-
次生细胞壁(以下简称“次生壁”)的形成是植物细胞在完成初生生长后的关键发育过程,这一过程主要发生在木质部纤维细胞、导管等支持性组织中,以赋予细胞更强的机械支撑和运输能力[1−2]。细胞通过纤维素合成、半纤维素合成和苯丙烷途径合成前体物质,如纤维素微纤丝、木聚糖、木质素单体等,这些物质通过细胞膜或囊泡运输至细胞壁,形成多层次的结构[3−4]。当细胞停止扩展时,木质化过程启动,木质素在细胞壁中沉积,形成坚硬的次生壁。次生壁通常分为S1、S2和S3共3层,其中S2层最厚,富含纤维素和木质素,是细胞壁的主要构成部分[5]。木质部的发育依赖于次生壁的形成,该过程对植物的正常生长、环境适应性及抗逆性起到至关重要的作用。近年来,随着分子生物学、生物信息学及基因编辑技术的快速发展,逐渐揭示了次生壁形成的分子调控网络,特别是转录因子、微RNA(microRNA,miRNA)和激素信号在这一过程中的作用。其中NAC和MYB转录因子在该过程中起到核心的调控作用,它们通过结合特定的启动子区域调控木质素、纤维素以及半纤维素相关基因的表达。同时,microRNA通过调节这些转录因子的表达,进一步精确控制细胞壁合成[6−8]。然而,次生壁的调控机制依然复杂多样,尤其是不同植物物种在次生壁形成过程中表现出的特异性和复杂性尚未完全被阐明。本研究旨在总结近年来植物次生壁形成的最新研究进展,重点探讨NAC、MYB和microRNA在这一过程中的作用及其相互调控机制。
-
NAC (NAM,ATAF1/2和CUC2)是植物特有的一类转录因子,参与多种重要的生物学过程,如生长发育、响应环境胁迫等[9−10]。在拟南芥Arabidopsis thaliana中,VND (vascular-related NAC domain)家族基因VND1~7编码一类重要的NAC转录因子,这些转录因子主要参与木质部导管细胞的分化和次生壁的形成,并在木质部细胞的程序性死亡中发挥重要作用[11−14]。其中,VND6和VND7是调控导管细胞分化及导管次生壁生物合成的关键因子,能直接结合靶基因启动子中19 bp的特定保守序列(T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T),即SNBE (secondary wall NAC-binding element)基序,调控与次生壁形成相关的基因表达。过表达VND6和VND7分别会使后生木质部和原生木质部的次生壁加厚[7, 15−16]。VND1~5在拟南芥木质部导管中特异性表达,与VND6和VND7功能冗余,可激活纤维素、木聚糖和木质素等次生壁合成基因的表达,共同调节导管中次生壁的生物合成和程序性细胞死亡(除VND2外)[13]。已有研究发现:VND转录因子之间存在相互调控关系。VND1~7蛋白可以结合VND7启动子上的SNBE基序,但VND1~7在体内过表达并未显著提升拟南芥内源性VND7的表达水平[17],这表明VND7的表达可能受到严格的内在调控。此外,VND1主要调控拟南芥次生壁合成的相关基因,并能低水平激活VND6和VND7的表达[18],表明VND1可能在木质部发育的早期阶段发挥重要作用,激活一部分关键基因的表达,进而调节次生壁的形成。VND1~7蛋白在调控木质部发育过程中扮演了复杂的角色,表达的调控受多种内在和外部因素的调控,这可能存在一个多层次的反馈调控网络。类似地,杨树Populus trichocarpa也存在VND型转录因子,其中PtrWNDs (wood-associated NAC domain),又称PtVNS (VND-,NST/SND- and SOMBRERO(SMB)-related proteins),是次生壁合成的关键调控因子,能够激活纤维素、木聚糖和木质素合成基因的表达,调控次生壁生物合成[19]。研究表明:杨树与拟南芥VND同源的8个基因(PtVNS01~08)在木质部发育和次生壁形成中起重要作用。如PtVNS7/PtrWND6A和PtVNS8/PtrWND6B与拟南芥VND7类似,发挥一级调控功能,在导管分化和次生壁增厚中作用显著[20−21]。杨树PdWND3A与拟南芥的VND4/5同源,过表达该基因的转基因杨树,导管细胞数量增加,木质素含量升高[22]。可见,VND家族转录因子在木质部发育、次生壁合成以及维管组织的调控中扮演着至关重要的角色,不仅直接调控次生壁合成基因,还通过相互作用和反馈调控维持木质部细胞的正常功能。
NST1 (NAC secondary wall thickening promoting factor 1)、NST2和NST3/SND1 (secondary wall-associated NAC domain protein 1)已被鉴定为拟南芥调控纤维细胞次生壁形成的关键转录因子,三者可以通过结合其靶基因SNBE基序来激活并调控次生壁的合成[16, 23]。NST1主要在花药内壁、茎束间纤维和木质部细胞中表达,T-DNA插入突变后会显著抑制拟南芥茎中木质部次生壁的增厚[24]。抑制SND1和NST1基因的表达会导致纤维细胞次生壁形成丧失,显示两者的功能重叠[25]。NST2与NST1共同调控花药内壁和茎束间纤维细胞次生壁增厚,当SND1、NST1和NST2基因同时突变时,纤维细胞次生壁增厚完全被抑制[23−24] ,表明NST2、SND1和NST1共同调节茎纤维细胞次生壁合成。在杨树中,PtVNS09和PtVNS10被鉴定为拟南芥NST1和NST2的直系同源基因,PtVNS11和PtVNS12与NST3/SND1同源,同样在木质部发育和次生壁合成中发挥重要作用。通过CRISPR/Cas9获得的四重突变体vns09 vns10 vns11 vns12中,木纤维、射线薄壁细胞和韧皮部纤维的次生壁出现缺陷,但部分导管周围的木纤维中仍有次生壁沉积,说明杨树中的NST/SND型基因在木质部纤维细胞次生壁形成中具有重要作用[20−21, 26] 。此外,在杨树中还发现了PtrWND1B在次生木质部纤维细胞中发生可变剪接,产生2种转录本:PtrWND1B-s和 PtrWND1B-l。PtrWND1B-s 编码完整的 NAC 蛋白,促进纤维细胞壁加厚;而 PtrWND1B-l 编码截断的 NAC 蛋白,抑制纤维细胞壁加厚[27]。可见,相对于拟南芥,木本植物在调控次生壁形成的过程更加复杂多变,这可能是在进化过程中树木调节纤维细胞壁形成的一种新的机制,以确保木材形成的稳态过程。NAC转录因子在棉花Gossypium hirsutum纤维细胞壁的增厚中同样发挥着重要作用。已有研究鉴定出一种转录因子GhFSN1(fiber secondary cell wall-related NAC1),它在棉花纤维次生壁增厚阶段特异性表达。通过酵母单杂和凝胶迁移率实验发现:GhFSN1 能够直接结合 GhKNL1、GhMYBL1、GhGUT1、GhDUF231L1和 GhIRX12 基因启动子中的SNBE基序,并激活其表达。同时过表达GhFSN1能够激活棉花GhCESA4、GhCESA7和GhCESA8等纤维素合成酶基因的表达,促进纤维素合成和沉积,导致纤维细胞壁增厚,但是GhFSN1是否通过直接靶向纤维素合成酶基因还未得到实验验证[28]。在棉花中还发现了与GhFSN1序列高度相似的GhFSN5转录因子, GhFSN5 是次生壁合成的负调控因子。拟南芥中过表达 GhFSN5 可导致次生壁合成相关基因的显著下调,致使茎和根中次生壁厚度明显降低[29]。由此可见,为了精确调控棉花纤维次生壁合成,棉花基因组中可能存在多个次生壁相关的NAC转录因子,它们在功能上有所分化,以适应棉花纤维次生壁合成的需求。NST/SND家族基因在植物木质部纤维细胞的次生壁形成中起核心作用,同样,这些基因在不同物种中高度保守,并通过协同作用调控纤维细胞次生壁的增厚。在不同植物物种中,VND、NST/SND同源基因发挥着类似的功能,都能够通过结合SNBE基序来调控其下游基因的表达,这种保守性反映了它们在植物生长和发育中的核心作用,表明这些基因是植物适应环境和维持生理功能的关键因素。因此,VND、NST/SND基因家族在不同植物中的相似功能体现了其在维管组织发育等基础生物学过程中的保守性和核心调控作用,也为其他非模式物种的遗传改良和功能研究提供了理论基础。
除次生壁合成的主导转录因子之外,一些NAC转录因子以及其他转录因子能够直接或间接地调控次生壁的形成。XND1 (xylem NAC domain 1)主要参与调控植物次生壁的形成并起到重要的负调控作用。XND1和其相关的转录因子在植物中通过多种途径负向调控次生壁的形成,影响木质部发育。研究表明:拟南芥XND1过表达则导致植株显著矮化,且伴随木质部导管细胞次生壁增厚的缺失[30−32]。随着深入研究,发现XND1可与VND6结合,将VND6引导至细胞质并阻碍其活性,从而抑制次生壁的沉积[33]。在杨树中,XND1的过表达会显著抑制杨树生长发育,表现出木质部次生壁形成受阻,细胞壁厚度变薄[34]。研究发现一些转录因子能与XND1产生相互作用。PagGRF12a (growth-regulating factor 12a)和PagGIF1b相互作用形成复合物,直接上调PagXND1a的表达,从而抑制木质部次生壁的形成[35]。Ⅰ类KNOX基因PagKNAT2 (knotted1-like homeobox gene 2)与 PagKNAT26b与拟南芥的KNAT2/6同源,能够下调杨树中的SND1、VND6转录因子表达,并直接激活PagXND1a,以抑制木质部细胞分化及次生壁沉积[36]。可见,PagKNAT2/6b是负调控多种NAC转录因子的木质部发育关键节点。在梨树Pyrus bretschneideri中,PbXND1同样抑制次生木质部的发育,其过表达会抑制木质部生长并显著降低植株高度。TCP4(TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL FACTORS 4)能够直接结合VND7启动子,激活次生壁合成和程序性细胞死亡,加速导管形成。PbXND1通过与PbTCP4蛋白互作,充当其转录抑制因子,影响PbTCP4的DNA结合能力,从而调控次生壁发育[37−38]。不同物种中的XND1同源基因在调节木质部发育和次生壁形成过程中扮演着关键角色,与其他转录因子的相互作用进一步完善了这一调控网络。VNI2 (VND-INTERACTING2)被鉴定为VND7的相互作用蛋白,其201~210 位氨基酸序列能显著抑制VND7的转录活性,并且该氨基酸序列的突变会导致 VNI2 对 VND7 的抑制能力下降。VND7也能结合VNI2启动子下调其表达,表明两者间的相互调节可维持维管组织的正常发育[39−41],但其具体的作用机制仍需深入研究。其他转录因子也能调控VND基因的表达,如LBD15 (lateral organ boundaries domain 15)通过直接结合VND7启动子正向调控VND7表达,促进次生壁的形成[42]; 而WRKY15能通过抑制VND7的上游基因表达来控制导管分化和次生壁形成[43]。作为木质部发育的负调控因子,VNI2、XND1以及KNAT2/6等转录因子之间也许存在一定的关系,深入研究这些转录因子之间的关系有助于了解和阐明木质部导管分化以及次生壁合成的复杂调控网络。表1总结了参与调控次生壁形成的NAC转录因子。
NAC转录因子 功能 参考文献 杨树 拟南芥 PtVNS7/PtrWND6A VND7 导管次生壁加厚 [20−21] PtVNS8/PtrWND6B VND7 导管次生壁加厚 [20−21] PtVNS09/PtrWND2A NST1 纤维细胞次生壁加厚 [26] PtVNS10/PtrWND2B NST2 纤维细胞次生壁加厚 [26] PtVNS11/PtrWND1B NST3/SND1 纤维细胞次生壁加厚 [26] PtVNS12/PtrWND1A NST3/SND1 纤维细胞次生壁加厚 [26] PdWND3A VND4/5 导管次生壁加厚 [22] PagXND1a XND1 抑制导管分化和
次生壁加厚[35−36] Table 1. NAC transcription factors involved in regulating secondary cell wall formation
-
MYB转录因子家族是植物中最大的转录因子家族之一,参与了细胞分化、细胞周期的调节以及对激素和环境因子的响应[44]。研究发现:MYB转录因子能通过促进或抑制木质素、纤维素和木聚糖的生物合成来调节次生壁的形成[45]。拟南芥MYB46和MYB83在植物木质部的发育中发挥着关键作用,在维管组织和木质部纤维中表达,抑制其表达会阻碍纤维细胞和导管次生壁的增厚。VND和NST/SND转录因子能通过直接靶向MYB46和MYB83调控次生壁的生物合成,构成主次级调控网络。因此,MYB46和MYB83作为次级开关,协调了次生壁合成的上下游基因表达[13, 46−47]。MYB46和MYB83可以靶向靶基因启动子上的SMRE (secondary wall MYB-responsive element)基序,即有7个核苷酸(ACC[A/T]A[A/C][T/C])的特异结合元件,进而激活一系列参与木质素生物合成的下游MYB转录因子,包括激活木质素合成的MYB20、MYB42、MYB58、MYB63、MYB85等以及抑制木质素合成的MYB4、MYB7和MYB32[45, 48−51]。此外,MYB46能够直接调控纤维素合酶基因CesA4 (cellulose synthase A 4)、CesA7、CesA8等和木聚糖合酶基因IRX8 (irregular xylem 8)、IRX9、IRX14等,以及一些木质素生物合成的相关基因表达来调控木质素、纤维素和木聚糖的合成[52−53]。除了MYB46和MYB83调控的网络之外,一些MYB转录因子也参与次生壁的合成。MYB103为SND1的直接靶标,是调控次生壁形成的关键因子之一,在木质素和纤维素合成中发挥作用[46, 54],但其是否受VND型转录因子调控仍需要进一步研究。MYB61也在木质部发育中起到正调控作用,其过表达型植株显示出更多的木质部导管,导管细胞壁增厚,木质素含量提高[55]。
杨树中的多个MYB转录因子通过激活或抑制不同基因的表达,精细调控次生壁的合成和木质部的发育。PtrMYB2/3/20/21被鉴定为拟南芥MYB46/83的同源基因,PtrMYB2 和 PtrMYB21 能够互补拟南芥 myb46 myb83 双突变体的表型缺陷。作为控制次生壁生物合成的主要转录开关,同样可以通过结合启动子SMRE基序来激活多个下游基因,如纤维素生物合成相关的PtrCesA4、PtrCesA7、PtrCesA8等和木质素生物合成相关的Ptr4CL1 (4-coumarate ligase 1 )、PtrCCoAOMT1 (caffeoyl-CoA O-methyltransferase 1 )等[56]。PtrMYB28、PtoMYB92与PtrMYB203分别与拟南芥中的MYB58/63、 MYB42/85和MYB3同源,它们能激活木质素生物合成的关键基因表达从而调控次生壁的形成[57−59]。值得注意的是,PtrMYB2/3/20 等MYB 转录因子可以激活 PtoMYB92 的同源基因 AtMYB42/85 的表达,因此,这些杨树 MYB 转录因子也可能通过类似的方式调控 PtoMYB92 的表达。PtoMYB216对杨树次生壁的形成也有重要作用,在木质素含量丰富的木质部中表达量最高,过表达会导致木质素异位沉积和次生壁增厚[60],但它如何通过结合下游基因启动子区域还没被完全揭示。同样,杨树中存在相应的次生壁合成的负调控因子。PdMYB221通过直接结合杨树中 PdCesA8、PdGT47C 和 PdCOMT2 等基因启动子中的AC元件(AC element)来抑制合成纤维素、木聚糖和木质素等关键基因的表达[61]。由于PdMYB221与AtMYB4同源,而目前发现AtMYB4只参与木质素合成,说明了在植物进化过程中MYB转录因子的功能在不断拓展。MYB转录因子也可与其他转录因子互作参与次生壁的形成。PaMYB199能直接抑制次生壁加厚相关的基因,如 PaIRX10 和 PaIRX15L-1等基因启动子的SMRE基序,这些基因在半纤维素的生物合成中起着重要作用。研究发现:PaC3H17 是一种CCCH 型锌指蛋白,能抑制 PaMYB199 的表达,并与其相互作用形成复合物,从而抑制 PaMYB199 对次生壁合成基因的作用[62]。一些MYB转录因子还受到次生壁合成转录网络一级开关的直接调控。如PtrMYB074直接受到SND1型转录因子的调控,能够反式激活与次生壁合成相关的靶基因。通过酵母双杂交筛选,发现PtrMYB074与PtrWRKY19相互作用形成二聚体,共同靶向PtrbHLH186基因进行激活,从而调控木材的形成过程[63−64],这是MYB间接调控次生壁合成的一种方式。此外,PagMYB128被发现是杨树次生壁生物合成的关键调控因子。PagMYB128通过激活纤维素、木质素和半纤维素合成相关的基因,如PagCesA4、PagPAL2/3 (phenylalanine ammonia-lyase 2/3)、PagFRA8 (fragile fiber 8)等的表达,并能与PagSND1形成正反馈回路,在木材形成中发挥重要作用[65]。最近的研究揭示了杨树中PagMYB31介导的调控网络。PagMYB31能通过结合靶基因的启动子直接抑制PagSND1、MYB3/20、PagVND6等转录因子的表达,来抑制木质部细胞的扩展和次生壁的增厚,从而调控木质部发育。与此同时PagMYB31 能够直接抑制一些与细胞扩张相关的基因,例如 PagEXPA4a/b、PagEXPA17、PagEXPA15a等。细胞扩张是次生壁形成的前提,因此 PagMYB31 通过抑制细胞扩张相关基因的表达,间接影响次生壁的厚度[66]。这可以将PagMYB31放置于杨树次生壁形成调控网络中的一个核心节点。PdMYB118不仅参与调控杨树花青素的合成,还能通过结合PagKNAT2/6b启动子中的AC元件来抑制其表达,促进木质素的沉积和木质部发育,而PagKNAT2/6b在上文中提到是次生壁合成的一个关键节点,这说明PdMYB118可能参与了次生壁合成的反馈调节[67]。MYB转录因子通过精细调控纤维素、木质素、半纤维素等生物合成途径,协同调控次生壁的形成,并参与木质部发育。这些转录因子通过相互作用形成复杂的调控网络,既有正向调控也有负向调控,共同影响木材的形态、结构和功能。表2总结了参与调控次生壁形成的MYB转录因子。
MYB转录因子 功能 参考文献 杨树 拟南芥 PtrMYB2/3/20/21 MYB46/83 纤维素、木质素和半
纤维素的合成[56] PtrMYB28 MYB58/63 木质素合成 [57] PtoMYB92 MYB42/85 木质素合成 [58] PdMYB221 MYB4 抑制木质素合成 [61] PtoMYB216 MYB61 木质素合成 [60] PaMYB199 − 抑制次生壁沉积 [62] PtrMYB074 − 木质素合成 [63] PagMYB31 MYB69 抑制纤维素、木质素和
半纤维素的合成[66] PdMYB118 MYB75 木质素合成 [67] PtrMYB203 MYB3 抑制木质素合成 [59] PagMYB128 MYB103 纤维素、木质素和
半纤维素合成[65] 说明:“−”表示目前还未发现拟南芥的同源基因。 Table 2. MYB transcription factors involved in regulating secondary cell wall formation
MYB和NAC转录因子在植物次生壁形成的调控网络中扮演着核心角色,两者的相互作用不仅是植物木质部发育的关键机制,也是植物适应环境变化和生长发育的精密调控系统。NAC转录因子通过作为主调控因子,启动或抑制下游MYB转录因子的表达,从而触发次生壁的合成途径。这一过程通过精细的层级调控确保木质素、纤维素和木聚糖的协同合成,优化植物的结构强度与功能。同时,MYB与NAC转录因子之间也存在负向调控机制,这种负向调控机制可能帮助植物在木质部发育过程中精细调整细胞壁的增厚程度,避免过度木质化。两者之间通过正负反馈机制交织在一起,形成了一个高度整合且灵活应对内外部信号的调控网络。这个网络不仅在拟南芥等模式植物中得到了验证,更在杨树、梨树等木本植物的次生壁形成和木材发育过程中展现出广泛的适应性与进化意义。
-
microRNA是一类约22个核苷酸长的非编码小RNA分子,广泛存在于真核生物中,主要通过调节基因表达来发挥作用[68]。microRNA通过靶定到目标mRNA的互补序列,来抑制蛋白质的合成或促进mRNA的降解。这些小RNA分子参与植物生长发育的多种过程,包括发育、细胞分化、增殖、胁迫响应等[69−70]。在木质部发育过程中,microRNA也发挥了重要作用。miR165和miR166能调控拟南芥的5种 HD-Zip Ⅲ [ATHB8、ATHB15、REV (Revoluta)、ATHB14 和ATHB9]转录因子的转录本的稳定性来控制后生和原生木质部的特异性。miR165a的过表达导致上述的HD-ZIP Ⅲ 基因的下调,进而使木质部细胞数量减少,并导致茎中的纤维次生壁变薄[71−72]。此外,microRNA还可以通过靶向一些MYB转录因子来调节木质部的次生发育过程。拟南芥miR858a可以调控植物中类黄酮和木质素生物合成网络来协调代谢通量的平衡。miR858a通过抑制其下游 MYB11、MYB12 和 MYB111 转录因子表达来抑制类黄酮的生物合成途径,从而使木质素生物合成相关基因上调,致使植株木质素含量增加[73]。
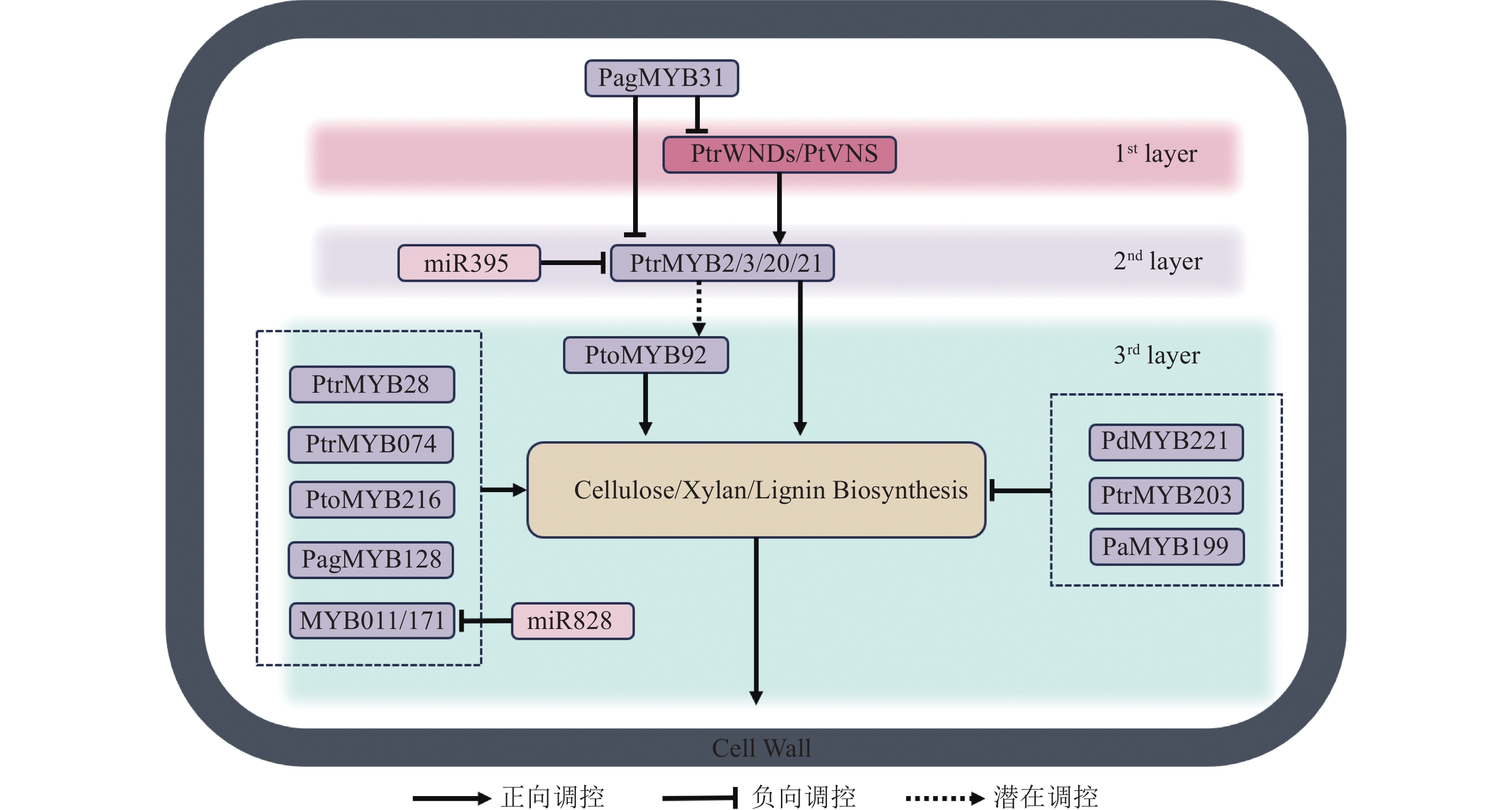
杨树miR828在木材发育过程中能够调节木质素生物合成和细胞壁的木质化,能通过直接抑制靶基因MYB011和MYB171,负向调控木质素生物合成相关基因的表达[74]。miR319a通过调控PtoTCP20间接影响杨树次生壁形成和木质部发育。具体而言,miR319a抑制PtoTCP20的表达,而PtoTCP20能激活PtoWND6表达,从而调控次生木质部分化和次生壁合成[75]。此外,PtoTCP20还调控miR396d-PtoGRF15模块,激活miR396d以抑制PtoGRF15的表达,后者的过表达会增加木质部产量并减少韧皮部生长[76−77]。这些调控关系表明miR319a在维持杨树次生壁形成的调控中具有重要作用。miR395c能调节硫酸盐代谢,影响杨属植物的次生木质部发育。miR395c的过表达会影响杨树中硫酸盐代谢途径,并抑制MYB46的表达,导致木质部导管增大和纤维次生壁厚度减少。漆酶在木质素的聚合过程中至关重要,miR397能够靶向下游漆酶合成基因PtrLAC (laccase)并负向调节其表达。通过构建杨树miR397过表达体系并进行转录组分析,发现过表达的杨树中多个PtrLAC基因表达下调,木质素含量也相应降低。此外,在过表达miR397的体系中,鉴定出一种bHLH家族的PtrUNE12 (ubiquitin E2 enzyme 12)转录因子,其过表达能促进次生木质部的发育,使次生壁比野生型杨树更厚[78−79]。近期,杨树中Pag-miR257、Pag-miR408等microRNA功能被揭示,它们能通过调控木质素生物的合成来影响杨树次生壁的厚度[80−81],但其具体机制和下游信号通路仍需进一步研究。microRNA通过调节木质素的合成和次生壁的形成,精细控制木材发育过程,主要通过调节关键转录因子(如MYB、HD-Zip等)及代谢途径,影响木质素合成、细胞壁木质化和次生木质部发育,进而影响植物的生长和木材的质量。然而,目前对于microRNA 通过直接调控NAC转录因子的次生壁形成的相关机制知之甚少,相关研究揭示了拟南芥miR164能够通过靶向NAC1转录因子的mRNA,同时过表达NAC1的植株比野生型植株更大,茎更粗,根系更发达。由此可见,miR164与NAC1也许也参与了木质部发育的相关进程,但它们对于次生壁形成相关具体的作用机制仍有待深入研究。图1展示了参与杨树次生壁形成的调控网络。
-
木质部发育的分子调控机制研究已经揭示了NAC、MYB等转录因子在细胞分化、次生壁合成及程序性细胞死亡中的关键作用。未来的研究将继续深化这些基因在不同植物中的功能,特别是在非模式植物和特殊环境下的调控差异性。通过基因编辑技术,如CRISPR/Cas9,对杨树乃至非模式植物的NAC、MYB以及一些其他转录因子的功能进行更加精细的研究,揭示它们如何在不同物种中调控木质部的分化与发育。在杨树中关于系统的次生壁形成网络还没有被完全揭示,拟南芥MYB46/83的同源基因PtrMYB2/3/20/21是否对下游PtrMYB28和PtoMYB92等基因的直接调控作用还没有得到实验验证,此外,PtrWNDs所有成员是否能直接调控PtrMYB2/3/20/21也未可知[56−58]。与此同时,木质部纤维与导管分子之间的信号传递机制,以及 NST/SND 基因与 VND 基因家族之间的关系还未阐明。例如在四重突变体中,杨树导管附近的纤维仍有次生壁的加厚,可见杨树VND型转录因子也许对NST/SND型起到了一定的补偿作用[21]。未来可以结合多组学技术(如单细胞测序、空间转录组学)以及分子生物学手段,全面解析木质部发育的分子网络,揭示转录因子家族之间的复杂交互,为木本植物提供优良性状并增加木材产量。
microRNA在木质部发育中的精细调控作用也亟待深入研究。microRNA参与了木质部发育调控因子之间的复杂网络,能够通过抑制特定转录因子的表达,精准调控木质部细胞的分化及次生壁的形成过程[71, 74−75]。尽管该调控机制已经初步被了解,但microRNA在不同发育阶段及不同环境条件下次生壁形成的具体机制仍未完全被揭示。目前,对于microRNA调控VND型转录因子的相关研究还未报道,未来的研究可以集中于揭示microRNA与VND、MYB转录因子之间的精确调控关系,它们三者的协同调控机制将在未来木本植物研究中占据核心地位。近期,WANG等[82]基于不同发育时期的杨树转录组和microRNA数据,识别出参与次生木质部形成的一些microRNA-靶基因模块。SHEN等[83]利用sRNA测序和降解组测序技术,揭示了马尾松Pinus massoniana次生木质部中microRNA的调控网络,发现了一些可能参与木材形成的microRNA-mRNA模块。这些都为理解植物木质部发育microRNA调控过程提供了新的见解。未来,通过结合全转录组和降解组测序等多组学技术,可以更全面揭示microRNA在木质部发育中的动态调控作用,尤其是microRNA与VND、MYB转录因子之间的精确调控关系,这不仅有助于更深入理解植物发育机制,也为提升植物的抗逆性提供了新的研究思路。
随着生物信息学、分子生物学以及测序技术的不断进步,研究人员可以通过数据整合进一步解析木质部发育的复杂网络。运用多种测序技术、表观遗传修饰和代谢物变化的多维度分析[84−86],以及深度学习[87]将帮助揭示木质部发育过程中的关键调控网络和节点,为优化植物木材产量和木质素代谢途径提供新的策略。这些调控网络的解析将为揭示木质部发育的多层次调控机制提供新的视角。
Regulatory roles of transcription factors and microRNA in the development of secondary cell walls in xylem
doi: 10.11833/j.issn.2095-0756.20240639
- Received Date: 2024-12-02
- Accepted Date: 2025-04-10
- Rev Recd Date: 2024-12-25
- Publish Date: 2025-08-01
-
Key words:
- secondary cell wall /
- transcription factors /
- microRNA /
- molecular regulation /
- xylem development
Abstract: The secondary cell wall of plants is crucial for maintaining structural integrity, providing mechanical support, and facilitating the transport of water and minerals. With advancements in molecular biology and bioinformatics, the molecular regulatory mechanisms underlying secondary cell wall formation have been gradually elucidated. The research has indicated that transcription factors (such as NAC and MYB) and microRNA constitute a multi-layered regulatory network, which plays vital roles in secondary cell wall formation. NAC and MYB transcription factors are directly involved in the biosynthesis of secondary walls by activating genes related to lignin, cellulose, and xylan synthesis. At the same time, microRNA achieve fine-tuned regulation of secondary cell wall development by targeting these transcription factors and their downstream genes. This review focuses on summarizing the recent research progress in secondary cell wall formation, especially the interplay and regulatory mechanisms of transcription factors and microRNA in this process. Moreover, it also discusses the current research challenges and future directions in this field. The systematic organization of these findings will enhance the understanding of the molecular basis of plant xylem development, providing theoretical support for wood improvement and bioenergy development. [Ch, 1 fig. 2 tab. 87 ref.]
| Citation: | ZHANG Jiannan, XIA Guohua. Regulatory roles of transcription factors and microRNA in the development of secondary cell walls in xylem[J]. Journal of Zhejiang A&F University, 2025, 42(4): 853−863 doi: 10.11833/j.issn.2095-0756.20240639 |









 DownLoad:
DownLoad: