-
水分是植物生长发育过程中不可缺少的环境因子。目前,人口增长与气候变化加剧了水资源短缺的程度,干旱已成为制约植物生长发育的关键因素之一[1]。了解植物在干旱胁迫下的反应对于提升农业生产力至关重要,这与作物品质和产量密切相关[2]。在应对干旱胁迫的过程中,植物能够通过调整其形态结构以及调控细胞内的生理生化过程,来增强自身的抗旱能力。这些适应性变化受基因表达的精细调控,不仅涉及抗旱相关基因的激活或抑制,还涉及复杂的信号转导途径和调控网络,以确保植物在干旱胁迫下的生存和生长[3]。
随着高通量测序技术和系统生物学的迅速发展,组学技术(包括基因组学、转录组学、蛋白质组学、代谢组学、微生物组学、表型组学等)在生命科学领域已经成为必不可少的研究手段[4]。组学技术可以在分子水平上理解植物胁迫耐受机制,并结合表型数据分析其在整体生长和表型特征上的影响[5]。基因组学有助于识别胁迫响应相关基因,推动基因工程和靶向育种策略实施,例如对番茄Solanum lycopersicum耐旱基因型的重测序发现:与热休克蛋白和离子转运蛋白相关的候选基因在耐旱响应中发挥关键作用[6]。转录组学通过分析基因动态表达,鉴定胁迫下差异表达基因(DEGs),揭示转录调控网络,例如对干旱敏感型和耐旱型2种黄瓜Cucumis sativus品种进行转录组测序,检测到的DEGs为揭示黄瓜响应干旱的关键通路提供了数据支持[7−8]。蛋白质组学通过对蛋白质的大规模研究,提供干旱胁迫期间激活的生化途径信息。代谢组学研究小分子和代谢物,提供植物细胞在压力下发生的生化变化图谱,进一步阐明参与干旱反应的代谢途径[6, 9]。
多组学整合技术通过捕获多个层级的数据,揭示各组分之间的复杂联系和调控机制,获得生物体对胁迫响应的整体解析,并确定参与植物非生物胁迫耐受反应的关键分子,比单一组学方法更全面地揭示应激反应相关途径和调控网络[10−11]。在对干旱敏感型水稻Oryza sativa和耐旱型水稻的研究中,转录组揭示了与干旱响应相关的基因表达变化,蛋白质组揭示了蛋白翻译机制、氧化还原平衡和碳代谢相关蛋白的表达响应,代谢组则明确了芳香族氨基酸和可溶性糖等代谢产物在渗透调节和活性氧(ROS)清除中的作用,三者的整合全面解析了水稻抗旱的分子机制,为作物遗传改良提供了范式[12]。此外,跨物种功能基因组学平台也展现出独特优势,例如通过酵母异源表达文库技术,证实了白车轴草Trifolium repens TrSAMDC1基因在干旱、盐和氧化胁迫耐受性中的正向调控作用,揭示了该基因通过多胺代谢增强细胞抗氧化能力的分子机制[13],多组学数据的互证机制为关键基因功能解析提供了高效验证体系。本文系统总结了关于园艺植物应对干旱胁迫机制的研究,重点关注多组学技术在其中的应用,以期为提升园艺作物抗旱性和稳定产量提供理论依据。
-
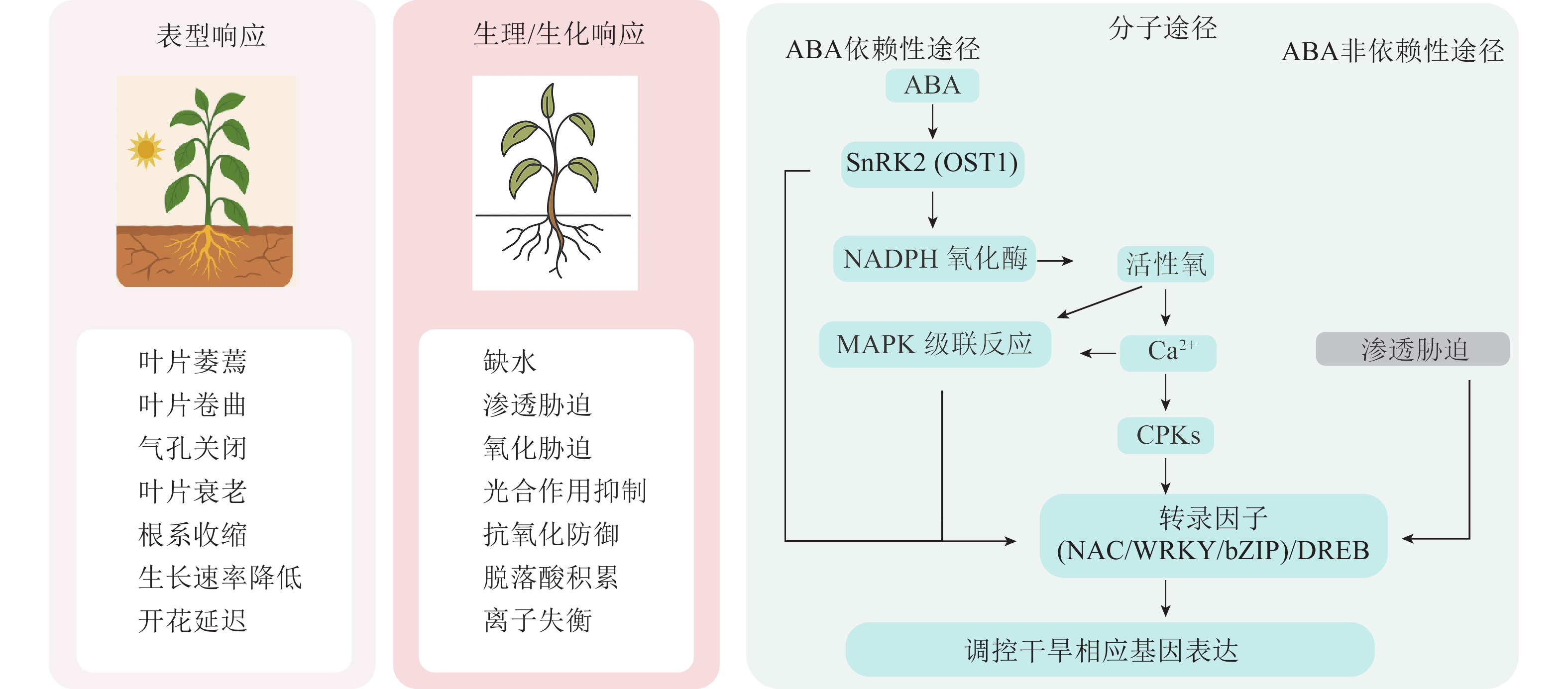
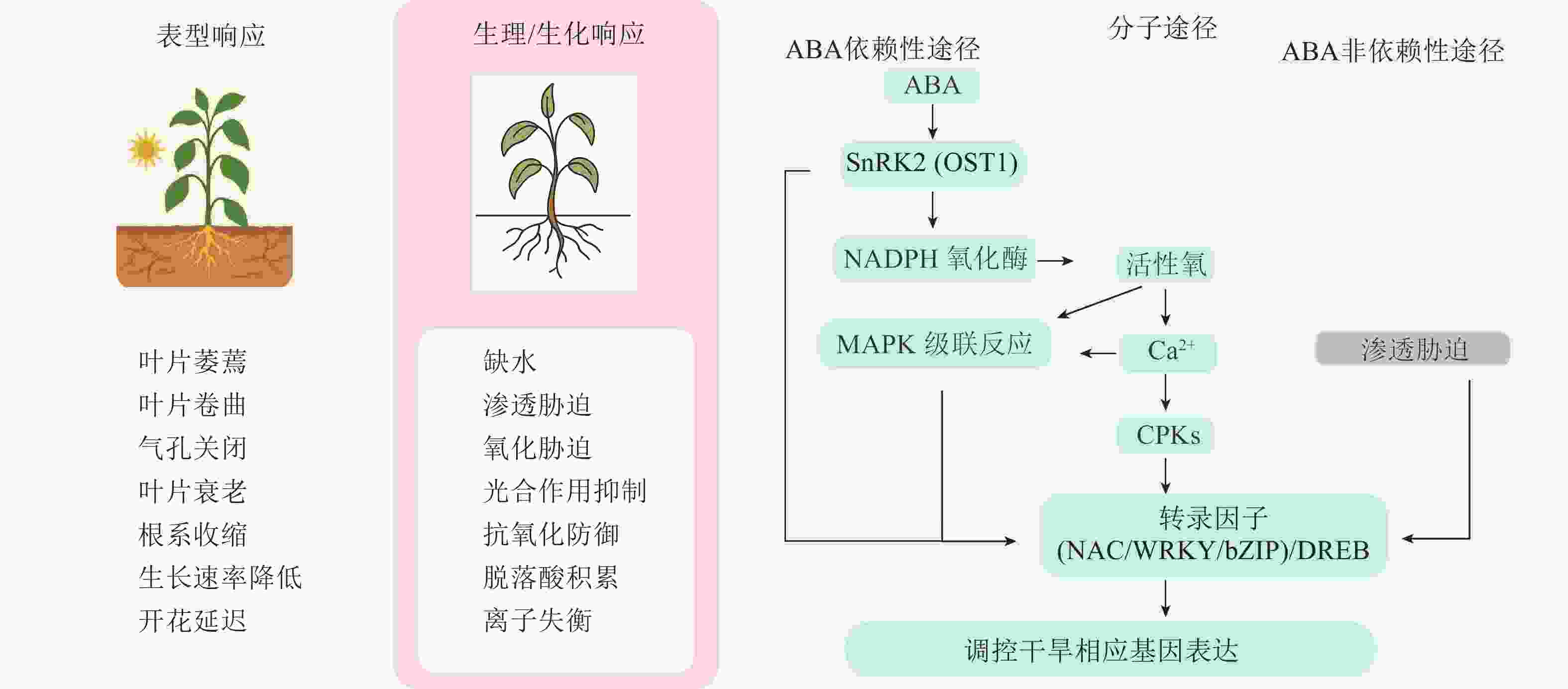
植物受到干旱胁迫后,会通过一系列形态结构上的适应性变化来提高水分利用效率和生存能力。根系是感知土壤水分变化的首要器官,在干旱胁迫下,植物根系会通过纵深生长、增加根冠比的方式吸收土壤深层水分。而地上部分则会通过关闭气孔和减少叶面积的方式来降低用水需求,以减少生长季末期因水分短缺而导致的生存风险[14]。例如,4种茶菊Chrysanthemum × morifolium幼苗的叶片数、叶面积、生物量和叶片相对含水量均随着干旱胁迫程度的增强而降低[15]。
在生理生化层面上,干旱胁迫会显著影响叶绿素合成、营养代谢、离子稳态和呼吸作用,这些都会导致光合作用速率降低[16]。另外,干旱胁迫还诱导ROS的产生和积累,在细胞、生化和分子水平上产生氧化应激,导致离子失衡、蛋白质变性和核酸的烷基化氧化[17−18],而植物体内的抗氧化防御系统可以一定程度上缓解这种伤害。以黄瓜为例,干旱胁迫显著抑制其光合作用和水分吸收,诱导ROS和丙二醛(MDA)积累,多种抗氧化酶活性发生变化,非酶类抗氧化物减少,表明干旱通过引发氧化胁迫而激活抗氧化系统,在抗旱过程中发挥关键作用[19]。
随着现代生物技术的发展,植物应对干旱胁迫的分子机制逐渐被揭示。干旱响应涉及信号感知、转导及多个基因和信号通路的协同调控[20]。许多与转录调控、激素调节相关的基因在干旱应答中发挥重要作用,其激活不仅调控下游基因表达,还可通过影响DNA甲基化、组蛋白修饰或非编码RNA等表观遗传机制,介导胁迫响应相关基因的持续或可遗传表达,进而增强植物抗旱能力[16]。研究表明:植物体内响应干旱的通路主要包括脱落酸(ABA)信号通路、钙离子(Ca2+)信号通路、丝裂原活化蛋白激酶(MAPK)信号通路[21]。ABA信号通路作为主调控因子,通过PYR/PYL/RCAR受体、PP2Cs和SnRK2s的级联反应,精确调控气孔运动、基因表达和根系结构,确保植物在缺水条件下的生存[22]。Ca2+作为通用第二信使,其独特的信号特征被蛋白激酶(MAPKs、CPKs/CIPKs、RLKs)解码,进而激活下游响应[23]。关键转录因子如脱水响应元件结合蛋白(DREB)、NAC、MYB作为基因表达的主调控因子发挥作用。DREBs主要通过ABA非依赖性途径调控渗透调节物质和抗氧化基因,NACs则在发育和胁迫适应中发挥多样化功能,MYBs则广泛影响气孔运动、蜡质沉积和次生代谢[16]。
上述多层次干旱响应机制构成了复杂的调控网络(图1),任何应激条件都会引起基因表达的变化,导致植物转录组、蛋白质组和代谢组组成发生变化[24]。通过整合基因组、转录组、蛋白质组和代谢组数据,为阐明复杂的植物抗旱分子机制提供了新思路。
-
基因组学的核心在于揭示遗传信息及其调控机制,对理解生命过程、疾病机制和生物多样性具有重要意义。2000年第1个完整的拟南芥Arabidopsis thaliana基因组序列被组装,为更全面地分析植物生命活动机制提供了基础[25]。随着高通量DNA测序技术的发展,目前已经有3 500多个完整的植物基因组被测序并公开,为作物功能基因挖掘与遗传改良奠定了基础[26]。
-
自拟南芥和水稻等早期参考基因组建立后,苹果Malus domestica、葡萄Vitis vinifera、番茄、黄瓜、西瓜Citrullus lanatus、梅花Prunus mume、蝴蝶兰Phalaenopsis等园艺作物的参考基因组相继公布[27],为园艺植物抗旱性状的精细定位和候选基因发掘提供了基础框架,尤其在数量性状基因座(QTL)定位和全基因组关联分析(GWAS)等研究中发挥着关键作用。二代测序(NGS)技术的快速发展加速了植物中数量性状位点和相关基因的定位,促进基因组和功能基因组研究[28]。由于其高通量、低成本等优点,NGS技术已经替代了表达序列标签和微阵列技术,成为主流表达研究工具。此外,NGS技术生成的大量分子标记显著提升了QTL定位和GWAS分辨率。简单重复序列(SSR)标记因多态性高、协同表现性强,也常用于抗旱QTL研究,这类标记在抗旱基因定位中发挥了重要作用[29],表1列出了部分与干旱胁迫相关的重要基因。
基因 植物种类 基因类型 基因功能 组学技术 文献 CgbZIP1 大花菊Chrysanthemum grandiflora bZIP转录
因子CgbZIP1通过上调抗逆相关基因的表达来增强植物的逆境适应性 转录组学 [40] CmbZIP9 蒙古菊Chrysanthemum mongolicum CmbZIP9过表达能上调抗逆相关基因表达水平增强植物耐旱性 转录组学 [41] RcNAC091 月季Rosa chinensis NAC转录
因子RcNAC091通过转录激活RcWRKY71正向调控ABA
信号通路响应干旱基因组学、转录组学 [42] NAC29 番茄Solanum lycopersicum 关键转录因子,调控水分状态响应 转录组学、代谢组学 [43] MdNAC29 苹果Malus domestica MdNAC29通过抑制抗逆相关基因的表达降低植物
对干旱的抵抗能力转录组学 [44] VaNAC17 葡萄Vitis vinifera VaNAC17通过调节内源茉莉酸(JA)生物合成和清除ROS增强抗旱性 转录组学 [45] EjWRKY17 枇杷Eriobotrya japonica WRKY转
录因子EjWRKY17显著上调多个ABA依赖型和非依赖型胁迫响应基因增强植物对干旱胁迫的响应 转录组学 [46] AfWRKY20 紫穗槐Amorpha fruticosa AfWRKY20响应干旱胁迫诱导的ABA信号传导的调节机制 转录组学 [47] PaWRKY 杏Prunus armeniaca PaWRKY11/14/48在植物耐旱响应中发挥关键作用 基因组学 [48] RcMYB8 月季Rosa chinensis MYB基因
亚家族RcMYB8通过上调相关基因促进ROS清除与脯氨酸积累,增强植物对盐和干旱胁迫的耐受性 转录组学 [49] MsMYBH 紫花苜蓿Medicago sativa 通过直接激活干旱响应相关基因的表达,帮助植
物在干旱后期通过生长抑制实现抗性适配基因组学、转录组学 [50] PeDREB2a 胡杨Populus euphratica DREB转
录因子PeDREB2a在植物中通过调控胁迫响应信号通路,增强植物对干旱和盐胁迫的适应能力 转录组学 [51] CBF4 葡萄Vitis vinifera 在植物叶片和根组织中均观察到CBF4在干旱和
盐胁迫下的上调表达基因组学 [52] NCED1、NCED4 茶树Camellia sinensis NCED基
因家族ABA生物合成的关键基因,促进气孔关闭 转录组学 [53] CsCYT75B1 甜橙Citrus sinensis 细胞色素
基因CsCYT75B1可以诱导转基因拟南芥产生耐旱性 转录组学 [54] Table 1. Drought resistant genes in horticultural plants
GWAS是重要的基因组学技术之一,它基于自然群体遗传变异,能够高效定位与复杂性状相关的QTL和候选基因,广泛应用于植物抗逆性状解析与育种改良研究[30−31]。董舒超等[32]通过测定301份脱水24 h后的番茄离体叶片相对失水率,利用GWAS挖掘出3个与番茄耐旱性显著关联的单核苷酸多态性位点(SNP),并鉴定出7个候选基因,这些位点可解释4%~19%的耐旱表型变异,为番茄抗逆性遗传改良奠定基础。
-
植物在遭受盐、旱、热、病原等逆境胁迫时,常通过DNA甲基化、组蛋白修饰和非编码RNA等表观遗传机制,在基因组水平上调控相关基因的表达[33]。这一调控方式具有可逆性、可塑性和记忆效应,使植物能够快速感应外界变化并增强适应能力,是抗旱机制研究的重要方向之一。
DNA甲基化是植物表观调控的核心机制之一,主要发生在CG、CHG和CHH位点,通常与基因转录沉默密切相关。例如,在苹果中发现的DNA甲基化转座元件是通过表观遗传调控参与植物逆境响应的重要机制[34]。组蛋白修饰通过调节染色质结构状态,影响基因的可及性和表达活性。常见的修饰形式包括乙酰化(acetylation)、甲基化(methylation)、磷酸化(phosphorylation)等。WANG等[35]发现拟南芥中的RPN1a-JMJ27模块通过动态调控H3K9me2组蛋白修饰,精准平衡染色质修饰可塑性,从而增强植物对干旱胁迫的适应性。非编码RNA包括微小RNA(miRNA)和长链非编码RNA(lncRNA)等,它们通过转录及转录后水平调控基因表达,参与胁迫响应的调控过程[36]。GUO等[37]发现miRNA在葡萄不同基因型中的特异性表达调控,是造成葡萄耐旱性差异的重要原因之一。此外,表观调控机制间存在协同作用,如DNA甲基化与miRNA表达的互作、组蛋白修饰对lncRNA基因表达的调节,未来需要进一步整合多组学技术系统探究其调控机制。
然而,关于转基因和突变植物抗旱性研究的分析显示:基因组数据和已识别基因数量的指数级增长并未按比例成功转化为抗旱栽培品种的数量[38],这意味着抗旱基因的发现并不等同于能提升植物整体抗性。一方面,干旱抗性的复杂遗传规律(多基因、上位效应和基因环境交互作用)从根本上限制了基因组学方法的预测准确性和全面应用[39],另一方面,干旱响应是一个高度动态的过程,涉及基因表达和调控的时间变化。基因组数据本身并不能捕捉这些动态的调控转变,也不能捕捉最终决定蛋白质功能和植物表型的转录后和翻译后修饰。因此基因组学预测在动态、真实世界的干旱情景中往往未能得到充分验证,迟滞了研究结果向作物改良应用转化。
-
转录组学主要分析特定条件下样品中所有类型的RNA转录本,包括信使RNA(mRNA)、miRNA和lncRNA[55]。植物对环境胁迫的生理和形态适应通常伴随广泛的转录变化,转录组学的广泛应用有助于解析不同植物基因型对干旱胁迫的响应机制,并筛选具有开发潜力的目标基因。转录组研究技术包括早期的基因表达序列分析技术(SAGE)、表达序列标签技术(EST)以及目前依赖NGS的高通量技术,如RNA测序(RNA-seq)和数字基因表达谱(DGE)等[11, 56]。其中,RNA-seq因具备高灵敏度、无参考基因组限制、检测范围广等优势,逐渐成为研究植物在干旱胁迫下基因表达变化的主流技术[57]。
RNA-seq能够检测DEGs,且通过京都基因与基因组百科全书(KEGG)等通路分析工具,可直观展示这些基因在代谢通路和信号网络中的动态变化,从而破译植物非生物胁迫耐受性的调控机制。SELVI等[58]利用Illumina平台的二代测序技术完成耐旱型和干旱敏感型甘蔗Saccharum spp.叶片转录组测序,通过差异表达分析鉴定出一系列耐旱候选基因,揭示了调控干旱响应的分子机制。在观赏植物方面,基于Illumina平台的菊花转录组测序共组装约9.8万个转录本,鉴定出8 558个脱水响应转录本,其中307个转录因子显著差异表达,为培育耐旱菊花品种提供了候选基因和分子标记[59]。GUO等[60]通过RNA-seq结合实时荧光定量PCR(RT-qPCR)验证,鉴定出81 725个参与牡丹Paeonia suffruticosa干旱响应的非冗余基因。
转录组研究已经揭示了大量的转录因子(TFs)正向调控植物耐旱性,如bZIP、NAC、AP2/ERF、MYB、WRKY等。JIA等[61]通过转录组测序和加权基因共表达网络分析(WGCNA),研究了月季在干旱和复水过程中的分子响应机制,筛选出42个在navajowhite1和blue模块中具有高连接度的转录因子,涵盖NACs、WRKYs、MYBs、AP2/ERFs、ARFs等家族,为分子育种提升月季抗旱性提供理论依据。DREBs主要以ABA非依赖性途径调控多个胁迫诱导基因的表达,将拟南芥DREB1A/CBF3基因导入丹参Salvia miltiorrhiza后,异位表达的AtDREB1A显著增强了丹参植株的耐旱性,表明诱导转录因子表达能有效调控胁迫响应网络[62]。
然而,研究发现:基因表达水平与对应蛋白质的积累量之间往往呈现较弱的相关性,说明在植物应对胁迫的过程中,许多关键的调控作用并非发生在转录阶段,而是通过翻译后修饰等方式实现的[38]。因此,仅依靠转录组数据无法全面、真实地反映蛋白质的实际丰度与功能活性,这也凸显了转录组分析在解析胁迫响应调控机制时的局限性。与此同时,高通量测序技术带来的海量数据对生物信息学分析能力提出了更高要求;且数据中存在的技术变异性(如测序批次差异等)作为干扰因素可能掩盖真实的生物学信号,进而影响研究结果的可靠性[63]。
-
蛋白质组学主要识别和定量蛋白质的种类和数量,并分析其功能、结构以及相互作用。在植物抗旱机制的研究中,蛋白质组学通过鉴定和定量分析干旱胁迫下的蛋白质变化,可以揭示植物在干旱条件下的生理响应和分子调控机制。当前的蛋白质组学技术主要包括质谱分析(MS)、二维凝胶电泳(2-DE)、液相色谱(LC)和同位素标记相对和绝对定量技术(iTRAQ)等技术[64]。质谱分析可以确定参与应激反应的蛋白质,突出增强耐受性的生理适应,如气孔调节和渗透调节[65]。除此之外,还有蛋白质印迹技术和蛋白质芯片技术,分别用于特定蛋白质的检验,以及高通量的蛋白质表达分析,共同推动蛋白质组学在生命科学研究中的应用。
在干旱胁迫下,植物会诱导多种蛋白质的表达,使其表现出上调或下调的趋势。这些蛋白质通过参与光合作用调节、胚胎发育晚期丰富蛋白(LEA蛋白)积累、ABA代谢调控以及氧化应激响应等生理代谢过程来增强抗旱性[66]。YE等[67]利用iTRAQ技术分析干旱处理20 d的柳枝稷Panicum virgatum叶片蛋白组,发现9-顺式环氧类胡萝卜素加氧酶(NCED)显著上调,该酶参与ABA合成,是干旱信号响应的关键酶类,表明柳枝稷可能通过ABA生物合成途径响应干旱胁迫。LI等[68]通过2-DE和基质辅助激光解吸电离串联飞行时间质谱(MALDI-TOF/TOF)技术结合数据库比对,对墨兰Cymbidium sinense和西藏虎头兰C. tracyanum在干旱胁迫和恢复状态下的差异蛋白进行分析发现:2种兰花应对干旱胁迫时存在不同的适应策略。
在利用蛋白质组学对植物干旱胁迫的研究中,植物组织固有的生化复杂性,特别是高丰度的干扰代谢物,始终制约着蛋白质的高效提取。这种根本性的方法学瓶颈直接损害了蛋白质样品的质量、纯度和代表性,从而限制了干旱胁迫研究中所有后续蛋白质组学分析的准确性和可靠性。其次,翻译后修饰(PTMs)作为分子开关,在酶活性调节、信号转导通路调控及分子相互作用介导中发挥关键作用,但植物中多样且复杂的PTMs仍未被充分研究,并且通常不清楚它们如何影响蛋白质及其功能[69],使得干旱响应调控的关键层面尚未被充分揭示。上述技术障碍的累积效应导致植物蛋白质组学落后于其他组学,也意味着蛋白质组学在揭示园艺植物抗旱机制方面的全部潜力尚未实现。
-
代谢组学主要涉及特定物种、特定发育阶段或特定器官、组织和细胞中的低分子量代谢物的定量和定性分析[70]。了解代谢组学对于解析园艺植物如何通过代谢途径适应干旱等环境胁迫至关重要。核磁共振(NMR)波谱和质谱等代谢组学技术的广泛应用,为了解干旱胁迫下园艺植物的生化响应提供了可能[71]。质谱因能提供快速、敏感和选择性的定性和定量分析发挥核心作用,通常与气相色谱(GC)和LC分离技术相结合[72]。随着分析技术(如LC-MS、GC-MS、NMR等)的改进以及公共数据库的持续完善,代谢组学在植物抗逆研究中的应用范围日益拓展[73]。
代谢组学揭示了植物在干旱胁迫下进行代谢的生化途径,如渗透保护机制、抗氧化防御系统、能量代谢调整等过程[74−75]。HOCHBERG等[76]通过代谢谱分析和基于相关性的网络分析发现,中心代谢特别是氮代谢在葡萄逆境响应中起着重要作用。ARGAMASILLA等[77]比较柑橘2种抗性不同的砧木在涝渍和干旱胁迫下的代谢物变化,发现这2种在脯氨酸、苯丙素及植物激素水平上的响应存在共性与差异,揭示了应对不同水分胁迫的特异性调控机制与代谢基础。
在代谢组学研究植物干旱胁迫时,首先面临的根本障碍是植物代谢物的化学多样性与结构复杂性,且缺乏单一全面的分析技术,难以实现真正的整体代谢组学分析[78]。其次,代谢物丰度变化无法直接反映代谢途径通量或基因/蛋白质功能——作为复杂酶促反应的终点,其变化由多种因素引起,难以直接推断潜在调控机制,这使得代谢组学数据更多揭示相关性而非因果性。最后,代谢响应的动态性决定了单点或少数时间点的分析无法完整呈现植物从瞬时适应到新稳态建立的全过程,可能导致对胁迫响应的理解碎片化。
-
综合应用基因组、表观基因组、转录组、蛋白组和代谢组等,可从不同层面高效挖掘与抗旱相关的关键基因,系统解析干旱调节因子之间的静态与动态互作,并识别关键的遗传与代谢通路。多组学协同分析不仅突破了单一层次研究的局限性,还结合机器学习算法(如WGCNA、随机森林)和共表达网络构建,挖掘出“基因-蛋白-代谢物”互作枢纽,解析从胁迫感知到生理适应的级联调控逻辑,为园艺作物抗旱分子育种的精准设计提供了理论支撑与技术路径[79]。
随着测序技术的不断进步,大量园艺植物已完成了全基因组高质量测序和组装[80],为推动多组学联合分析在环境胁迫研究中的应用奠定了坚实基础[11]。转录组揭示基因表达的动态变化,帮助识别调控植物抗逆性及表型转变的关键因子;蛋白质组反映基因产物在翻译及翻译后修饰过程中的实际功能执行情况;代谢组则呈现基因表达变化对下游代谢通路及终产物形成的具体影响[81−82]。这些组学的联合分析不仅能构建从基因表达到代谢物变化的因果关系网络,而且能更深入地解析复杂的调控过程,为提高抗旱性的基因工程和育种计划提供靶标。
将转录组学和代谢组学数据相结合,可以揭示植物适应干旱的响应机制,突出渗透调节与能量代谢等维持细胞稳态的关键过程[83−85]。例如,在西瓜的干旱胁迫研究中,转录组与代谢组联合分析表明,胁迫后6 h为响应关键期,主要涉及糖代谢、激素信号转导及光合作用等通路的调控[83]。CUI等[86]对干旱胁迫后的东方百合进行转录组与代谢组联合分析,发现差异基因与代谢物主要共同富集于糖类代谢通路,表明干旱胁迫降低了东方百合的糖代谢水平,鉴定这些途径中的候选基因为提高百合抗旱性的基因工程和分子育种提供了靶标。LIU等[87]通过对转基因苹果植株的表型观察、理化指标测定、转录组和代谢组联合分析,发现MdPYL9基因的过表达可增强苹果抗旱性,其机制可能与调控类黄酮合成相关基因(CHS、CHI、4CL)的表达并促进apigenin-7-O-glucoside积累有关。ZHANG等[88]对贴梗海棠Chaenomeles speciosa进行研究,通过整合转录组、蛋白质组和代谢组数据,分析干旱胁迫下花青素生物合成途径相关基因、蛋白质及代谢物的变化,揭示了其干旱条件下花色的变化机制。多组学联合分析通过结合这些不同层次的信息,有助于全面理解植物在干旱胁迫下的复杂响应机制。
基因组编辑技术,特别是成簇规律间隔短回文重复序列及其相关蛋白(CRISPR/Cas)系统,为验证抗旱基因功能和培育抗旱品种提供了广阔机会[89]。通过QTL分析,在番茄中鉴定出多个与抗旱性相关的QTLs。进一步对干旱敏感型和耐旱型自交系开展转录组与代谢组联合分析,共鉴定出332个在干旱处理下2种材料均显著差异表达的核心基因,以及553种代谢物,这些核心基因和代谢物主要参与氨基酸、类黄酮及ABA等代谢途径,为构建番茄干旱与复水过程中的基因-代谢调控网络提供了关键支撑[43]。最近,整合转录组、全基因组亚硫酸氢盐测序和染色质免疫共沉淀测序(ChIP-seq)的多组学研究揭示了苹果在干旱胁迫下的表观基因组特征[90]。这项研究首次绘制了苹果干旱响应的全基因组表观遗传图谱,揭示了组蛋白密码与DNA甲基化的时空动态规律,为利用表观遗传育种提升作物抗逆性提供了理论依据和分子靶点。
-
多组学技术是揭示分子过程、识别生物标志物、基因工程靶点和调控网络方面的强大工具[11]。然而,识别这些候选要素仅仅是第一步。为确认候选基因的生物学功能并建立分子变化与抗旱表型的因果关系,开展完善的功能验证至关重要。而要证明某基因在抗旱中的作用,需通过改变其表达或活性,观察对应的表型变化。过表达研究可提供功能获得性证据,证实基因/蛋白质水平升高能增强植物抗旱性。基因编辑(如CRISPR-Cas9技术)和基因沉默(RNAi)则提供了功能缺失性证据,表明降低或消除基因活性会损害抗旱性。例如,在番茄中通过对SlbHLH96基因开展功能获得与功能缺失研究,已掌握了强有力的因果关系证据[91]。HAN等[92]通过CRISPR-Cas9技术敲除草莓Fragaria×ananassa中的FvICE1基因,发现突变体在冷和干旱胁迫下表现出更低的耐受性,而FvICE1过表达植株则表现出更强的耐寒和耐旱能力,表明该基因作为正调控因子在草莓抗逆性中发挥关键作用。在菊花中,通过RNA干扰沉默CmNF-YB8基因显著增强了植株的抗旱能力,明确其通过调控气孔开闭和角质层厚度影响耐旱性[93]。因此,对候选基因的全面功能验证,理想策略应同时包含过表达与基因编辑/沉默2种方法,以最终明确其在抗旱性中的作用。
此外,可以结合生理生化指标进行评估,并利用RT-qPCR或RNA-seq分析下游基因表达以明确调控路径。还可通过亚细胞定位、蛋白互作实验(如酵母双杂交、免疫共沉淀技术、双分子荧光互补技术)探究分子互作机制,结合代谢组学验证代谢通路的改变,或在酵母、拟南芥等模式物种中进行异源表达快速验证生物学功能。进一步的功能确认常通过启动子活性分析或突变体互补实验完成,从多个层面佐证候选基因在植物胁迫响应中的功能。
-
多组学技术的飞速发展,为深入理解园艺植物抗旱机制提供了全新的视角和工具。通过这些技术,研究人员不仅能够分析植物在干旱胁迫下的基因表达、蛋白质变化及代谢物的积累模式,还能够揭示不同生物层级之间的相互作用。特别是在干旱条件下,基因组数据为抗旱相关基因的定位提供了基础,转录组数据揭示了基因表达的时空动态变化,蛋白质组和代谢组则帮助理解这些变化如何在细胞和代谢途径层面上体现。
然而,目前多组学技术的应用仍然面临诸多瓶颈。首先,多源异构数据整合的挑战依然突出,不同组学平台产生的数据标准化、去批次效应以及有效的跨组学数据融合算法仍需进一步完善。其核心挑战不仅在于收集数据,还在于开发必要的复杂计算和概念框架,将来自高度复杂生物系统(庞大且注释不良的基因组、多细胞器、多样化代谢物)的多样化数据类型合成为连贯、可操作的数据体系[94]。其次,候选基因和信号通路的功能验证仍依赖传统的稳定转化体系,周期长、效率低,尤其对于种类繁多的园艺植物更为明显。此外,高质量参考基因组的缺乏,特别是在复杂基因组或多倍体园艺植物中,严重阻碍了基因的准确注释和后续深层机制的解析。尽管如此,随着数据分析方法(如WGCNA、随机森林等机器学习算法)和多组学数据集成平台的不断开发和应用,将极大地提升对干旱应答机制的理解。
随着基因组学、转录组学、蛋白质组学和代谢组学技术的持续发展,以及与空间转录组和单细胞转录组等前沿技术的结合,能够更加系统地挖掘植物抗旱的核心基因、关键蛋白以及枢纽代谢途径。未来研究应聚焦在以下4个方面:第一,应优先创新和完善专门用于整合异构植物多组学数据的新机器学习算法和人工智能驱动平台。这些工具对于提高预测建模能力和完善表型分析准确性至关重要,从而加速研究和育种周期[95]。第二,应该将重点放在通过基因工程、基因沉默和过表达研究对多组学筛选出的候选基因的功能验证方向。完善的功能验证是多组学研究成功的核心,它将基础科学发现转化为可操作的生物技术策略,从而开发出适应气候变化的园艺作物新品种。同时,积极探索将模式植物研究成果应用于实际园艺作物的策略,克服物种间功能保守性差异的挑战。第三,推动多组学数据收集、处理和分析协议的标准化。建立和推广开放数据存储库和共享平台,促进数据互操作性和数据更新,从而实现更广泛的比较分析和元分析,加速知识发现。最后,应持续对单细胞和空间组学、表观基因组学、脂质组学等新兴技术的研究和开发,以提供更深层次的细胞和分子机制见解,为精准育种和作物改良提供新的靶点。通过多组学的整合分析,能够更加全面地揭示植物在干旱胁迫下的分子调控网络,为抗旱育种提供更加有力的理论依据和技术支撑。这不仅将推动全球园艺作物的可持续发展,也为应对全球气候变化提供切实可行的解决方案。
Multi-omics approaches for drought resistance research in horticultural plants
doi: 10.11833/j.issn.2095-0756.20250385
- Received Date: 2025-07-23
- Accepted Date: 2025-08-26
- Rev Recd Date: 2025-08-25
- Available Online: 2025-10-29
- Publish Date: 2025-10-20
-
Key words:
- horticultural plants /
- drought stress /
- multi-omics /
- regulation mechanism /
- research progress
Abstract: Drought stress, one of the main abiotic stresses, is a primary limiting factor for plant growth, development, yield, and quality, posing a significant threat to the sustainable development of global agriculture. Drought resistance, a complex quantitative trait controlled by multiple genes, involves multi-level regulatory mechanisms, making it challenging for traditional research methods to systematically elucidate its molecular basis. In recent years, the rapid development of multi-omics approaches has provided support for systematically revealing the mechanisms of plant responses to drought stress. This review summarizes the advancements of technologies such as genomics, transcriptomics, proteomics, and metabolomics, as well as their applications in drought resistance research in horticultural plants. This review highlights the research progress of multi-omics joint analysis in key areas such as the mining of drought-resistance genes, the analysis of signaling pathway, and the development of drought resistance molecular markers. Furthermore, this review summarizes the challenges and limitations of multi-omics approaches in plant drought resistance applications. It prospects the application of new technologies like spatial omics, single-cell omics, and AI-driven multi-omics data mining in plant drought resistance research, aiming to provide theoretical support for drought resistance mechanism research and stress-resistant breeding. [Ch, 1 fig. 1 tab. 95 ref.]
| Citation: | ZHU Shifan, WU Jinyue, LIANG Xia, et al. Multi-omics approaches for drought resistance research in horticultural plants[J]. Journal of Zhejiang A&F University, 2025, 42(5): 875−887 doi: 10.11833/j.issn.2095-0756.20250385 |









 DownLoad:
DownLoad: