分析(图 1~2),可知PvJ3蛋白的N端有J蛋白家族典型的J结构域,同时具有锌指结构域,C末端有法尼基化信号CAQQ序列。用ClustalX2软件对不同物种的J3同源蛋白的氨基酸进行序列比对,结果发现:J3蛋白在各类物种中的相似性很高,与拟南芥J3蛋白的一致性可达80.90%(图 2)。采用临近法构建系统进化树,结果显示:雷竹J3蛋白与单子叶植物高粱Sorghum bicolor和玉米Zea mays的亲缘关系最近,水稻Oryza sativa,大麦Hordeum vulgare和小麦Triticum aestivum次之,与双子叶拟南芥和亚麻荠Camelina sativa的亲缘关系最远(图 3)。
-
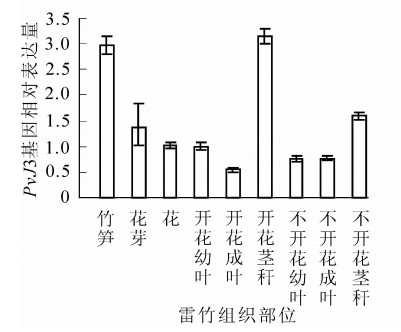
通过qRT-PCR技术,对同一竹鞭上开花和不开花雷竹中PvJ3基因在不同组织部位的表达水平进行分析。结果表明:PvJ3基因在同一竹鞭的笋、花芽、花,开花与不开花的茎秆、成叶、幼叶中均有表达。在开花雷竹的茎秆中PvJ3基因的表达量最高,茎、花芽、花、幼叶和成叶中的表达依次降低。在不开花雷竹的茎秆中PvJ3基因的表达量同样也是最高,幼叶、成叶中表达量基本一样(图 4)。
-
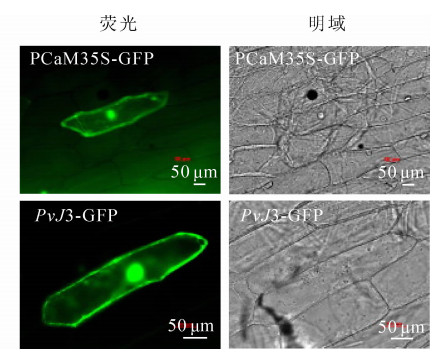
用基因枪轰击洋葱表皮细胞,通过荧光显微镜观察瞬时表达载体PvJ3-GFP绿色荧光蛋白的信号,确定PvJ3蛋白的定位情况(图 5)。pCaM35S-GFP对照在整个洋葱细胞里均有荧光信号,而PvJ3-GFP在细胞质核细胞核里有荧光信号,因此推断PvJ3蛋白主要定位在细胞质和细胞核中。
-
构建过表达载体pC1301-PvJ3转化野生型拟南芥收取T0代种子,潮霉素筛选抗性植株,以抗性植株基因组DNA为模板,经PCR检测得到T1代拟南芥共37株。观察统计T1代植株中长势较好的30株转基因植株的表型,发现PvJ3基因的过表达植株开花时间明显早于野生型(图 6B)。统计结果显示:转基因植株开花天数比野生型植株显著提早(P=6.51E-15<0.01),平均提早3.50 d(图 6C),开花时莲座叶数目比野生型植株显著减少(P=9.24E-09<0.01),平均减少4.43叶(图 6D),且表现出多主茎现象(图 6A)。
2.1. 基因的克隆和序列分析
2.2. 组织特异性表达分析
2.3. 亚细胞定位结果分析
2.4. PvJ3基因在拟南芥中的过量表达分析
-
高等植物成花过程存在多种诱导途径,大量基因参与这些途径的调控,成花关键基因的研究对于开花过程及花发育调控有着重要意义[38]。拟南芥中研究发现,SOC1和FT是感知不同开花途径信号的整合因子,SVP是直接调控FT和SOC1的转录调控因子[8, 39],J3蛋白通过与SVP直接相互作用而衰减SVP与SOC1和FT调控序列的结合从而上调SOC1和FT的表达量,参与拟南芥的成花过程,这一发现使得拟南芥成花调控网络更加完善[15]。本研究成功从雷竹中克隆得到J3同源基因并命名为PvJ3,雷竹PvJ3所编码的氨基酸序列与拟南芥J3蛋白氨基酸序列一致性达80.90%,对其蛋白结构域分析可知,PvJ3拥有相对保守的J结构域以及锌指结构域,可以说明PvJ3属于J家族中的一员。
为了研究雷竹PvJ3蛋白的功能,本研究通过qPCR技术和亚细胞定位分析该蛋白在雷竹中的表达模式和表达部位。对雷竹PvJ3基因进行了不同组织部位特异性表达分析,结果显示雷竹PvJ3基因在同一竹鞭的不同组织部位都有表达,已发现双子叶植物拟南芥J3基因同样在根、茎、叶、花芽、花和果荚等不同组织中表达[29],说明J3基因在单双子叶植物中均表现出全株表达的现象。在相对表达量上,PvJ3基因在开花雷竹茎秆中表达量最高,在花芽和花中的表达量降低,这与已报道的拟南芥中J3基因在花中表达量最高[15]的结果存在差异。PvJ3蛋白亚细胞定位结果显示该蛋白定位在细胞质和细胞核,与拟南芥中J3蛋白的亚细胞定位结果相一致[15],拟南芥中和J3蛋白具有互作关系的SVP,SOC1和FT蛋白都是核蛋白,说明雷竹PvJ3可能在细胞核中与这些开花整合因子相互作用,从而对开花途径进行调控。
将PvJ3基因导入野生型拟南芥植株验证其功能,T1代转基因植株的表型统计结果表明:转基因拟南芥植株开花时间明显早于野生型植株,开花时莲座叶数量明显减少(图 6 C,图 6D),而拟南芥J3基因的过表达植株和野生型植株的开花时间并没有明显差别[15],这说明雷竹PvJ3基因可以显著影响花期,可能是一个开花促进因子。值得注意的是,转雷竹PvJ3基因拟南芥植株同时表现出多主茎现象(图 6A),这一现象在拟南芥J3过表达植株中并没有被观测到[15]。以上结果表明,雷竹PvJ3基因不仅在成花调控中的功能和拟南芥J3基因存在一定差异,同时还可能参与茎的发育等其他生长发育过程,在基因功能上比拟南芥同源基因要复杂的多。
目前,J3基因功能的有关研究主要以模式植物拟南芥为研究对象,单子叶植物中还未见报道。通过对比J3基因在雷竹和拟南芥中的表达模式不难发现:雷竹PvJ3基因和拟南芥J3基因都是全株表达,同时编码蛋白都定位在细胞质和细胞核,这表明J3基因在不同植物中的表达部位不存在明显差异,并且可能是一个核内作用蛋白。在相对表达量上,雷竹PvJ3基因和拟南芥J3基因存在一定差异,同一竹鞭上开花和不开花雷竹中PvJ3基因在茎秆中表达量最高,而拟南芥中J3基因表达量在花中最高,这可能与雷竹PvJ3基因参与茎的发育有关。转基因实验的结果表明:雷竹PvJ3基因不仅可以显著提早拟南芥开花时间,同时还促使拟南芥出现多主茎现象,这表明雷竹PvJ3基因很可能参与除了开花调节以外的其他生长发育过程的调控。由于拟南芥和竹子之间的差异很大,雷竹PvJ3基因在竹子中是否可以促进植物提早开花,同时还参与哪些生物学过程,需要后续研究进一步验证。
















 DownLoad:
DownLoad: