-
近年来,全球气候变暖,降雨量减少以及不适当的灌溉,导致干旱以及半干旱地区土壤盐渍化愈加严重,土壤盐渍化成为全球农业生产中面临的重大难题[1-2]。目前,中国可耕地面积中约1/5的盐渍化土地,总面积高达0.98亿hm2[2]。中国是世界人口大国,人均耕地面积不足0.1 hm2,如何利用盐渍化的土地,促进粮食产量的增加是艰巨的课题[3],因此,研究植物的耐盐性和生理机制,筛选及培育耐盐作物品种成为农业研究领域的热点之一。
在长期的进化过程中,植物进化出多种应对盐胁迫的防御机制,其中报道较多的主要有4种:①渗透调节平衡机制。分为有机渗透调节和无机渗透调节。无机渗透调节是植物细胞通过吸收钠离子(Na+)和钾离子(K+)等无机离子作为渗透调节剂,将Na+在细胞内区隔化,从而使根和地上部积累较多的Na+,但不会出现明显的毒害症状[4-6]。有机渗透调节则是指植物通过自身合成并积累一些可溶性无毒的有机小分子物质,如糖类、氨基酸类和甜菜碱等来维持细胞渗透平衡[7-8]。如盐胁迫下,苹果Malus pumila通过积累可溶性有机溶质和无机盐离子来降低胞内渗透势[9-10],番茄Lycopersicon esculentum叶片中可溶性糖含量升高[11]。②离子的区域化与pH调节机制。植物细胞利用跨膜运输将Na+和K+等无机离子转运至液泡中,使之与细胞质隔绝,降低渗透势,避免细胞器遭受毒害[12]。同时,植物自身可以分泌有机酸从而调节根细胞中的pH稳态,缓解盐胁迫对根系的毒害[13]。③抗氧化防御系统机制。在高盐胁迫下,植物细胞质膜因受到水分胁迫或者离子胁迫而受损,从而导致脂质过氧化,使细胞内积累大量的活性氧,当积累的活性氧超过了细胞自身的清除能力就会对细胞造成损伤[8, 14-15]。在长期的进化过程中,植物自身会产生活性氧清除系统,包括一些抗氧化酶类等抗氧化物质,如超氧化物歧化酶(SOD)、过氧化物酶(POD)、过氧化氢酶(CAT)、谷胱甘肽过氧化物酶(GPX)等,它们是衡量植物抗逆性的重要指标[6,16-17]。④内源激素和信号转导途径。研究发现植物内源激素在盐胁迫反应中起着重要作用,如脱落酸(ABA)、乙烯(ET)、茉莉酸(JA)等[18-20]。同时,蛋白激酶途径、ABA途径(ABA依赖型、ABA不依赖型)、盐超敏感(SOS)途径等[21-24]信号转导途径也参与了植物对盐胁迫的响应,并在其中发挥重要作用。
植物激素信号调控网络是植物应对各种非生物胁迫所必须的[25]。茉莉酸(jasmonic acid,JA)作为重要的植物逆境激素在调控植物耐盐性方面发挥着重要的作用。耐盐水稻Oryza sativa品种内源茉莉酸含量高于盐敏感品种,外源茉莉酸甲酯处理能够提高水稻幼苗耐盐性和大豆Glycine max幼苗对盐胁迫的抗性[26-27]。盐胁迫下,水稻JA信号调控基因OsJAZ9与OsbHLH062互作介导离子稳态或通过增强抗氧化能力,从而增加对盐胁迫的耐受性[28-29];内源茉莉酸通过维持番茄体内活性氧稳态来提高番茄的耐盐性[30]。但是,目前关于JA信号如何调控植物的耐盐性仍鲜有报道。茉莉酰氨基酸结合物合成酶(jasmonoyl amino acid conjugate synthase,JAR1)催化JA形成JA-Ile,AtJAR1基因发生突变后可导致JA信号传递中断。因此,本研究运用CRISPR/Cas9基因编辑技术创建拟南芥Arabidopsis thaliana AtJAR1基因突变体,并考察突变体对盐和ABA胁迫下的种子萌发和根系生长影响,以及盐胁迫下突变体对K+的吸收转运和AtHAK5表达量的影响,从而解析JA信号通路在植物耐盐性中的可能生理机制。
-
CRISPR/Cas9靶序列的设计和载体的构建参照WANG等[31]和朱丽颖等[32]的方法。以拟南芥茉莉酰氨基酸结合物合成酶AtJAR1为靶基因,选取鸟嘌呤和胞嘧啶含量高、特异性较强的2个关键片段作为靶序列,获得双靶点CRISPR/Cas9基因编辑载体。以哥伦比亚野生型拟南芥为材料,使用农杆菌Agrobacterium tumefaciens侵染法进行拟南芥遗传转化[33-34]。
-
CRISPR/Cas9编辑载体具有mCherry荧光蛋白报告基因,因此,本研究参考朱丽颖等[32]的方法,挑选T1代转基因阳性种子,提取转基因阳性植株的基因组DNA,在目标基因靶序列2端设计引物进行PCR扩增(约200 bp),利用质量浓度8%的非变性聚丙烯酰胺凝胶(PAGE,丙烯酰胺与甲叉双丙烯酰胺的质量比为29∶1)对PCR产物进行检测[32]。
-
取播种后21 d的植株整个地上部,称其鲜质量。每个株系设置5个生物学重复,用单因素方差分析对株系间的差异显著性进行分析。
-
1 mL 体积分数为75%乙醇溶液清洗3 min,弃乙醇加入1 mL 体积分数为35%的漂白水和体积分数为0.05% 吐温20混合溶液,轻轻摇晃混匀5 min,弃混合溶液;最后加入1 mL灭菌超纯水,清洗5次,置于4 ℃避光春化2 d。
-
在1/2 MS植物培养基上添加不同浓度NaCl,分别配成0、75、100、125 mmol·L−1 NaCl的1/2 MS植物培养基。将ABA溶于体积分数为70%的乙醇中,配置成25 mmol·L−1的母液,参考STASWICK等[35]的浓度。用体积分数为70%的乙醇进行稀释,分别配成含0、0.25、0.50、1.00、5.00、10.00、20.00 μmol·L−1ABA的1/2 MS植物培养基。
-
将消毒后的种子均匀点在添加有不同浓度NaCl(0、75、100、125 mmol·L−1)和ABA (0、1、5、10、20 μmol·L−1)的1/2 MS植物培养基上,置于22 ℃、14 h光照/10 h黑暗的温室中培养。隔24 h统计1次萌发率,连续6 d,以拟南芥胚根穿透种皮时即认定萌发。每个浓度重复3次。萌发率=试验时间(d)的萌发种子总数/供试种子总数×100%。
-
将消毒春化后的种子单粒点于1/2 MS植物竖直培养基上,于22 ℃、16 h光照/8 h黑暗的条件下竖直培养5 d后,移栽在添加不同浓度NaCl(0、50、100 mmol·L−1)和ABA(0、0.25、0.50、1.00 μmol·L−1)的1/2 MS植物竖直培养基上,继续培养7 d,以根长来评估它们的生长状况。
-
将1/2 MS植物培养基上生长7 d幼苗,移栽至装有1/5 Hoagland培养液的蓝色花盆中(配方见表1),培养14 d后对每个株系分别进行盐处理和对照处理。盐处理更换外源添加50 mmol·L−1NaCl的培养液,在处理0和24 h时,分别取每个株系的根部样品用于基因实时表达量的检测。处理2 d后,将每个株系的地上部和根部用去离子水润洗后分别转入做好标记的信封中,放入恒温60 ℃烘箱连续烘6 h以上进行杀青。
组成 母液浓度/
(mol·L−1)母液添加体
积/(mL·L−1)水培液中浓
度/(mmol·L−1)组成 母液浓度/
(mol·L−1)母液添加体
积/(mL·L−1)水培液中浓
度/(mmol·L−1)KNO3 1.0 1.25 1.25 KH2PO4 0.5 1.00 0.50 Ca(NO3)·4H2O 1.0 1.00 1.00 Fe盐① 1.00 MgSO4·7H2O 0.4 1.00 0.40 微量元素② 0.10 说明:①1 000倍铁盐母液的配置:称取5.56 g七水合硫酸亚铁(FeSO4·7H2O)放入100 mL烧杯中,边加水边搅拌;将7.50 g 二水合乙 二胺四乙酸二钠(EDTA·2Na·2H2O)放入1 L烧杯中加水煮沸,缓慢加入FeSO4溶液,于微波炉中煮沸2 min;避光放入60 ℃烘 箱烘2 h以上;室温静置,冷却后定容至1 L,即为浓度为20 mmol·L−1的母液。使用时稀释1000倍。②10 000倍微量元素母液的 配置:称取H3BO3 6.18 g;MnCl2·4H2O 0.99 g;CuSO4·5H2O 1.25 g;ZnSO4·7H2O 1.44 g;H2MoO4 0.08 g;NaCl 0.20 g溶解 于水中,冷却后定容至1 L。使用时稀释10 000倍 Table 1. 1/5 Hoagland formula for hydroponic culture solution
-
将待测样品研磨后,称取50 mg左右样品用称量纸送至洁净干燥的消解管底部,每个样本设置3次生物学重复,依次加入7 mL浓硝酸、1 mL 质量分数为30%的过氧化氢水溶液,室温放置2 h,最后放入微波消解仪消解。利用石墨赶酸仪于190 ℃下蒸发反应液至2 mL左右。冷却后将反应液倒入离心管内,用超纯水将反应液定容至15 mL。
-
使用钾和钠的氯化物分别配制标准曲线。离子的质量浓度梯度为0、0.2、0.4、0.6、0.8、1.2、1.6、2.0 mg·L−1。利用原子吸收光谱法测定K+和Na+的质量摩尔浓度。
-
具体的计算公式如下:C = [(Cn –C′) × 0.015/(M×10−3)]/m。其中:C为1 g干物质样本中该离子的物质的量(μmol·g−1);Cn为各样品消解液中该离子的质量浓度(mg·L−1);C′为空白对照消解液中该离子的质量浓度(mg·L−1);M为待测元素的相对分子量(g·mol−1)。m为样品的干物质质量(g)。
-
采用Trizol法提取拟南芥根中总RNA,之后进行基因组DNA的去除、反转录,以反转录后的cDNA为模板进行实时荧光定量PCR (qRT-PCR)检测基因的相对表达量。从拟南芥信息资源库中查找以编码拟南芥肌动蛋白基因Actin2为内参基因,根据已知的AtVSP1、AtVSP2和AtHAK5基因(Gene ID: AT5G24780、AT5G24770和AT4G13420)序列,设计qRT-PCR引物(表2)。采取
$2^{-\Delta \Delta C _{{t}}} $ 法计算待测基因的相对表达量。基因 上游引物(5′→3′) 下游引物(5′→3′) AtActin2 GTCGTACAACCGGTATTGTGCT TGTCTCTTACAATTTCCCGCTCT AtVSP1 TGGATCTTTGACCTAGACGACA GAGTTCCAAGAGGTTTTCGTA AtVSP2 TGACCTAGATGATACCCTCCTCTC CAATCCCGAGCTCTATGATGTT AtHAK5 TCTGCATCACTGGGACGGAG CAGTATAACGGATCAGGGATTGA Table 2. Sequence of related primers
-
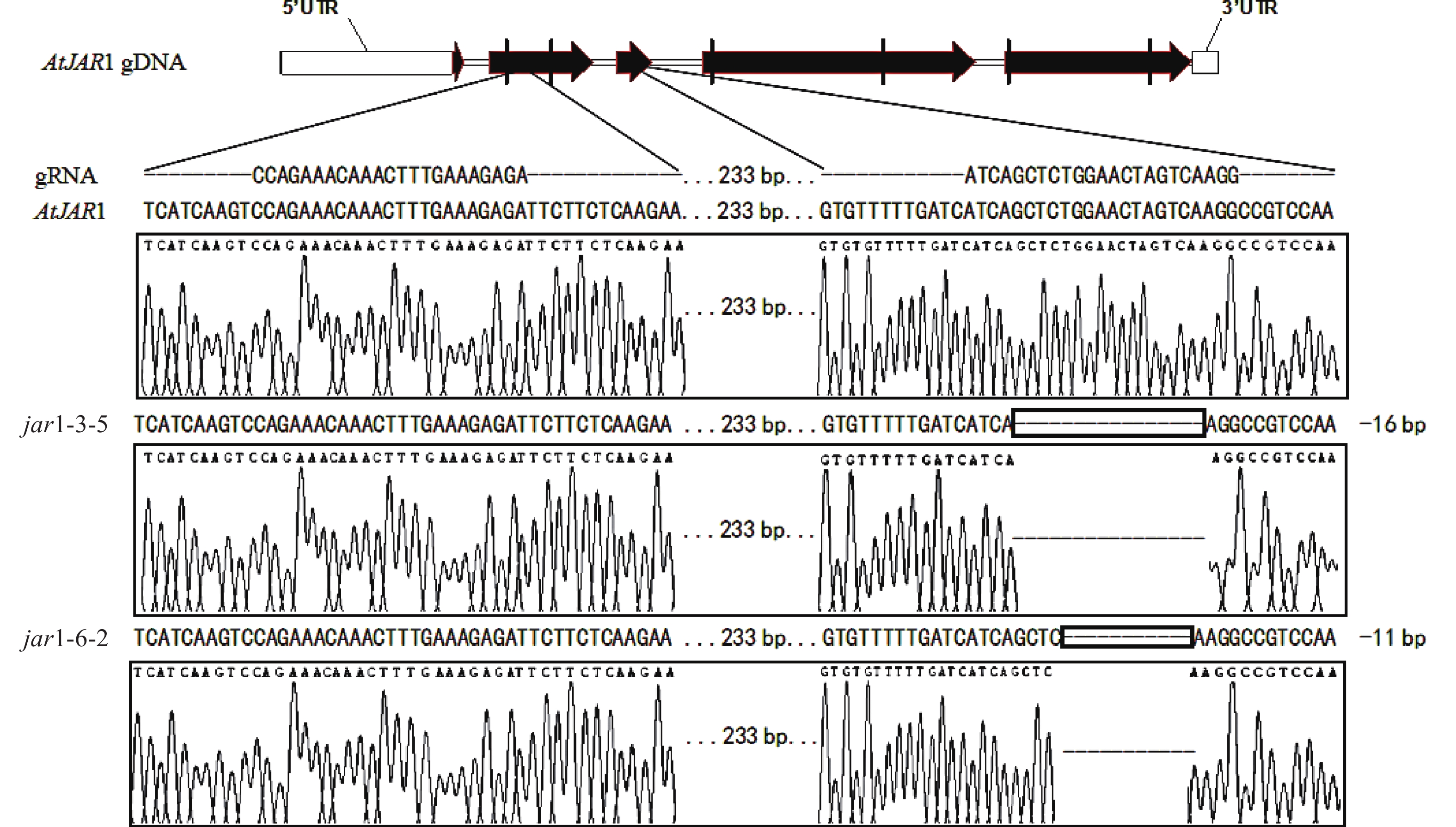
在早期的基因功能研究中,常采用反向遗传学手段,构建基因突变体。因技术水平的限制,前人构建突变体的方法多为化学诱变法。然而,这种方式构建的突变体多只在目标基因发生点突变,使氨基酸残基发生变化,基因功能未必完全丧失。例如,有研究发现:使用甲基磺酸乙酯(EMS)诱变创建的ATS1基因突变体存在严重的基因渗透现象[36]。因此,本研究利用CRISPR/Cas9基因编辑技术构建AtJAR1功能完全丧失型突变体。经多代筛选鉴定,从转基因后代中获得了2个纯合突变体,分别将其命名为jar1-3-5和jar1-6-2。对其靶位点附近序列进行测序分析,结果表明:jar1-3-5和jar1-6-2突变体在第2个靶位点,即第3个外显子处,分别发生了16和11 bp缺失的突变(图1),导致编码序列(CDS)发生了相应变化,氨基酸序列发生移码突变。在转录后翻译过程中,jar1-3-5的第111位氨基酸残基由丝氨酸突变为异亮氨酸(I),并在继续错误翻译12个氨基酸(SGTSQGRPKFIP-KAVQSLFLSLMN)后,引入终止密码子UAA,提前终止翻译;jar1-6-2的第113位氨基酸残基由甘氨酸突变为精氨酸,并在继续错误翻译8个氨基酸(TSQGRPKF-PSKVYSFH)后,引入终止密码子UGA,提前终止翻译。因此,构建的突变体可判断为功能完全丧失型突变体。
-
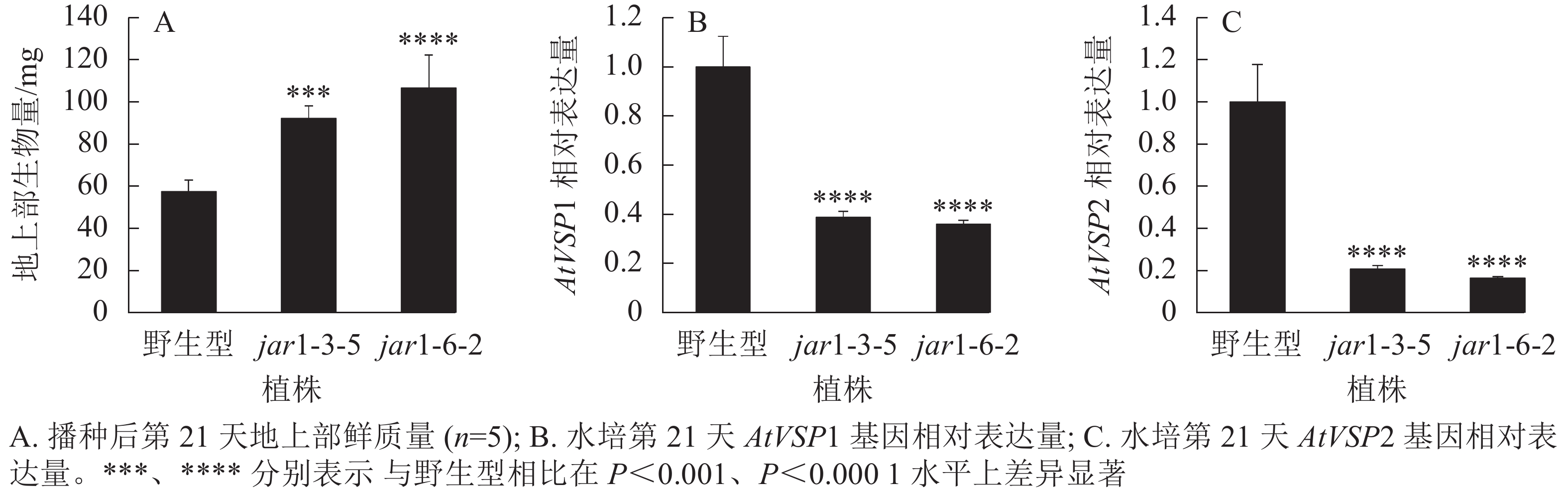
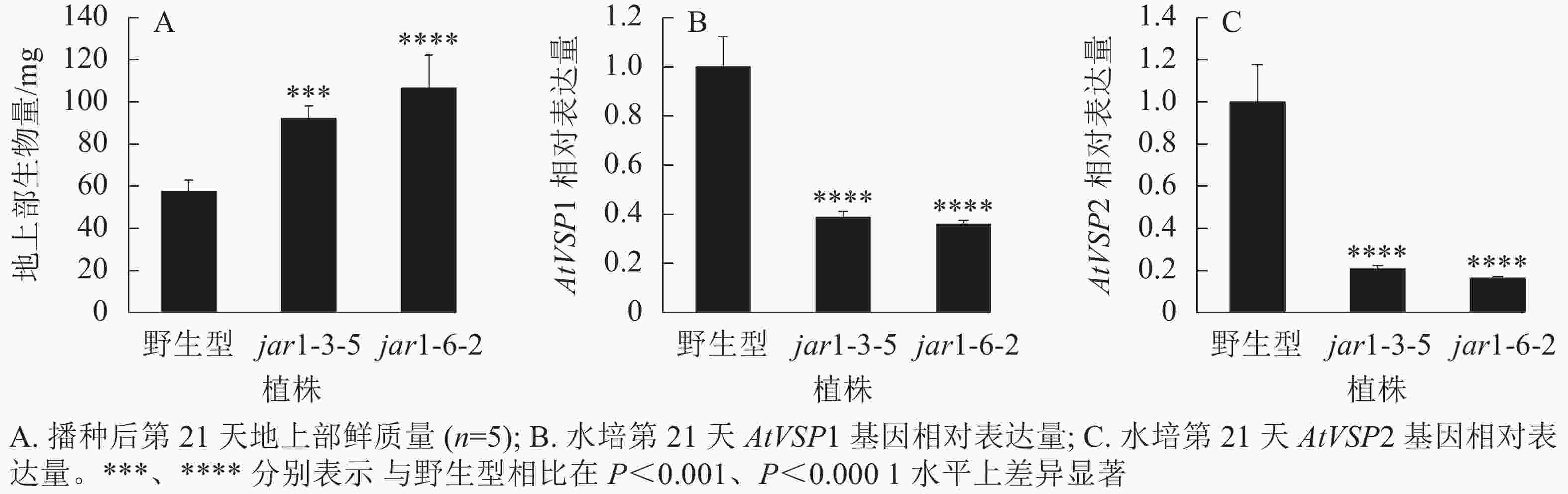
将筛选的突变体(jar1-3-5和jar1-6-2)和野生型种子进行播种,在生长前期观察并比较两者的表型差异。结果显示:前21 d时,突变体植株可以正常生长,甚至其地上部明显大于野生型。进一步对生长21 d的植株地上部生物量进行统计分析发现:突变体极显著(P<0.001)高于野生型(图2A)。之后,野生型和突变体之间的生长差异逐渐减小,并且在生长32 d时突变体植株叶片开始出现萎蔫的现象。
此外,AtJAR1可以催化JA形成JA-Ile,激活JA信号途径,AtJAR1基因突变会影响植物体内茉莉酸信号强度。因此,对茉莉酸信号标记基因AtVSP1和AtVSP2进行表达量分析显示:AtJAR1突变后,AtVSP1和AtVSP2基因在根中的表达量极显著(P<0.0001)减少,其中AtVSP1表达量下降了60%左右(图2B),AtVSP2表达量下降了80%左右(图2C)。表明获得的这2个突变体中茉莉酸信号强度明显减弱。
-
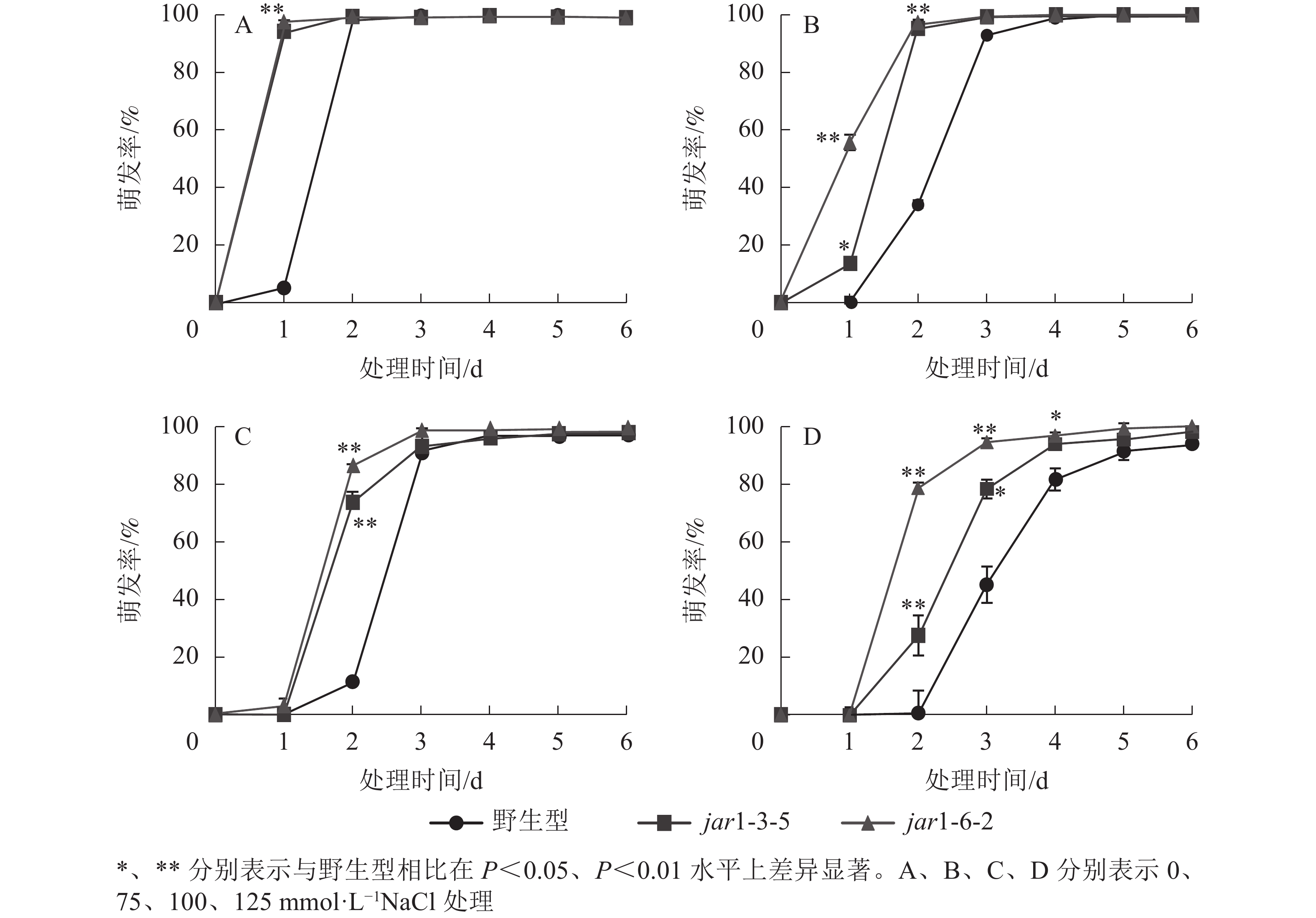
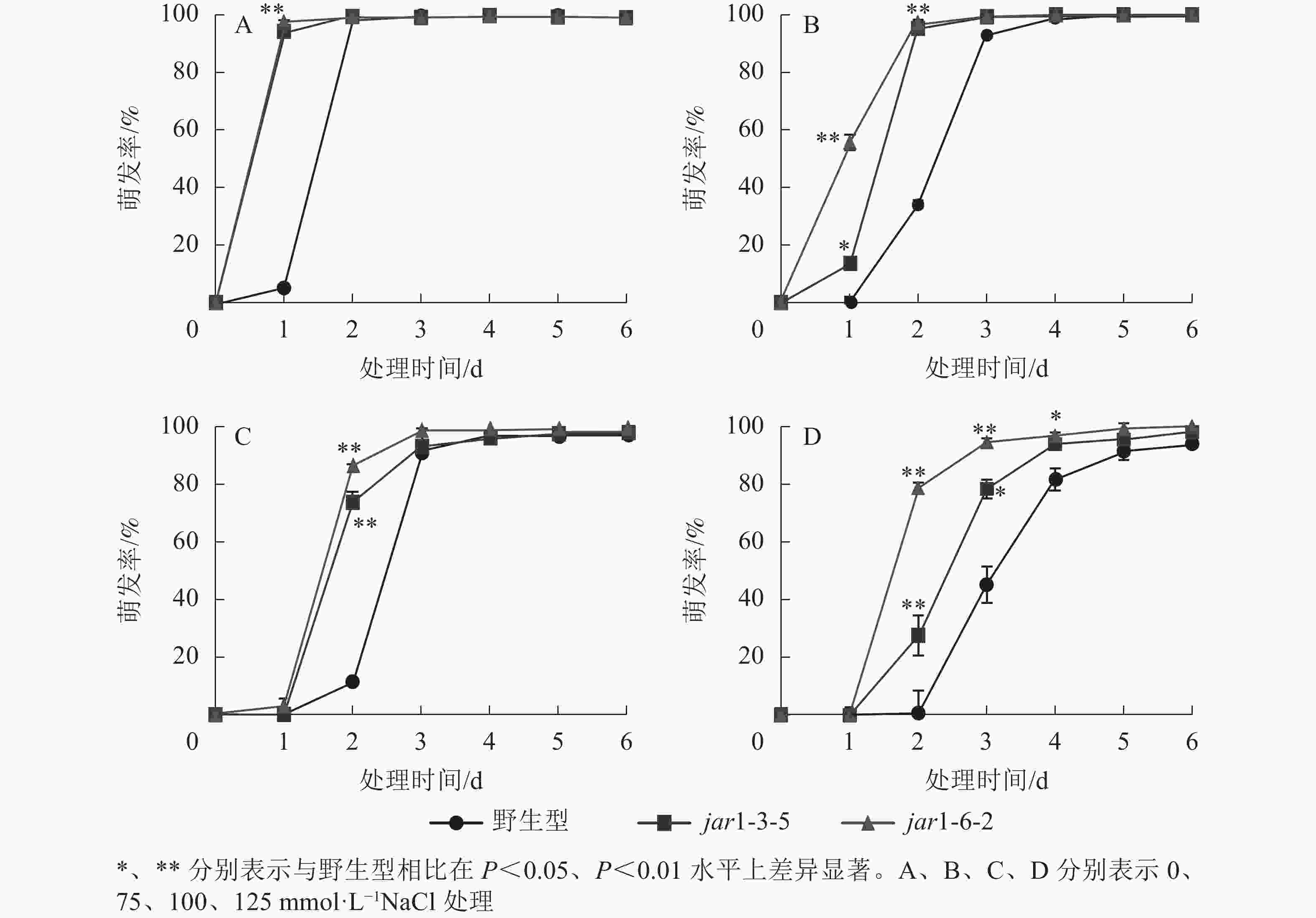
种子萌发是植株个体发育的重要阶段。已有研究表明:盐胁迫能够抑制植物种子萌发,同时,JA作为重要的植物逆境激素在调控植物耐盐性方面发挥着重要的作用[37-38]。然而JA信号通路在盐胁迫下的种子萌发过程中发挥着何种作用尚不明确,因此,对盐胁迫下野生型和突变体的种子萌发率(图3)进行统计分析可知:在不含NaCl的条件下,第1天突变体种子的萌发率极显著(P<0.01)高于野生型,第2天时各株系基本全部萌发,没有显著差别。在不同浓度NaCl处理时,虽然突变体和野生型最终都能达到或者接近于正常生长条件下的最大萌发率,但是达到最大萌发率的时间都随着NaCl处理浓度的增加而有所推迟,且突变体均早于野生型;在处理的前3 d中突变体的萌发率虽随着NaCl处理浓度的增加有所下降,但均高于野生型,并且随着NaCl处理浓度的增加,这种差别逐渐显著。表明AtJAR1突变加速了种子的萌发,降低了拟南芥在幼苗建成时对盐的敏感性。
-
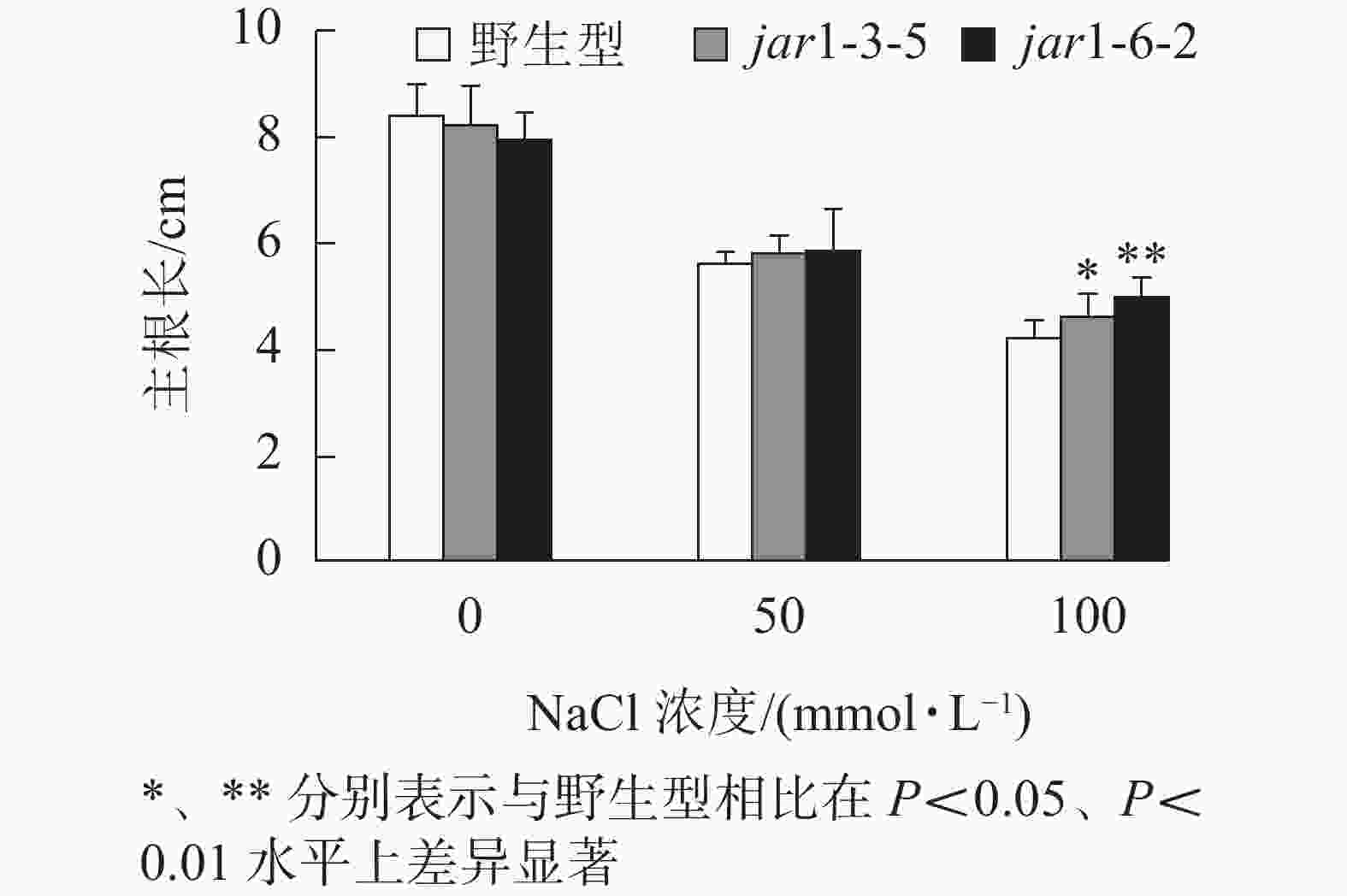
幼苗建成是植株个体发育的另一个重要阶段。一直以来盐胁迫对幼苗的影响也是进行抗逆研究的重要方向之一,因此,本研究比较了在盐胁迫条件下野生型和突变体幼苗根的生长情况(图4)。将在正常培养基上生长5日龄的幼苗转移至含有不同浓度NaCl的竖直培养基上继续培养7 d后,发现在未用NaCl条件下,突变体的主根长与野生型相比无显著差异。随着NaCl浓度的增加,两者的主根长度都随之下降,但是,突变体的主根长度逐渐大于野生型,在100 mmol·L−1 NaCl处理时,突变体的主根长显著(P<0.05)大于野生型,表明AtJAR1突变降低了拟南芥根对盐的敏感性。
-
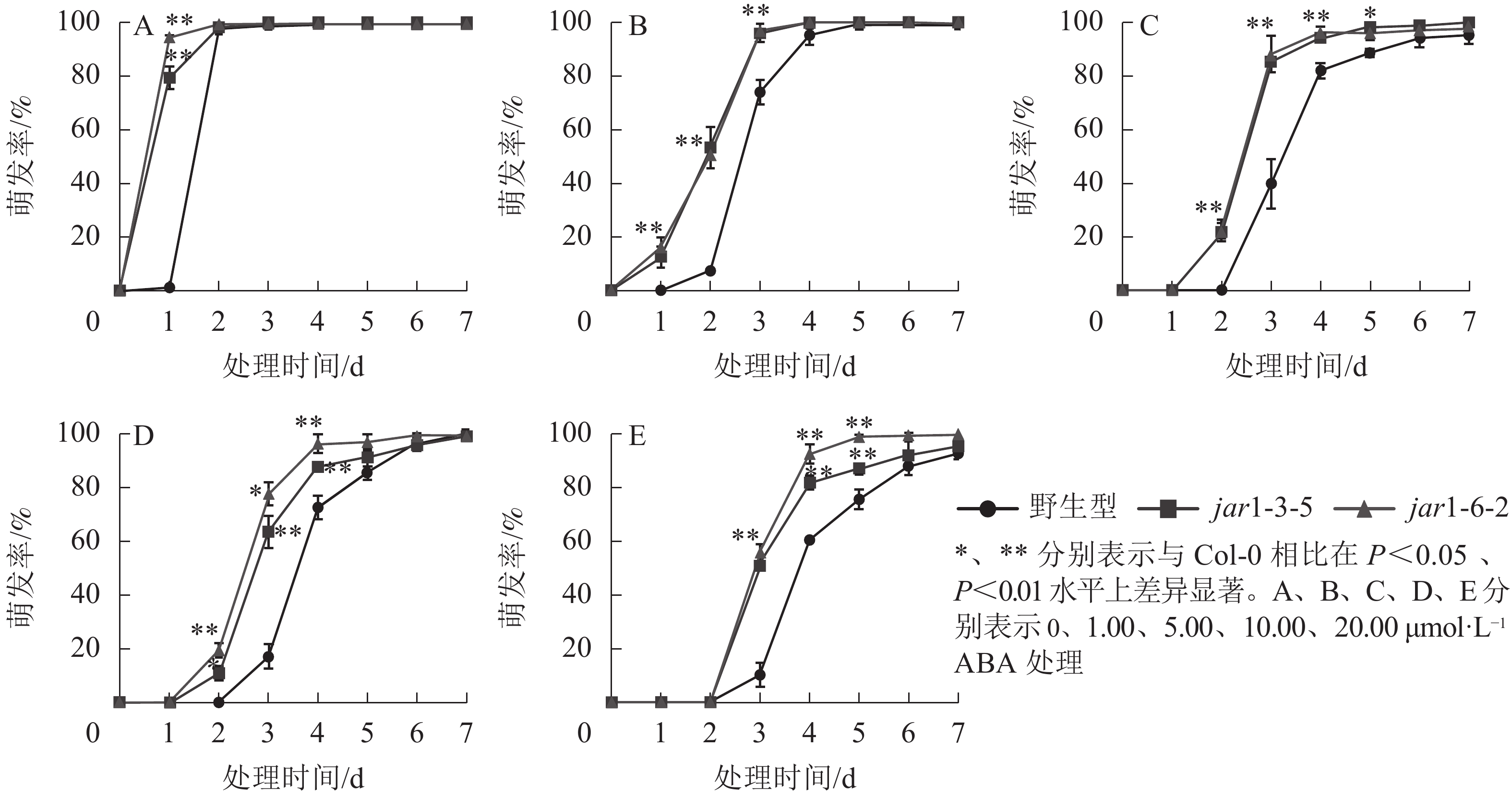
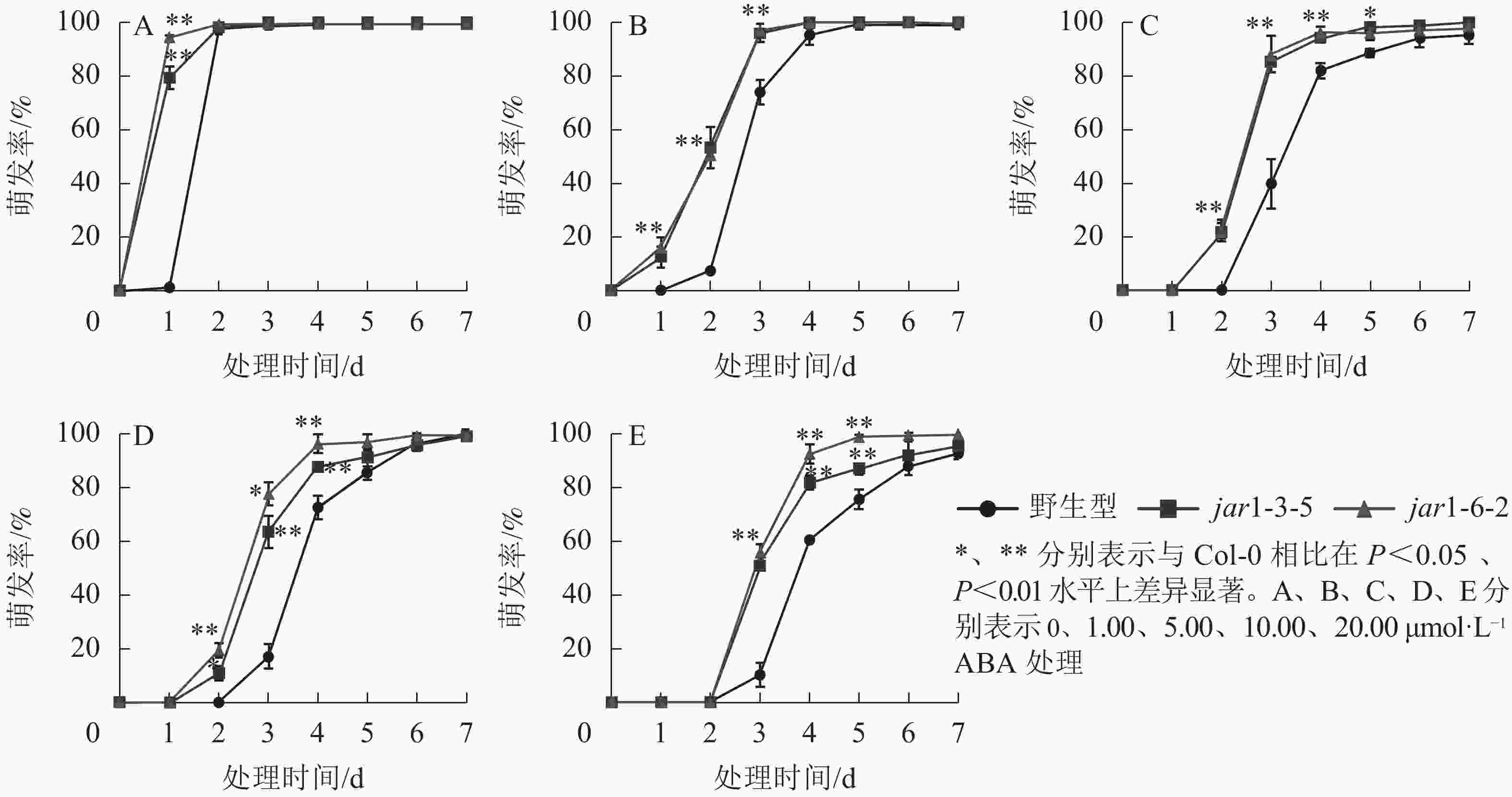
ABA作为重要的植物激素,常与其他激素一起协同调控植物对逆境胁迫的应答响应,但是,目前并不明确JA信号途径与ABA交互作用共同调控植物抗逆性的机制。本研究对jar1突变体在ABA胁迫下的种子萌发率进行了统计(图5)。结果表明:随着ABA浓度的增加,野生型和突变体种子的萌发时间都相应推迟,萌发率随之下降,但是突变体的萌发时间均要早于野生型,萌发率也均显著(P<0.05)高于野生型,表明AtJAR1突变缓解了ABA对种子萌发的抑制作用。
-
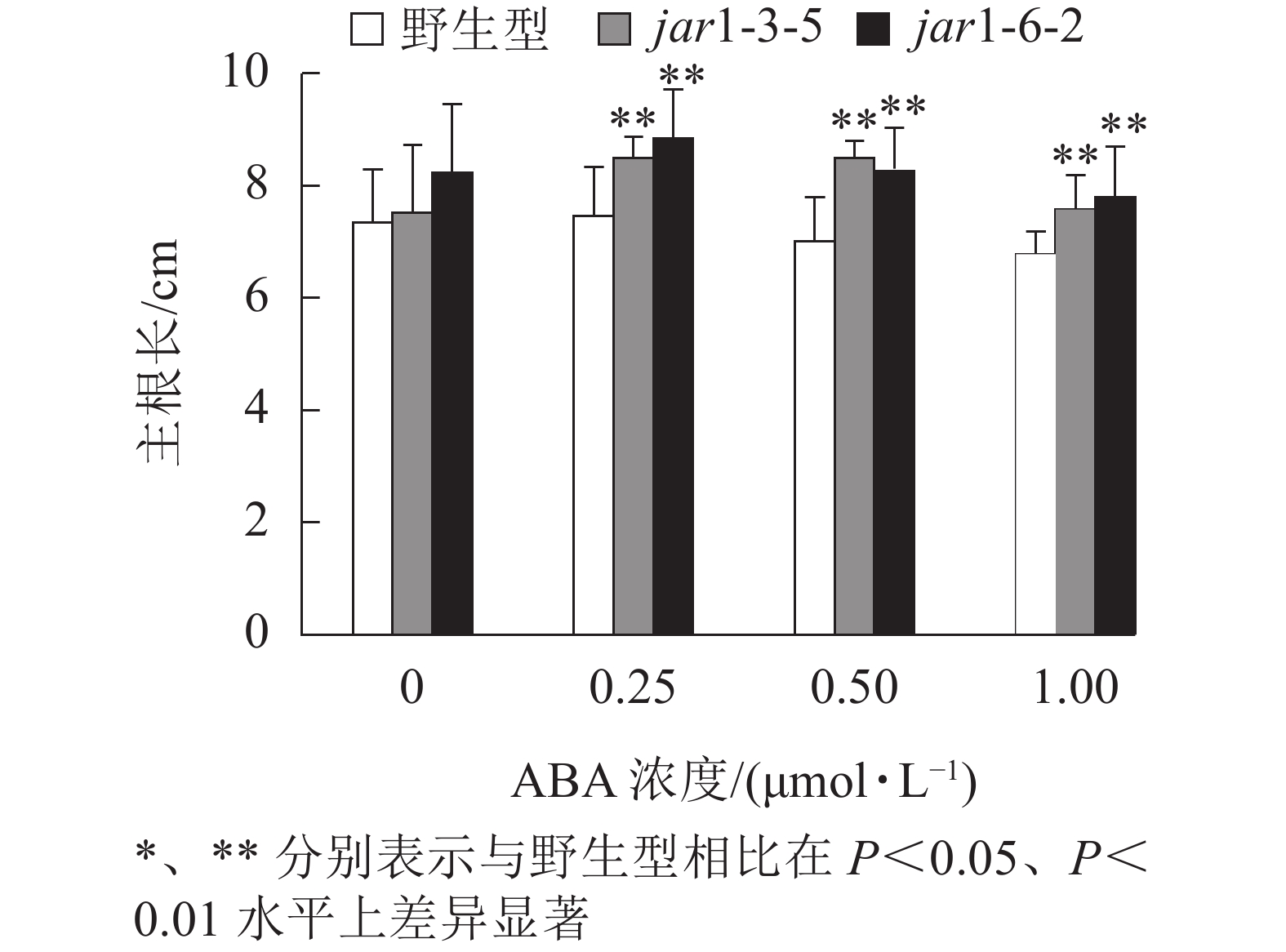
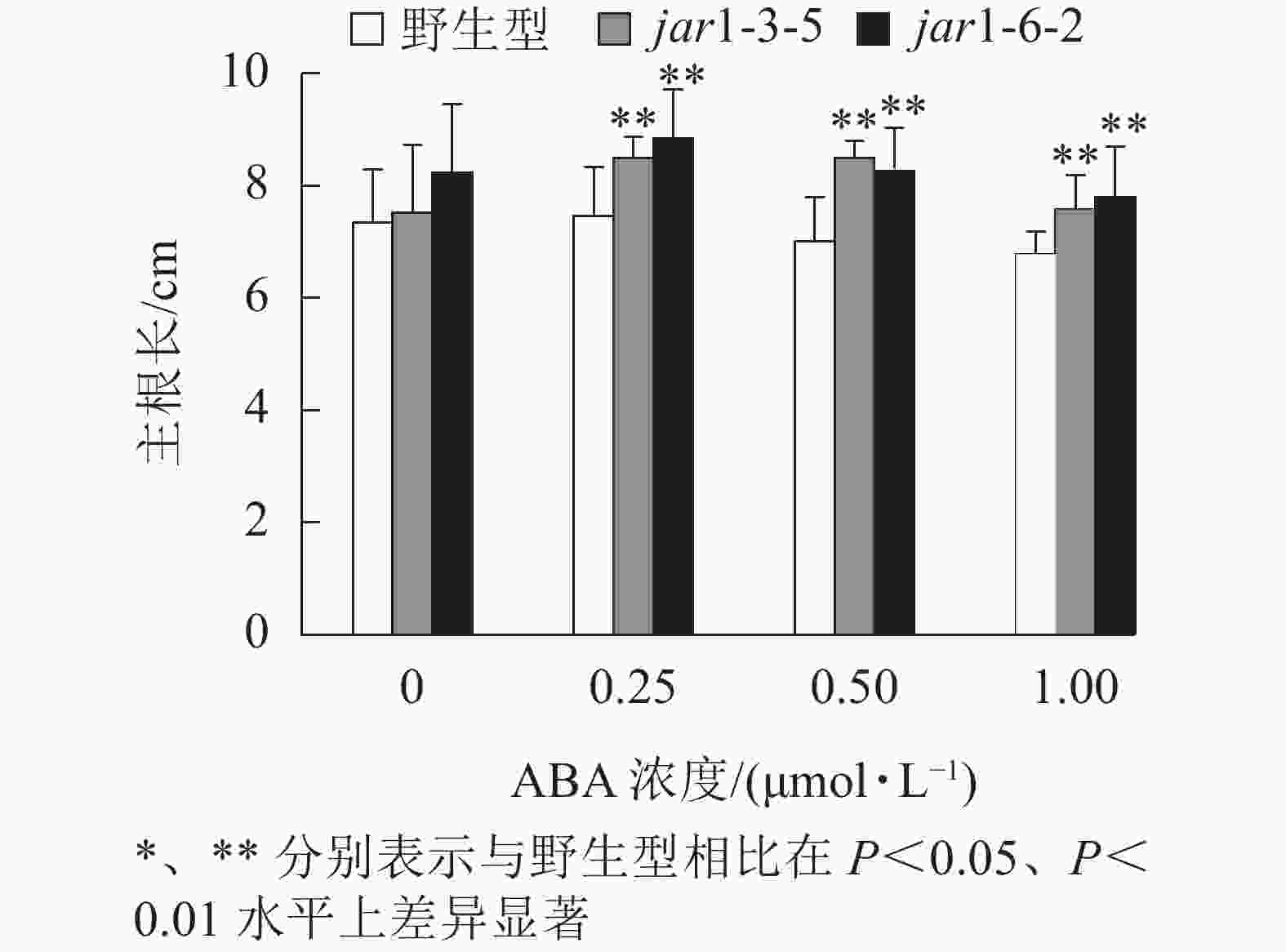
ABA胁迫下野生型和突变体幼苗根的生长情况见图6。将正常培养基上的5日龄幼苗转移到含有不同浓度ABA的竖直培养基上继续培养7 d后发现:在未用ABA处理时突变体的主根长与野生型相比无显著差异,但是,当用各浓度ABA处理时,突变体的主根长均极显著(P<0.01)大于野生型,表明AtJAR1突变降低了拟南芥根对ABA的敏感性。
-
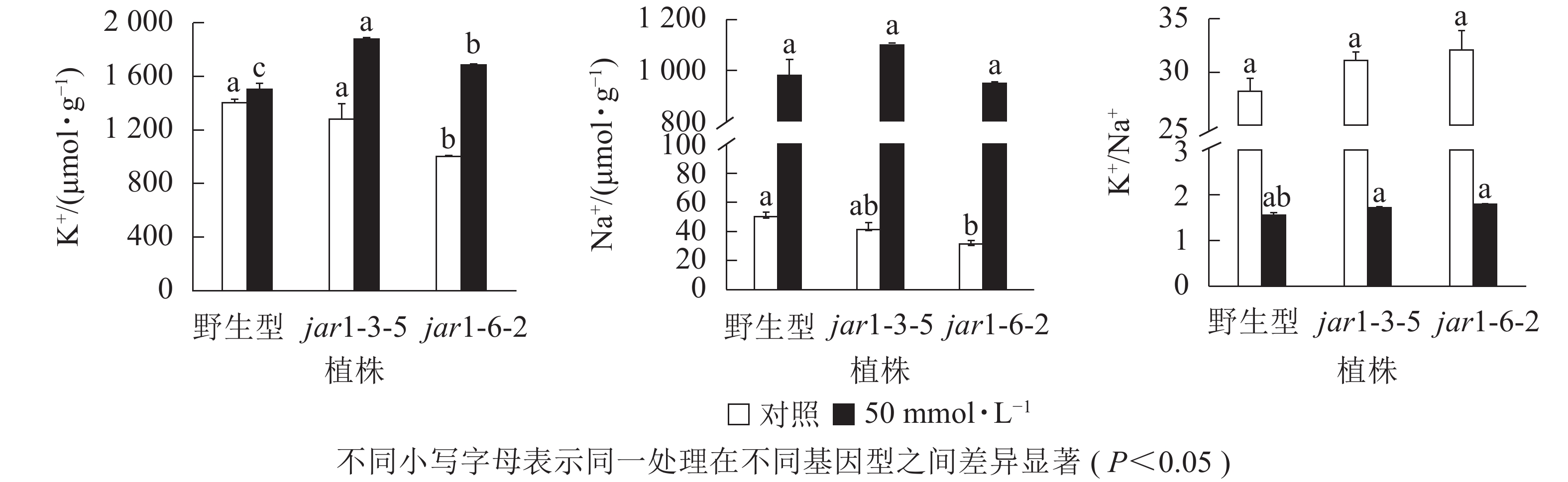
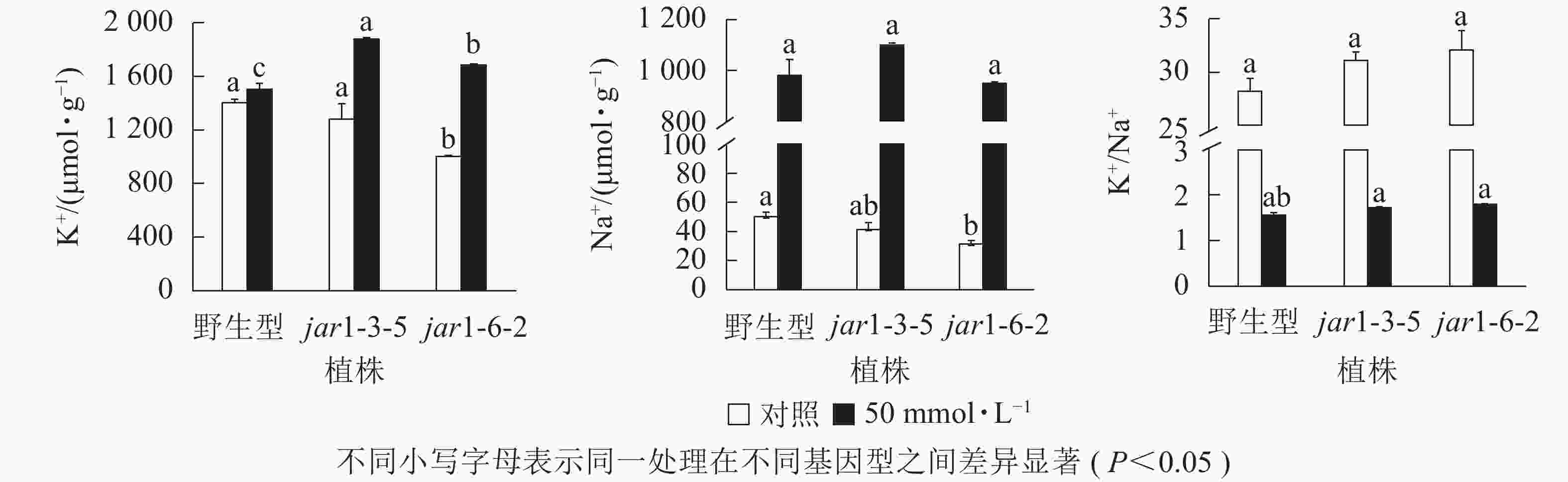
盐胁迫下Na+不断的积累,导致细胞K+质量摩尔浓度降低。为避免高盐条件下植物细胞中离子紊乱,维持K+稳定吸收,植物细胞胞浆中的代谢酶必须保持低Na+、高K+的浓度水平[39],因此,细胞中K+/Na+是盐胁迫下保证植物正常生长的关键。本研究利用原子吸收光谱法测定了正常和盐胁迫条件下突变体和野生型根中的Na+、K+质量摩尔浓度(图7),发现在正常生长条件下,突变体根中的Na+、K+质量摩尔浓度较野生型都有所下降,特别是jar1-6-2下降显著,但是K+/Na+并没有发生显著变化。当用50 mmol·L−1NaCl处理后,突变体根中的K+质量摩尔浓度较未处理时提高约40%,且显著(P<0.05)高于相同浓度NaCl处理下的野生型。NaCl处理后野生型和突变体根中的Na+质量摩尔浓度都急剧上升,但两者并没有显著差别。相应地,盐处理后野生型和突变体的K+/Na+都急剧下降,并且突变体比野生型高11%以上。表明AtJAR1突变,可能增加了盐胁迫下植物对K+的吸收和转运,影响了植物体内的钾钠平衡,从而增强了拟南芥的耐盐性。
-
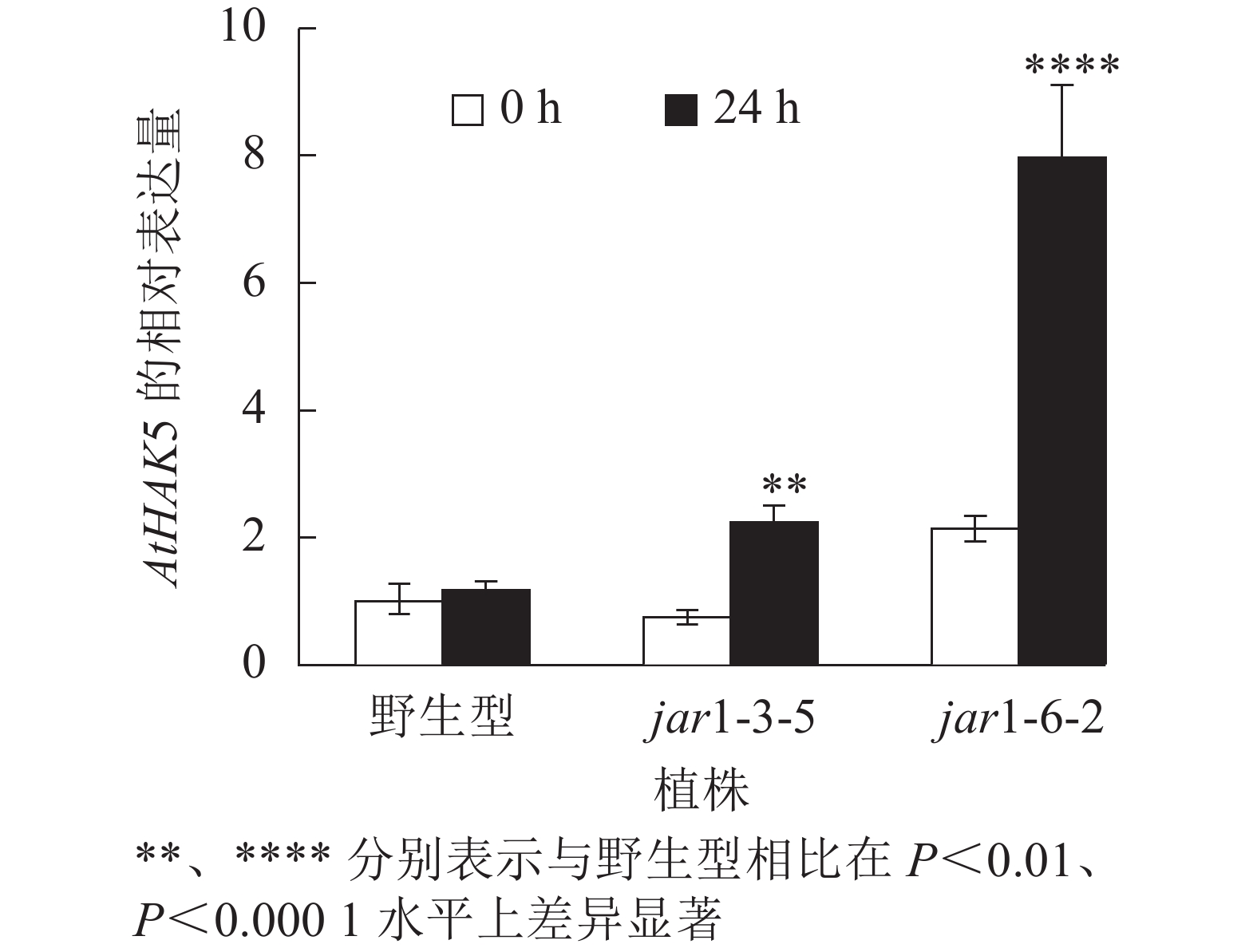
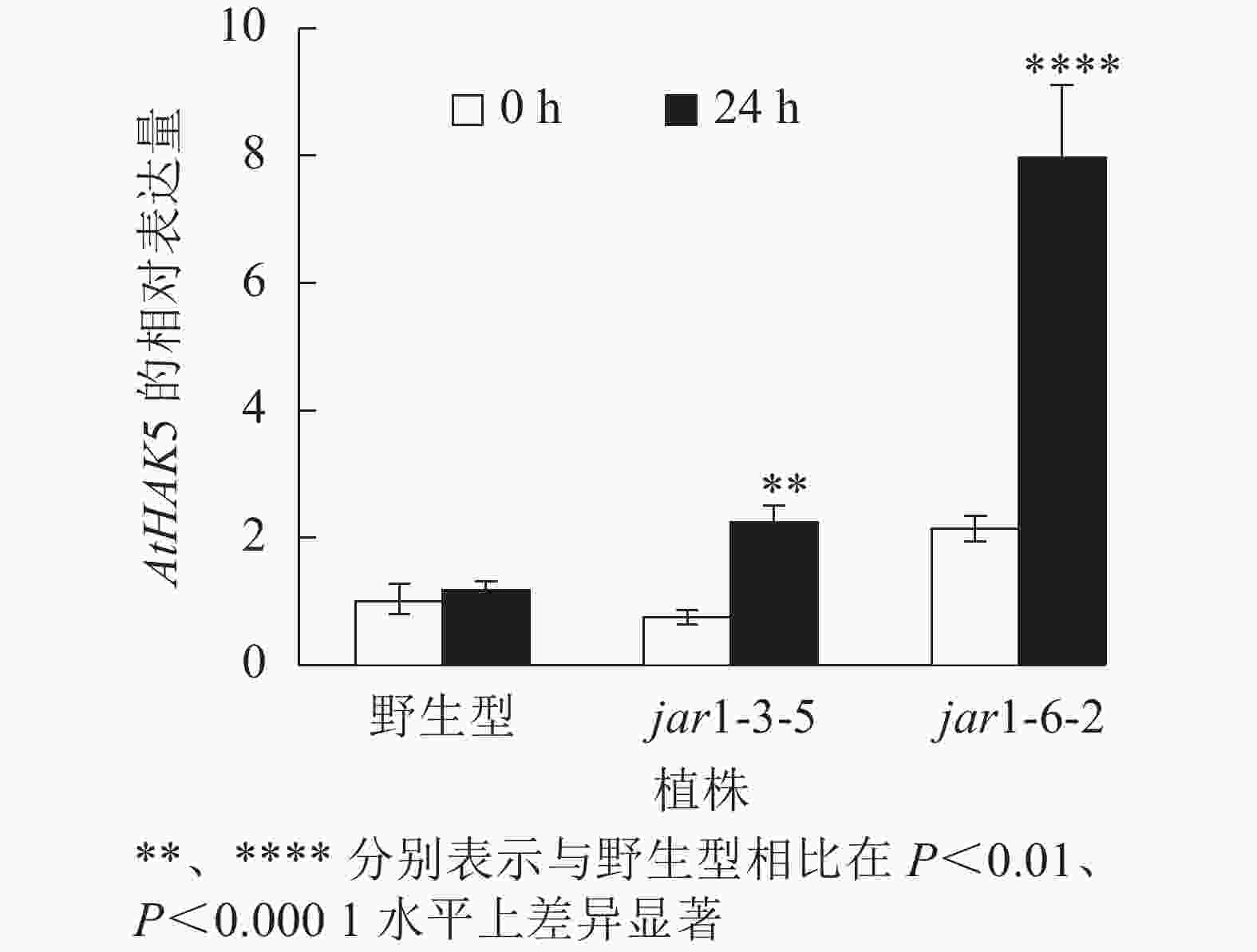
在植物获得K+与维持K+/Na+稳定的过程中钾转运蛋白发挥着重要作用,而KT/HAK/KUP转运体是植物中最重要的K+转运体家族之一,其中的HAK转运体可能在维持盐胁迫下的钾钠稳态过程中发挥重要的作用[39-42]。目前,拟南芥中,AtHAK5是研究最为广泛和深入的高亲和性K+吸收系统成员[43],因此,本研究通过对盐胁迫下jar1突变体中AtHAK5的表达量进行分析,探究JA信号通路在植物耐盐性中的调节机制。图8显示:在正常条件下野生型和突变体根中的AtHAK5表达量无显著差异;而50 mmol·L−1NaCl处理24 h后,野生型AtHAK5表达量无显著变化,而突变体中AtHAK5表达量上升3倍左右,极显著(P<0.01)高于野生型。表明盐胁迫下,AtJAR1突变影响了植物体内K+的吸收和转运。
-
JA是植物生长发育和防御过程中的关键信号分子,JA合成后经AtJAR1催化形成JA-Ile,JA-Ile介导F-box蛋白与COI1-JAZ蛋白相互作用,释放MYC2和其他转录因子(TFs)从而诱导早期JA应答基因的表达。本研究利用CRISPR/Cas9基因编辑系统创制的2个jar1突变体,其JA信号标记基因AtVSP1和AtVSP2的表达量极显著下降,说明AtJAR1基因功能丧失,从而影响了JA信号传导通路。同时,创制的这2个jar1突变体植株还表现出前21 d地上部生物量极显著大于野生型,之后这种差异逐渐减小并出现叶片萎蔫的表型,这在早期的研究中并未提及[34]。此外,研究发现:通过基因编辑手段使AtJAR1基因突变可以缓解ABA对种子萌发的抑制作用,而这与早期的研究结果正好相反[34]。出现这些表型的差异极有可能是由于早期研究中的突变体jar1-1是通过EMS诱变的方法获得,其目的蛋白仅第101位的丝氨酸(Ser)残基突变为苯丙氨酸(Phe)[44]。在这种突变体中,AtJAR1基因功能可能未完全丧失,而本研究通过基因编辑手段构建的突变体发生了大片段基因序列的缺失,从而引起氨基酸序列发生移码突变以及翻译提前终止等情况,是基因功能完全丧失型突变体。因此,获得的这2个jar1突变体将有助于彻底研究AtJAR1基因的功能。同时,本研究也为利用CRISPR/Cas9基因编辑技术创制基因功能完全丧失型突变体,从而为进一步研究基因功能提供借鉴和参考。
植物对盐胁迫的耐受性与植物激素有着密切关联。JA作为一种植物内源激素,一方面能够参与根系的生长与再生、下胚轴的生长、种子萌发、气孔的开闭等多种生长发育过程,另一方面可以激活植物的防御机制,以应对盐害、低温、干旱和病虫害等逆境胁迫造成的伤害[45]。其中,在茉莉酸调控植物应对逆境胁迫的过程中,首先被位于细胞表面的受体所识别,合成JA-Ile,而JA-Ile又能与E3泛素连接酶SCFCOI1和26S蛋白酶体相互作用,加快JAZ蛋白降解,从而缓解了与JAZ相互作用的转录因子的抑制作用。而这些转录因子又参与调控各种胁迫耐受性的生理输出响应[46-47]。所以,本研究运用基因编辑手段使AtJAR1基因功能丧失,JA-Ile的合成受阻,对盐胁迫耐受性的生理输出响应受到影响,表现为盐胁迫对种子萌发和根系生长的抑制作用有所缓解。此外,许多研究表明:在植物响应盐胁迫的过程中,JA和ABA存在交互作用,而与JAZ蛋白相互作用的转录因子MYC2是JA和ABA交互作用的关键点[38]。ABE等[48]和LORENZO等[49]研究指出:MYC2过表达和突变体植株分别表现出对ABA敏感性的增加和降低。ABE等[48]和IWASAKI等[50]则指出:MYC2可以促进盐胁迫和ABA胁迫响应基因RD22的表达。因此,本研究中jar1突变体表现出对ABA敏感性的下降可能是由于MYC2激活受阻所导致的。
在盐胁迫下,Na+会争夺植物钾通道上K+结合位点,促进K+的外排,降低K+的吸收利用,植物表现出对高盐胁迫的敏感现象[51]。本研究中,jar1突变体根系在盐胁迫下K+质量摩尔浓度显著增加,K+/Na+较野生型有所提高,这可能就是突变体对盐胁迫的敏感性下降的原因之一。在植物获得K+与维持K+/Na+稳定的过程中钾转运蛋白发挥着重要作用,T-DNA插入突变体研究发现:拟南芥中AtHAK5是HAK家族中主要的K+转运体之一[52]。在盐胁迫下jar1突变体中的AtHAK5表达量极显著增加,表明AtJAR1突变可以促进AtHAK5的表达,从而增加植株根系对K+的吸收,提高K+/Na+。此外,AtHAK5还被证实参与植物对盐胁迫及ABA的响应[51],说明突变体中AtHAK5表达的变化可能是通过JA信号转导途径与ABA的交互作用实现的。
综上所述,利用CRISPR/Cas9基因编辑技术创建AtJAR1基因功能完全丧失型突变体不仅有助于完善对AtJAR1基因功能的认识,同时为进一步研究植物激素与植物盐胁迫提供研究材料和研究基础。此外,JA信号通路可能通过MYC2与ABA交互作用,影响下游HAK5的表达,从而改变植物根系对钾的吸收,使K+/Na+发生变化,最终影响植物对盐胁迫的耐受性。
Functional analysis of AtJAR1 gene in salt tolerance of Arabidopsis thaliana
doi: 10.11833/j.issn.2095-0756.20210742
- Received Date: 2021-11-11
- Rev Recd Date: 2022-04-24
- Available Online: 2022-09-22
- Publish Date: 2022-10-20
-
Key words:
- Arabidopsis thaliana /
- CRISPR/Cas9 /
- jasmonal amino acid conjugate synthase /
- salt tolerance /
- K+/Na+ balance
Abstract:
| Citation: | LI Dandan, LIN Rong, LI Xinguo, et al. Functional analysis of AtJAR1 gene in salt tolerance of Arabidopsis thaliana[J]. Journal of Zhejiang A&F University, 2022, 39(5): 998-1009. DOI: 10.11833/j.issn.2095-0756.20210742 |










 DownLoad:
DownLoad: