-
花器官的研究一直以来是植物发育学研究中的一大热点。1991年Coen等[1] 在拟南芥Arabidopsis thaliana和金鱼草Antirrhinum majus的花器官同源异型突变体的遗传学研究基础上,首先提出了“ABC”模型,随后Theiβen等[2]又陆续提出了有关植物花发育的新模型,即“ABCD”模型、“ABCDE”模型及四元体模型。目前,对于酵母MCMI-拟南芥AG-金鱼草DEFA-动物血清应激因子SRF(MADS)转录因子的研究主要集中在它们的遗传学功能[3]及其植物新陈代谢调控方面[4]。通过对拟南芥突变体的研究,证明SEPALLATA-like基因对于花粉的成熟及花粉管的形成有着重要作用[5]。SEP-like基因是编码MADS蛋白的转录因子,在双子叶植物中基因的功能表现出不同程度的冗余性和亚功能化[5],E类开花基因的同源蛋白在花器官的发育形成方面比其他A/B/C 类转录因子更重要[6-7]。SEP3属于E类的拟南芥AGL9型基因,编码蛋白在MADS box家族形成四聚体复合物过程中起着重要作用[6]。MADS蛋白在开花过程中主要通过与其他蛋白互作[8],参与了许多调控开花过程[9]。SEP3蛋白是否只是在结合其他转录因子后形成异聚复合物[10]时才发挥作用,还是在形成同源多聚体后也有生物学功能,目前尚不清楚。已有的研究证明:MADS蛋白与特异的DNA结合形成同源多聚体并行使其功能[11],但SEP3蛋白同源多聚体形成的作用机制及其与功能之间的关系,至今还没有明确的答案。本研究尝试通过体外表达纯化可溶性SEP3蛋白的途径加以分析,将不同结构域的AtSEP3基因,克隆到原核表达载体pET-15b上,诱导表达后筛选获得可溶性表达,纯化获得高质量的AtSEP3蛋白。这项工作为通过AtSEP3蛋白的结构功能解析来分析植物花形成分子机制提供了重要基础。
-
哥伦比亚(Columbia,Col)野生型拟南芥由浙江农林大学亚热带森林培育国家重点实验室培育基地实验室栽植。大肠埃希菌Escherichia coli DH5α感受态细胞,表达菌株E. coli‘Rosetta’以及表达质粒pET-15b为浙江农林大学亚热带森林培育国家重点实验室培育基地保存;RNA提取试剂盒购自北京鼎国昌盛有限公司;DNA聚合酶、各种DNA限制性内切酶及DNA购自宝生物工程(大连)有限公司;DNA连接酶购自New England公司;DNA纯化试剂盒、胶回收试剂盒购自天根生化科技(北京)有限公司;引物由生工生物工程(上海)股份有限公司合成;其余各试剂均为国产分析纯。
-
用Trizol法提取拟南芥花芽组织RNA,反转录得到cDNA。根据拟南芥的SEP3基因的cDNA保守序列设计2个不同结构域(包括MIKC域,IKC部分)的引物,聚合酶链式反应(polymerase chain reaction,PCR)扩增后得到的产物连接到pMD18-T载体并转化到大肠埃希菌DH5α后取样测序。将测序验证正确的菌株扩大提取质粒后,用NcoⅠ和BamHⅠ双酶切提取的质粒和pET-15b载体,DNA胶回收试剂盒回收酶切产物后将片段和载体16 ℃过夜连接后转化大肠埃希菌DH5α并涂布氨苄抗性的LB平板上,挑菌培养后菌液PCR检测,并送菌样品测定碱基序列,克隆得到不同结构域片段的AtSEP3原核表达载体。
-
不同的诱导温度对于原核表达蛋白的可溶性有密切的关系,高温条件下菌株快速表达目的蛋白容易使蛋白形成不正确的构象折叠最后以包涵体的形式存在,不利于获得可溶性的蛋白。测序鉴定正确的重组质粒转化于表达宿主菌株E. coli ‘Rosetta’中,37 ℃培养15.0 h,挑取单菌落转接到100.0 mL的含50.0 mg·L-1卡那霉素的LB液体培养基中,在37 ℃,培养3.0 h,菌液浓度在吸光度D(λ)600为0.4~0.6时,加入终浓度为0.1 mmol·L-1 的异丙基硫代半乳糖苷(IPTG),分别在37 ℃,5.0 h 和22 ℃,15.0 h 不同的温度及时间条件下诱导培养。确定适合诱导的培养温度。
-
上述纯化得到的重组蛋白经聚丙烯酰氨凝胶电泳(SDS-PAGE)分离检测,考马斯亮蓝染色后,用手术刀小心切取目的条带,置于1.5 mL离心管中(加入小量双蒸水ddH2O),由北京信科奥达科技有限公司做肽质量指纹图谱鉴定分析。
-
使用Ni-TNA树脂初步纯化后用凝胶过滤层析柱纯化可溶性的融合蛋白。具体步骤如下:5 000 g离心收集菌液沉淀,加入10.0 mL 0.5 mol·L-1氯化钠,20.0 mmol·L-1三羟甲基氨基甲烷-盐酸(Tris-HCl)pH 8.0的裂解液,冰水下功率为200 W ,5 s开8 s关超声10.0 min,静置30.0 min 后4 ℃条件17 000 r·min-1离心40.0 min。收集上清液,平衡好Ni-NTA柱,将上清液上柱,在冰上用摇床缓慢结合30.0 min后,收集未特异性结合Ni-NTA柱的杂蛋白,分别用10个柱体积的不同咪唑浓度梯度的缓冲液(4/20/80 mmol·L-1咪唑,0.2 mol·L-1 氯化钠,20.0 mmol·L-1 Tris-HCl pH 8.0)洗脱非特异性结合的杂蛋白,最后用10倍体积的洗脱溶液(200.0 mmol·L-1咪唑,0.2 mol·L-1 氯化钠,20.0 mmol·L-1 Tris-HCl pH 8.0)冲洗收集目的蛋白。使用凝胶过滤层析柱纯化目的蛋白。将收集的洗脱液用超滤浓缩柱4 000 g离心浓缩至500.0 μL体积后上样,以洗脱缓冲液(0.2 mol·L-1 氯化钠,20.0 mmol·L-1Tris-HCl pH 8.0),0.5 mL·min-1流速分离纯化目的蛋白。取小量蛋白样品加入2倍蛋白上样缓冲液,煮沸10.0 min,离心沉淀再次超声后取10.0 μL加入10.0 μL上样缓冲液煮沸10.0 min后点样体积分数为15% SDS-PAGE检测,观察目的蛋白的表达纯化情况。
-
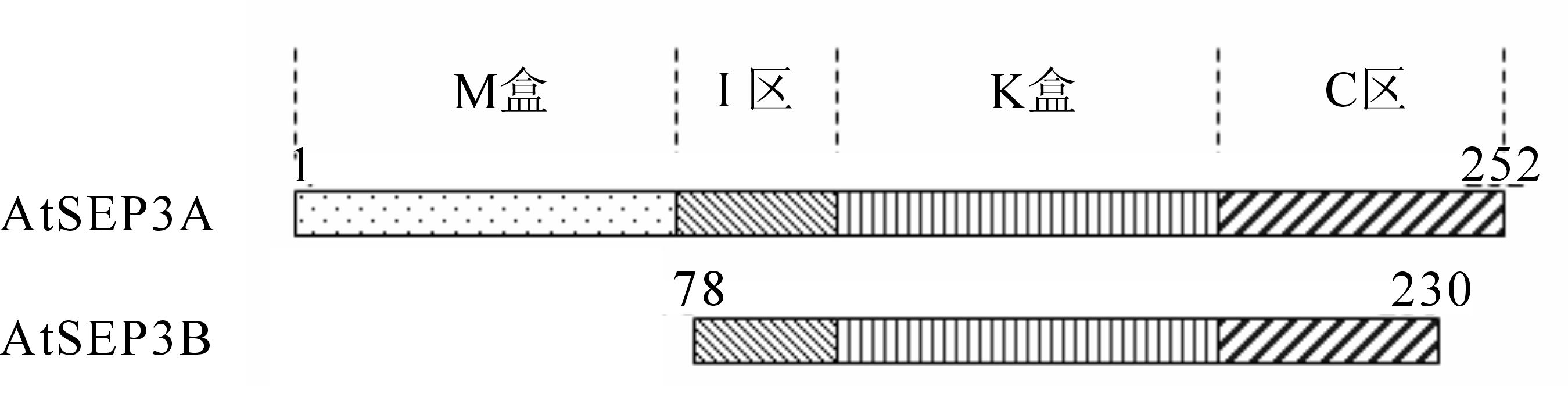
测序验证得到2个不同结构域的AtSEP3结构域的序列,构建不同大小结构域的载体(图 1)[11]。转化成功的菌株提取质粒进行PCR鉴定和双酶切鉴定均得到目的片段,成功构建得到不同结构域的pET15b-AtSEP3原核表达载体。
-
将构建好的原核表达菌株扩大培养,用裂解液溶解蛋白,SDS电泳图中(图 2)箭头所指是目的蛋白,检测发现AtSEP3A(30 kDa)以包涵体形式表达,AtSEP3B(18 kDa)为可溶蛋白。实验表明,包含部分IKC区的AtSEP3B以pET-15b为原核表达载体,在22 ℃条件下终浓度0.1 mmol·L-1 的IPTG诱导表达15.0 h,得到高表达的可溶蛋白。
-
为了检测原核表达蛋白,对SDS-PAGE考马斯亮蓝染色后显示的目的蛋白进行肽质量指纹图谱鉴定。北京信科奥康科技有限公司提供的初始检索结果显示(检索程序是Mascot: http://www.matrixscience.com,数据库为SwissProt):目的蛋白与蛋白NP_850953.1,鉴定得分为92分,检测结果中有18条肽段与目的蛋白序列相匹配(图 3A),红色部分表示与目的蛋白所匹配的多肽片段,匹配部分占目的蛋白的42%。Mascot Score直方图(图 3B),蛋白得分大于53分具有显著性差异(P<0.05),阴影区域为不可信任结果,阴影外红色表示匹配的结果,与数据库比对得到验证。Mascot Score直方图所示如下,左侧的小片段相匹配的肽段不具备显著性差异,没有说明意义,右侧红色柱状图表示与数据库蛋白相匹配的肽段,且具有显著性差异,说明纯化的融合蛋白是AtSEP3蛋白。
-
纯化各阶段产物的电泳结果如下图 4A所示。作为标准的小牛血清蛋白(BSA)溶液以单体和二聚体形式存在,大小分别是67 kDa和134 kDa。 黑色实线表示AtSEP3B,峰值位置接近BSA单体67 kDa。图 4B中SDS-PAGE检测蛋白凝胶过滤层析后样品纯度,箭头所指为目的蛋白,说明纯化得到了高纯度的AtSEP3B蛋白,其单肽链为18 kDa,图 4A中峰值大小靠近 67 kDa左右,表明纯化的AtSEP3B是以四聚体形式存在。
-
虽然植物开花分子机制的研究在模式植物上已经取得了一定的进展,但是随着研究的深入凸现了一些新的问题,例如植物控制花器官形成过程中的分子机制,MADS盒蛋白的结构及其功能机制均不明确。本研究以拟南芥成花过程中重要的调控基因SEP3为例,设计了不同结构域的载体片段,纯化得到了可溶的AtSEP3蛋白。
SEP3基因以高级复合物的形成存在,在开花不同环节中都扮演着重要作用[12],在 ABCE 模型形成的各个阶段都不可缺少。SEP3蛋白结合特异性的DNA片段后行使其转录因子的作用,还与其他特定的蛋白相互作用从而激活或抑制下游基因蛋白的功能[13]。Immink等[9]在2009年提出的SEP3四元体模型,其中MADS盒的M结构域结合DNA并稳定四聚体。本研究通过对AtSEP3蛋白结构域的分析表明,SEP3蛋白在形成四聚体时并不需要M结构域。换言之,结合DNA对四聚体的形成是不必要的。
大肠埃希菌原核表达系统适合多种原核基因和真核基因的蛋白表达。但外源蛋白的表达以及是否可溶与多种因素有关,包括表达温度、原核表达载体、结构域构建以及表达菌株。温度对于SEP3表达的可溶性有重要的影响。降低诱导温度,延长表达时间明显有利于SEP3外源蛋白的正确折叠和表达产量。可溶性与目的蛋白的性质也密切相关,极性的均一性影响着蛋白的可溶表达。生物信息学预测表明SEP3蛋白M区是亲水性,其他IKC区是疏水性。本研究筛选了不同的片段和原核表达体系,最终获得了AtSEP3B结构域片段的可溶性表达。这些结构域以四聚体形式存在,疏水区域有可能构成单体相互作用界面,稳定了四聚体,其余部分构成四聚体亲水表面,促进了蛋白可溶性。进一步通过结构生物学的方法分析SEP3蛋白四聚体形成以及空间结构与功能之间的关系正在进行中。
Arabidopsis thaliana SEPALLATA3 protein in a prokaryotic system
-
摘要: SEPALLATA3(SEP3)属于花器官建成ABCE模型中的E类基因,为了揭示SEP3蛋白是如何形成多聚体的,以及多聚体与其生物学功能之间的联系,开展了拟南芥Arabidopsis thaliana AtSEP3蛋白质体外可溶性表达实验,构建了原核表达载体,并在大肠埃希菌Escherichia coli细胞中进行重组蛋白的诱导表达。从蛋白不同的结构域、诱导时间和诱导温度等方面对蛋白的可溶性表达进行了分析。最后以pET-15b为表达载体,大肠埃希菌表达菌株E. coli ‘Rosetta’为宿主菌株,22℃诱导表达15 h,获得不同片段的AtSEP3可溶性蛋白。通过镍柱及分子层析色谱柱分离纯化,表明AtSEP3蛋白以四聚体形式存在。Abstract: The transcription factor of SEPALLATA3 (SEP3) belongs to the E class gene of the ABCE model of flower development. SEP3 has been shown to mediate complex formation and, therefore,a special attention was paid to its structure-function relationship. To determine how SEPALLATA3 protein oligomers were formed, the oligomeric status was studied by obtaining soluble protein expression in vitro. The SEP gene(AtSEP3) was constructed as a fusion protein and expressed in prokaryotic cells. After determining an optimal expression strategy, nickel chelating resin was used to purify the protein which was displayed as a tetramer in size exclusion chromatography(SEC). To get the high efficiency of soluble protein,different structural domains and expression conditions were screened,and 0.1% IPTG inducing at 22℃ for 15 h produced an optimal expression strategy. These preliminarily results provided an important basis for further study of the structure-function relationship. Overall,these results indicat that the SEP3 ‘glue’ protein in mediating multimerization is based on homomeric interactions.
-
Key words:
- botany /
- SEP3 protein /
- prokaryotic system /
- protein purification /
- polymer
-
-
[1] COEN E S,MEYEROWITZ E M. The war of the whorls:genetic interactions controlling flower development[J]. Nature,1991,353:31-37. [2] THEIβEN G. Development of floral organ identity:stories from the MADS house[J]. Curr Opin Plant Biol,2001,4(1):75-85. [3] SOLTIS D E,LEEBENS-MACK J H,SOLTIS P S. Developmental Genetics of the Flower:Advances in Botanical Research 44[M]. New York:Academic Press,2006. [4] ACKERMAN C M,YU Qingyi, KIM S,et al. B-class MADS-box genes in trioecious papaya:two paleoAP3 paralogs, CpTM6-1 and CpTM6-2,and a PI ortholog CpPI[J]. Planta,2008, 227:741-753. [5] CUI Rongfeng,HAN Jiakun, ZHAO Suzhen,et al. Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa)[J]. Plant J,2010,61(5):767-781. [6] MELZER R,VERELST W, THEIβEN G. The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro[J]. Nucl Acids Res, 2009,37(1):144-157. [7] CASTILLEJO C,ROMERA-BRANCHAT M,PELAZ S. A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression[J]. Plant J,2005,43(4):586-596. [8] IMMINK R G H,KAUFMANN K,ANGENENT G C. The ‘ABC’ of MADS domain protein behaviour and interactions[J]. Sem Cell & Dev Biol,2010,21(1):87-93. [9] IMMINK R GH,TONACO I AN,de FOLTER S,et al. SEPALLATA3:the ‘glue’ for MADS box transcription factor complex formation[J]. Genome Biol,2009,10(2):R24. doi:10.1186/gb-2009-10-2-r24. [10] TSAFTARIS A, PASENTSIS K, MAKRIS A,et al. The study of the E-class SEPALLATA3-like MADS-box genes in wild-type and mutant flowers of cultivated saffron crocus (Crocus sativus L.) and its putative progenitors[J]. J Plant Physiol,2011,168(14):1675-1684. [11] HUANG Hai, TUDOR M,SU T,et al. DNA binding properties of two Arabidopsis MADS domain proteins:binding consensus and dimer formation[J]. Plant Cell,1996,8:81-94. [12] ADAMCZYK B J,FERNANDEZ D E. MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis[J]. Plant Physiol,2009,149(4):1713-1723. [13] SMACZNIAK C, IMMINK R G H,MUIÑO J M,et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development[J]. Proc Nat Acad Sci USA,2012,109(5):1560-1565. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.2014.01.003







 下载:
下载: