-
断奶是反刍动物饲养过程中必须经历的一个阶段,而早期断奶是为提高生产效率所采用的技术手段之一。断奶前瘤胃的充分发育为断奶成功实施和缓解断奶应激有着重要的意义[1]。近年来,瘤胃上皮细菌的变化受到研究学者的关注。有研究认为:不同于瘤胃内容物相关的细菌群落[2],瘤胃上皮细菌群落在幼龄反刍动物中具有某些特定的功能(宿主代谢、免疫应答和瘤胃发育)[3−4]。瘤胃内容物相关细菌受到诸多宿主因素影响,如日粮、年龄、抗生素摄入以及宿主的健康;此外,这些菌群也受到环境因素的影响,如地理位置、季节和饲养方式等[5]。然而上述因素对瘤胃上皮相关菌群影响还未见报道。因此,准确认识瘤胃上皮细菌在反刍动物生长过程中的演化,有利于进一步通过营养手段调控瘤胃上皮细菌的组成,促进瘤胃上皮细菌的尽早成熟,为调控幼龄反刍动物的发育提供新思路。鉴于此,本研究以湖羊Ovis aries羔羊为研究对象,探讨了2个日龄羔羊瘤胃上皮细菌群落结构以及多样性的差异,为幼龄反刍动物实施早期断奶技术提供理论依据。
-
选用10只出生体质量相近[(3.60±0.37) kg]的初生湖羊羔羊,于日龄5 d时饲喂代乳粉,每天3次,自由采食;并于日龄10 d开始饲喂开食料及羊草。代乳粉、开食料和羊草的营养水平见表1。羔羊分别于日龄30 d (D30)和45 d (D45)屠宰5只(n=5)。屠宰后,立即将瘤胃组织用无菌磷酸盐缓冲盐水(pH 7.4)冲洗3次,以清除不能附着的细菌,先置于液氮中,后迅速转移至−80 ℃保存。
表 1 饲料的化学成分组成
Table 1. Chemical composition of the feeds
饲料类别 饲料成分干物质质量分数/(g·kg−1) 干物质 粗蛋白 中性洗
涤纤维酸性洗
涤纤维钙 磷 粗灰分 代乳粉 945.0 184.0 36.0 9.0 7.2 6.3 41.0 开食料 914.0 174.0 169.0 73.0 12.1 5.9 73.0 羊草 929.0 84.9 659.0 384.0 2.4 1.0 60.0 说明:开食料为每100.0 g干物质中,由58.5 g玉米、10.0 g小麦粉、27.0 g豆粕、1.1 g碳酸氢钙、1.4 g石灰石、1.0 g氯化钠和1.0 g维生素矿物质组成的预混料。 -
羔羊瘤胃上皮样品送至上海锐翌生物科技有限公司进行测序及微生物多样性分析。提取羔羊瘤胃上皮细菌总DNA后,用341F/806R通用引物(上游为5′-ACTCCTACGGGRSGCAGCAG-3′;下游为5′-GGACTACVVGGGTATCTAATC-3′)扩增16S rDNA保守区域V3~V4区,并对产物进行纯化,定量和均一化形成测序文库,测序平台为Illumina HiSeq PE250。测序完成后,对原始数据进行全质控,通过97%相似度的标准聚类,得到操作分类单元(OTU),每个OTU被认为可代表1个物种。将具有代表性的OTU序列与数据库比对,得到每个OTU对应的物种分类信息以及从门到属水平各样品的群落组成情况[6]。原始数据提交到美国生物技术信息中心(NCBI),序列号为PRJNA857329。
-
数据采用Excel 2010进行整理,结果采用SAS 8.0软件Duncan’s方差分析对各处理组进行多重比较。
-
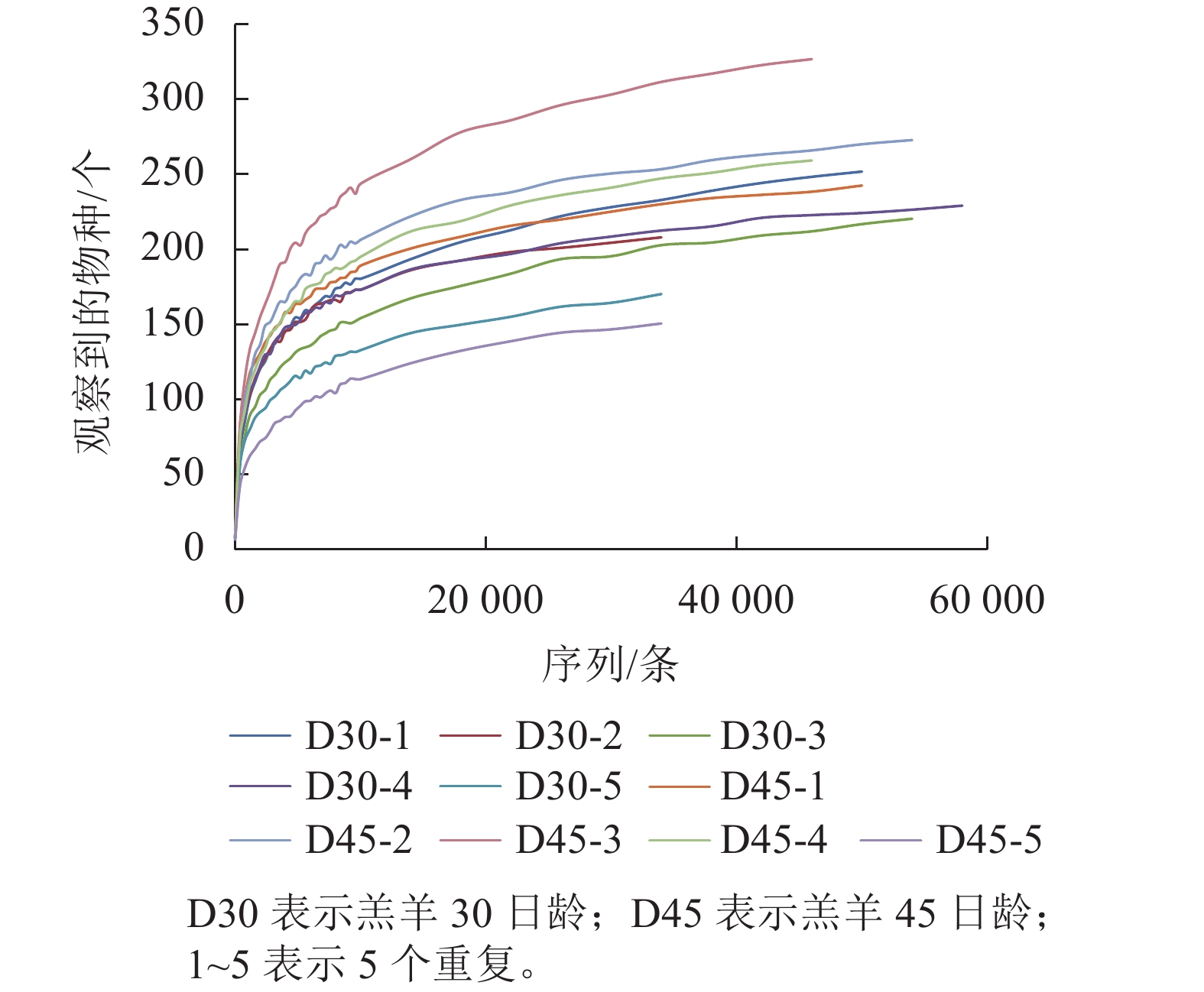
下机序列经拼接、优化、质控后共得到460 073条优质序列,平均每个样品为(57 509±7 989)条。共获得OTU为503个,平均每个样品OTU有(252±38)个。2组瘤胃上皮细菌的稀疏曲线见图1,虽然没有得到完全的平缓,但斜率在不断的减小,说明趋向于饱和。
从α多样性指数(表2)可知:D45组羔羊瘤胃上皮细菌的OTU个数显著多于D30组羔羊(P<0.05),Chao1指数D45组有增加的趋势(P=0.07),但是Shannon指数和Simpson指数2组间均差异不显著(P>0.05),Coverage指数2组均在99%以上。因此,说明测序深度能准确反映湖羊瘤胃上皮微生物组成。
表 2 不同日龄羔羊瘤胃上皮细菌多样性指数
Table 2. Ruminal epithelial bacterial diversity of Hu lambs at different ages
项目 OTU数 Chao1
指数Shannon
指数Simpson
指数Coverage
指数D30 171 b 243±28 a 4.98±0.16 a 0.93±0.02 a 99.90±0.04 a D45 211 a 290±33 a 5.32±0.39 a 0.93±0.04 a 99.80±0.02 a 均值标准误 11.100 15.400 0.148 0.014 0.000 P 0.040 0.070 0.152 0.932 0.338 说明:同列不同字母表示差异显著(P<0.05)。 从韦恩图(图2)可见:不同日龄湖羊羔羊瘤胃细菌共有的OTU数为271个,D45组羔羊特有的OTU为169个,而D30组羔羊特有的OTU数较少,为63个。

图 2 不同日龄湖羊羔羊瘤胃细菌OTU韦恩图
Figure 2. Venn diagram showing the unique and shared OTUs at different ages in bacterial communities of Hu lambs
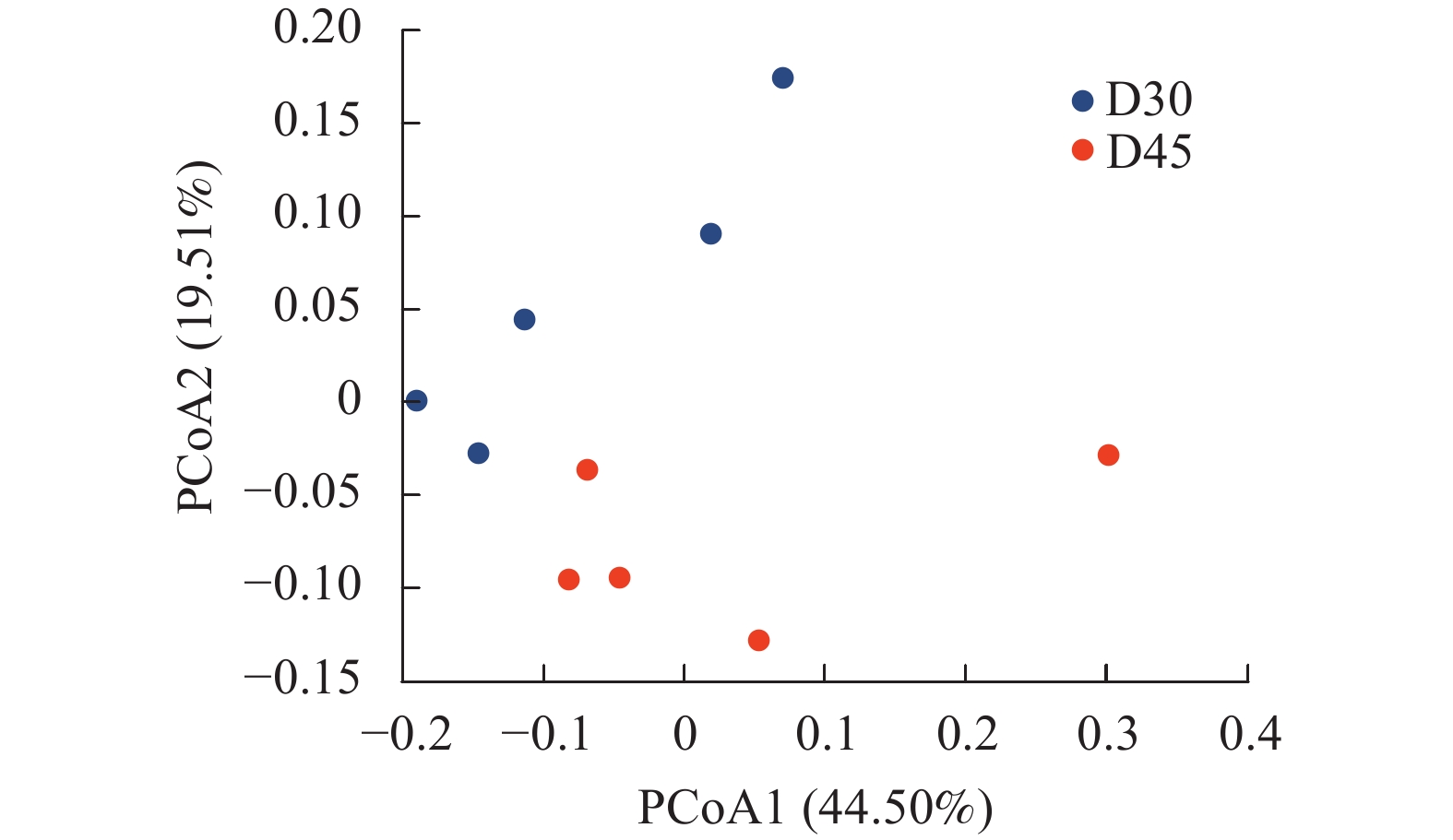
基于UniFrac的加权主坐标分析其第1主成分和第2主成分的贡献率分别为44.50%和19.51%(图3)。两组羔羊个体差异较为显著,分布较为涣散,然而D30组和D45组分离较远,说明随着日龄的增长羔羊瘤胃上皮细菌分布存在差异。
-
瘤胃上皮细菌门分类图(图4)显示:瘤胃上皮细菌主要归于厚壁菌门Firmicutes、拟杆菌门Bacteroidetes、变形菌门Proteobacteria和放线菌门Actinobacteria四大类。D45组(14.10%)放线菌门的相对丰度较D30组(4.36%)高。
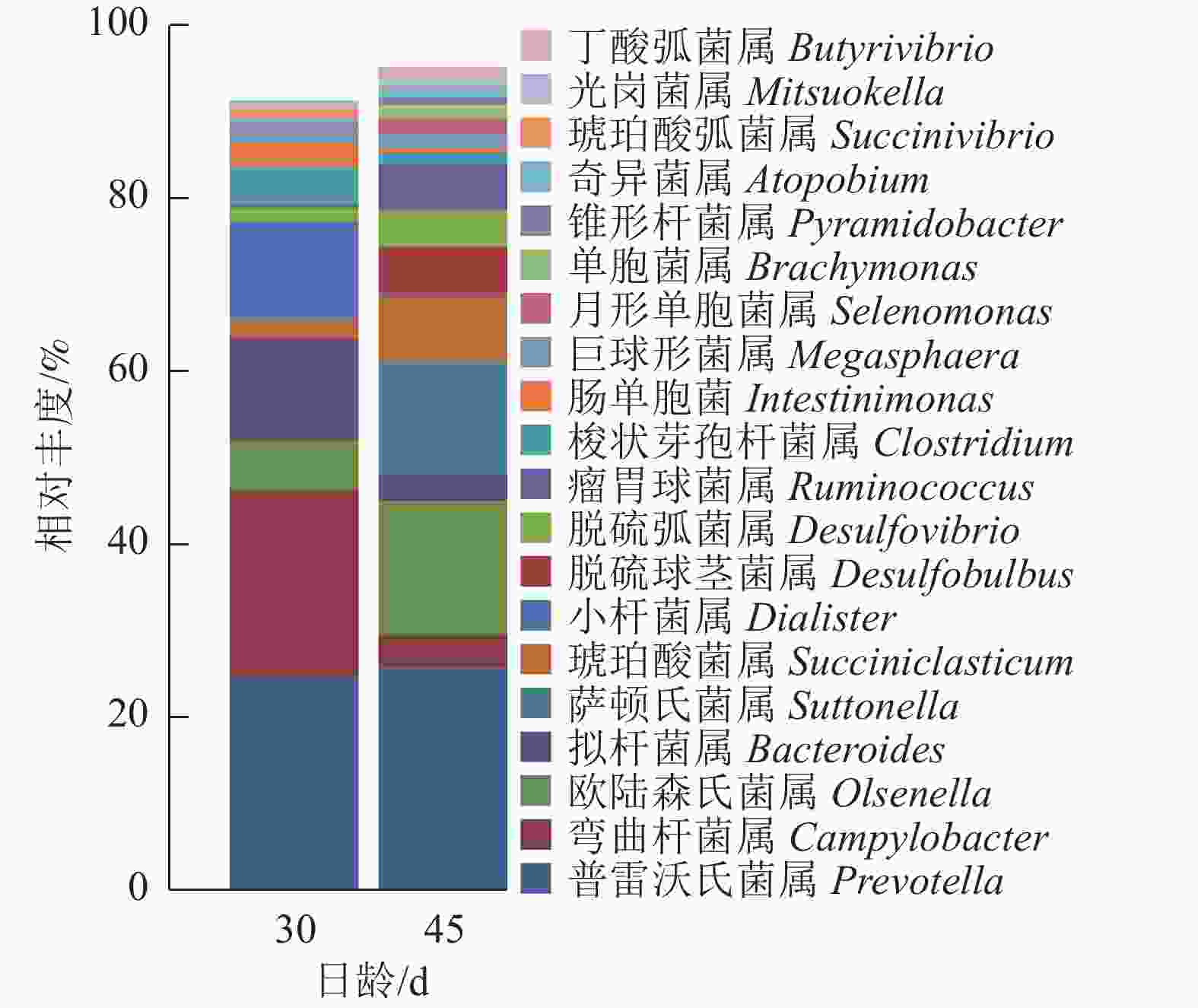
由图5可知:羔羊瘤胃上皮细菌中,普雷沃氏菌属Prevotella、弯曲杆菌属Campylobacter、欧陆森氏菌属Olsenella和拟杆菌属Bacteroides为优势菌属,其中普雷沃氏菌属Prevotella所占比例为25%。
Kruskal-Wallis非参数检验获得的组间总体差异结果显示(图6):与D30组羔羊相比,D45组羔羊瘤胃上皮弯曲杆菌属和肠单胞菌Intestinimonas的相对丰度显著降低(P<0.05),拟杆菌属和小杆菌属Dialister的相对丰度有降低的趋势(P=0.083);而萨顿氏菌属Suttonella、脱硫弧菌属Desulfobulbus和单胞菌属Brachymonas的相对丰度显著增加(P<0.05),琥珀酸菌属Succiniclasticum有增加的趋势(P=0.083)。
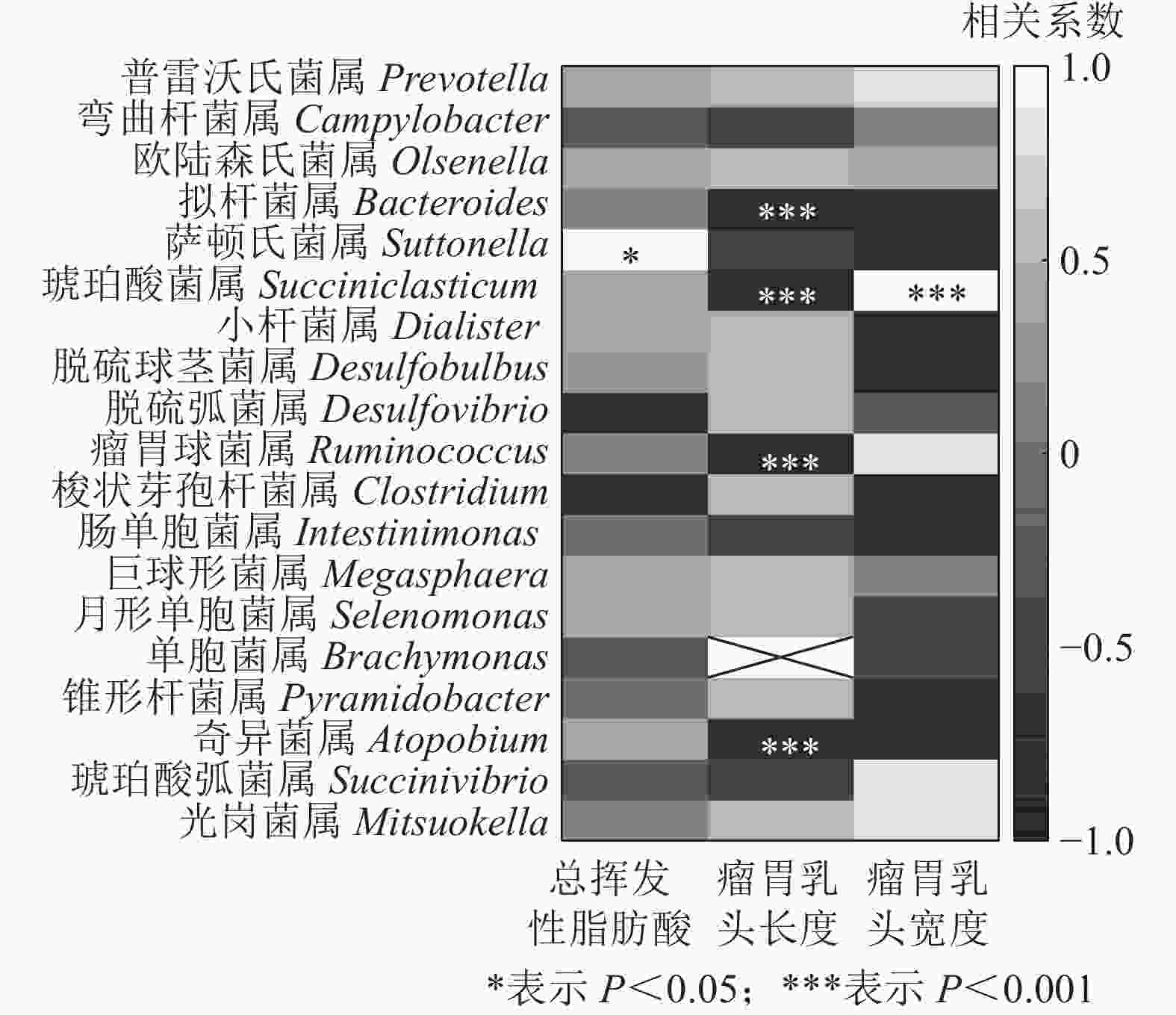
结合瘤胃挥发性脂肪酸、瘤胃乳头长度和宽度[7],与瘤胃上皮微生物进行相关性分析发现(图7):萨顿氏菌属与瘤胃挥发性脂肪酸呈正相关(P<0.05),拟杆菌属、琥珀酸菌属、瘤胃球菌属Ruminococcus和奇异菌属Atopobium与瘤胃乳头长度呈负相关(P<0.001),琥珀酸菌属与瘤胃乳头宽度呈正相关(P<0.001)。
-
反刍动物瘤胃内细菌可分为3个不同的亚群,包括瘤胃内容物细菌、饲料颗粒细菌和瘤胃上皮细菌,这3种细菌种群在瘤胃内相互影响相互作用[8]。然而,与另外2个种群相比,瘤胃上皮细菌因其较低的密度(占总菌数的1%~2%)而研究较少[9]。据报道:瘤胃上皮相关菌群在分类鉴定和功能上与瘤胃内容物细菌群存在明显差异[2, 10]。SADET-BOURGETEAU等[11]研究发现:瘤胃上皮菌群在饲喂高精料和粗饲料间不存在显著差异,推断与瘤胃内容物细菌群相比,瘤胃上皮菌似乎更加稳定。HU等[12]研究限饲对怀孕母羊影响时,发现瘤胃上皮菌群较为稳定,不受限饲影响。新生动物瘤胃上皮发育特征在宿主代谢、免疫应答和瘤胃发育中起着重要作用,因此瘤胃上皮细菌定植对微生物群与宿主的关系具有重要意义,可影响宿主的早期发育和健康[13],甚至对成年后的表现和健康产生持久的影响[14−15]。杨斌[1]研究表明:日龄10 d羔羊瘤胃上皮已有丰富的微生物定植,微生物多样性已经建立。本研究发现:随着日龄的增加,日龄45 d组羔羊瘤胃上皮细菌的多样性较日龄30 d组更为丰富,且各自具有独特的菌群,这一结果与JIAO等[13]的结论一致。JIAO等[13]探究了5个日龄(0、7、28、42和70 d)山羊瘤胃上皮菌群的定植情况,结果发现每个日龄组的山羊瘤胃上皮都有其独特的菌群,且高日龄组的山羊瘤胃上皮菌群较低日龄组的更为多样化,这与瘤胃内容物的微生物定植过程相似[16−17]。MAO等[17]研究指出:日龄45 d羔羊瘤胃内容物细菌的OTU数和系统发育多样性指数均高于日龄30 d羔羊。
厚壁菌门、拟杆菌门和变形菌门三大菌门是湖羊瘤胃上皮的主要菌门,这与SCHÄREN等[18]、PETRI等[19]、JIAO等[13]报道的反刍动物瘤胃上皮微生物的主要菌门一致。ANDERSON等[20]研究指出:厚壁菌门、拟杆菌门和变形菌门在门水平上相对丰度可达79.70%。尽管某些细菌的丰度随年龄的变化而变化,但它们在所有日龄中均存在,说明这些细菌是湖羊瘤胃上皮微生物区系的核心菌群。LIU等[21]研究发现:饲喂开食料显著降低了变形菌门的相对丰度,这一菌门的许多成员主要负责蛋白质降解和固氮的作用[22]。JIAO等[13]研究发现:随着日龄的增加,山羊瘤胃上皮中变形菌门的相对丰度显著降低,可能与母乳的摄入降低和精料开食料的投入有关。本研究中,2个日龄组变形菌门的相对丰度并不存在显著差异,可能原因是2组羔羊的饲喂方式相同,且采食量无显著差异[6]。同样,瘤胃内容物菌群结果也显示:4个主要菌门在2个日龄组不存在显著差异[17]。
属水平上,普雷沃氏菌属、弯曲杆菌属和欧陆森氏菌属三大菌属是湖羊瘤胃上皮的优势菌属。这一结果与LIU等[21]的研究结果一致,且发现弯曲杆菌属在日龄45 d组相对丰度显著降低。弯曲杆菌属是革兰氏阴性菌,在奶牛、绵羊以及山羊的肠道中定植[23]。JIAO等[13]发现:日龄42 d的山羊羔羊瘤胃上皮弯曲杆菌属占优势,其相对丰度可达20.33%。INGLIS等[24]和KRUEGER等[23]均研究指出:与饲喂精饲料相比,饲喂粗饲料,可从牛的瘤胃中分离出较多的弯曲杆菌属。本研究发现:随着日龄的增加,小杆菌属相对丰度有下降趋势。小杆菌属能够分解糖类,所包含的菌种常在口腔内被检测到[25−26],同时,小杆菌属也在瘤胃中出现[27−28]。研究表明:口腔中小杆菌属含量的增加与唾液减少有关[29],这提示瘤胃上皮小杆菌属相对丰度的减少可能与瘤胃的缓冲能力和液体周转有关[1, 28]。JIAO等[13]研究指出:日龄70 d羔羊瘤胃上皮中脱硫弧菌属是优势菌属,相对丰度达3.78%。有研究认为:脱硫弧菌属与硫的还原和丙酸的生成有关[30],琥珀酸菌属虽然不能发酵碳水化合物和氨基酸,但能将琥珀酸盐转化为丙酸[31],然而本研究中脱硫弧菌属和琥珀酸菌属的丰度与丙酸水平[6]呈相反的变化。
本研究表明:萨顿氏菌属与瘤胃挥发性脂肪酸呈正相关,拟杆菌属、琥珀酸菌属、瘤胃球菌属和奇异菌属与瘤胃乳头长度呈负相关,而琥珀酸菌属与瘤胃乳头宽度呈正相关,说明这些菌可能参与挥发性脂肪酸的代谢以及瘤胃组织的发育。此外,也有研究认为:特异的微生物通过促炎和抗炎途径及上皮细胞介导的信号来调节宿主的免疫反应[32]。
-
日龄45 d组羔羊瘤胃上皮细菌群落多样性较日龄30 d组更为丰富,且2个日龄组有各自不同的微生物区系,提示不同日龄羔羊的瘤胃上皮细菌定植存在差异,且通过相关性分析表明:这些菌群参与了瘤胃挥发性脂肪酸的代谢与瘤胃乳头的发育。
Differences of ruminal epithelial bacterial diversity of Hu lambs at different ages
-
摘要:
目的 比较不同日龄湖羊Ovis aries羔羊瘤胃上皮细菌多样性及菌群组成差异,为幼龄反刍动物早期断奶提供理论依据。 方法 选用10只出生体质量相近的初生湖羊羔羊,分别于日龄30 d (D30)和45 d (D45)各屠宰5只以获得瘤胃上皮组织,对其进行测序及微生物多样性分析。 结果 D45组羔羊瘤胃上皮细菌的操作分类单元(OTU)个数显著多于D30组羔羊(P<0.05),Chao1指数D45组有增加的趋势(P=0.07)。与D45组相比,D30组羔羊瘤胃细菌弯曲杆菌属Campylobacter和肠单胞菌Intestinimonas的相对丰度显著降低(P<0.05),拟杆菌属Bacteroides和小杆菌属Dialister的相对丰度有降低的趋势(P=0.083);而萨顿氏菌属Suttonella、脱硫弧菌属Desulfobulbus和单胞菌属Brachymonas的相对丰度显著增加(P<0.05),琥珀酸菌属Succiniclasticum有增加的趋势(P=0.083)。相关性分析发现:萨顿氏菌属与瘤胃挥发性脂肪酸含量呈正相关(P<0.05),拟杆菌属、琥珀酸菌属、瘤胃球菌属Ruminococcus和奇异菌属Atopobium与瘤胃乳头长度呈负相关(P<0.01),琥珀酸菌属与瘤胃乳头宽度呈正相关(P<0.01)。 结论 不同日龄羔羊的瘤胃上皮细菌定植存在差异,这些菌群参与瘤胃挥发性脂肪酸代谢及瘤胃乳头发育。图7表2参32 Abstract:Objective This study, with a comparison of the diversity and composition of rumen epithelial bacteria among Hu lambs (Ovis aries) of different ages, is aimed to provide basic data for early weaning of young ruminants. Method Ten male Hu lambs, with similar birth weight, were used in this study. At 30 days of age (D30) and 45 days of age (D45), five lambs were randomly selected and slaughtered to obtain rumen epithelial tissues (n=5), respectively. Sequencing and microbial diversity analysis were performed on them. Result The Operational Taxonomic Unit (OTU) number in D45 group was larger than that in D30 group (P<0.05), with the Chao1 index in D45 group showing a tendency to increase (P=0.07). The relative abundances of Campylobacter and Intestinimonas declined (P<0.05) with age, while the relative abundances of Suttonella, Desulfobulbus and Brachymonas increased (P<0.05) with age. The relative abundances of Bacteroides and Dialister tended to decrease (P=0.083), whereas Succiniclasticum tended (P=0.083) to increase. The abundance of Suttonella was positively (P<0.05) correlated with rumen total volatile fatty acid concentration, while the abundances of Bacteroides, Succiniclasticum, Ruminococcus and Atopobium were negatively (P<0.01) correlated with ruminal papillae length, and the abundance of Succiniclasticum was positively (P<0.01) correlated with rumen papillae width. Conclusion It was revealed that colonization by ruminal epithelial bacteria is age related and might participate in the metabolism of volatile fatty acid and ruminal papillae development of the rumen. [Ch, 7 fig. 2 tab. 32 ref.] -
Key words:
- age /
- Hu lambs /
- rumen epithelial /
- ruminal bacteria
-
表 1 饲料的化学成分组成
Table 1. Chemical composition of the feeds
饲料类别 饲料成分干物质质量分数/(g·kg−1) 干物质 粗蛋白 中性洗
涤纤维酸性洗
涤纤维钙 磷 粗灰分 代乳粉 945.0 184.0 36.0 9.0 7.2 6.3 41.0 开食料 914.0 174.0 169.0 73.0 12.1 5.9 73.0 羊草 929.0 84.9 659.0 384.0 2.4 1.0 60.0 说明:开食料为每100.0 g干物质中,由58.5 g玉米、10.0 g小麦粉、27.0 g豆粕、1.1 g碳酸氢钙、1.4 g石灰石、1.0 g氯化钠和1.0 g维生素矿物质组成的预混料。 表 2 不同日龄羔羊瘤胃上皮细菌多样性指数
Table 2. Ruminal epithelial bacterial diversity of Hu lambs at different ages
项目 OTU数 Chao1
指数Shannon
指数Simpson
指数Coverage
指数D30 171 b 243±28 a 4.98±0.16 a 0.93±0.02 a 99.90±0.04 a D45 211 a 290±33 a 5.32±0.39 a 0.93±0.04 a 99.80±0.02 a 均值标准误 11.100 15.400 0.148 0.014 0.000 P 0.040 0.070 0.152 0.932 0.338 说明:同列不同字母表示差异显著(P<0.05)。 -
[1] 杨斌. 早期补饲苜蓿调节幼龄湖羊生长和瘤胃发育的机制研究[D]. 杭州: 浙江大学, 2016. YANG Bin. Mechanism in Growth and Rumen Development Alteration by Early Alfalfa Supplementation in Hu Lambs [D]. Hangzhou: Zhejiang University, 2016. [2] ZHANG Ruiyang, YE Huimin, LIU Junhua, et al. High-grain diets altered rumen fermentation and epithelial bacterial community and resulted in rumen epithelial injuries of goat [J]. Applied Microbiology and Biotechnology, 2017, 101(18): 6981 − 6992. [3] MALMUTHUGE N, LI Meiju, CHEN Yanhong, et al. Distinct commensal bacteria associated with ingesta and mucosal epithelium in the gastrointestinal tracts of calves and chickens [J]. FEMS Microbiology Ecology, 2012, 79(2): 337 − 347. [4] MALMUTHUGE N, GRIEBEL P J. Taxonomic identification of commensal bacteria associated with the mucosa and digest throughout the gastrointestinal tract of preweaned calves [J]. Applied and Environmental Microbiology, 2014, 80(6): 2021 − 2028. [5] STEWART C S, FLINT H J, BRYANT M P. The rumen bacteria [M]//HOBSON P N, STEWART C S. The Rumen Microbial Ecosystem. 2nd ed. New York: Springer-Verlag, 1997: 10 − 72. [6] CAPORASO J G, KUCZYNSKI J, STOMBAUGH J, et al. QIIME allows analysis of high-throughput community sequencing data [J]. Nature Methods, 2010, 7(5): 335 − 336. [7] MAO Huiling, WANG Chong, YU Zhongtang. Weaning ages do not affect the overall growth or carcass traits of Hu sheep [J/OL]. Animals, 2019, 9(6): 356[2022-07-20]. doi: 10.3390/ani9060356. [8] CHENG K J, McCOWAN R P, COSTERTON J W. Adherent epithelial bacteria in ruminants and their roles in digestive tract function [J]. The American Journal of Clinical Nutrition, 1979, 32(1): 139 − 148. [9] MUYZER G, de WAAL E C, UITTERLINDEN A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA [J]. Applied and Environmental Microbiology, 1993, 59(3): 695 − 700. [10] CHO S J, CHO K M, SHIN E C, et al. 16S rDNA analysis of bacterial diversity in three fractions of cow rumen [J]. Journal of Microbiology and Biotechnology, 2006, 16(1): 92 − 101. [11] SADET-BOURGETEAU S, MARTIN C, MORGAVI D P. Bacterial diversity dynamics in rumen epithelium of wethers fed forage and mixed concentrate forage diets [J]. Veterinary Microbiology, 2010, 146(1/2): 98 − 104. [12] HU Fan, XUE Yanfang, GUO Changzheng, et al. The response of ruminal fermentation, epithelium-associated microbiota, and epithelial barrier function to severe feed restriction in pregnant ewes [J]. Journal of Animal Science, 2018, 96(10): 4293 − 4305. [13] JIAO Jinzhen, HUANG Jinyu, ZHOU Chuanshe, et al. Taxonomic identification of ruminal epithelial bacterial diversity during rumen development in goats [J]. Applied and Environmental Microbiology, 2015, 81(10): 3502 − 3509. [14] HEINRICHS A J, HEINRICHS B S. A prospective study of calf factors affecting first-lactation and lifetime milk production and age of cows when removed from the herd [J]. Journal of Dairy Science, 2011, 94(1): 336 − 341. [15] YÁÑEZ-RUIZ D R, ABECIA L, NEWBOLD C J. Manipulating rumen microbiome and fermentation through interventions during early life: a review [J/OL]. Frontiers in Microbiology, 2015, 6: 1133[2022-07-20]. doi: 10.3389/fmicb.2015.01133. [16] REY M, ENJALBERT F, COMBES S, et al. Establishment of ruminal bacterial community in dairy calves from birth to weaning is sequential [J]. Journal of Applied Microbiology, 2014, 116(2): 245 − 257. [17] MAO Huiling, ZHANG Yanfang, YUN Yan, et al. Weaning age affects the development of the ruminal bacterial and archaeal community in Hu lambs during early life [J/OL]. Frontiers in Microbiology, 2021, 12: 636865[2022-07-20]. doi: 10.3389/fmicb.2021.636865. [18] SCHÄREN M, KIRI K, RIEDE S, et al. Alterations in the rumen liquid-, particle- and epithelium-associated microbiota of dairy cows during the transition from a silage- and concentrate-based ration to pasture in spring [J/OL]. Frontiers in Microbiology, 2017, 8: 744[2022-07-20]. doi: 10.3389/fmicb.2017.00744. [19] PETRI R M, SCHWAIGER T, PENNER G B, et al. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis [J]. Applied and Environmental Microbiology, 2013, 79(12): 3744 − 3755. [20] ANDERSON C J, KOESTER L R, SCHMITZ-ESSER S. Rumen epithelial communities share a core bacterial microbiota: a meta-analysis of 16S rDNA gene illumine miseq sequencing datasets [J/OL]. Frontiers in Microbiology, 2021, 12: 625400[2022-07-20]. doi: 10.3389/fmicb.2021.625400. [21] LIU J, BIAN G, SUN D, et al. Starter feeding altered ruminal epithelial bacterial communities and some key immune-related genes' expression before weaning in lambs [J]. Journal of Animal Science, 2017, 95(2): 910 − 921. [22] 阮继生. “伯杰氏系统细菌学手册(第2版)”第5卷与我国的放线菌系统学研究[J]. 微生物学报, 2013, 53(6): 521 − 530. RUAN Jisheng. Bergey ’s Manual of Systematic Bacteriology (Second Edition) Volume 5 and the study of actinomycetes systematic in China [J]. Acta Microbiologica Sinica, 2013, 53(6): 521 − 530. [23] KRUEGER N A, ANDERSON R C, KRUEGER W K, et al. Prevalence and concentration of Campylobacter in rumen contents and feces in pasture and feedlot-fed cattle [J]. Foodborne Pathogens and Disease, 2008, 5(5): 571 − 577. [24] INGLIS G D, MCALLISTER T A, BUSZ H W, et al. Effects of subtherapeutic administration of antimicrobial agents to beef cattle on the prevalence of antimicrobial resistance in Campylobacter jejuni and Campylobacter hyointestinalis [J]. Applied and Environmental Microbiology, 2005, 71(7): 3872 − 3881. [25] DOWNES J, MUNSON M, WADE W G. Dialister invisus sp. nov., isolated from the human oral cavity [J]. International Journal of Systematic and Evolutionary Microbiology, 2003, 53(6): 1937 − 1940. [26] ISABELA N, RÔÇAS I N, SIQUEIRA J F. Characterization of Dialister species in infected root canals [J]. Journal of Endodontics, 2006, 32(11): 1057 − 1061. [27] BROADWAY P R, CALLAWAY T R, CARROLL J A, et al. Evaluation of the ruminal bacterial diversity of cattle fed diets containing citrus pulp pellets [J]. Agriculture,Food and Analytical Bacteriology, 2012, 2(4): 297 − 308. [28] MYER P R, SMITH T P L, WELLS J E, et al. Rumen microbiome from steers differing in feed efficiency [J/OL]. PLoS One, 2015, 10(6): e0129174[2022-07-20]. doi: 10.1371/journal.pone.0129174. [29] HAYASHI Y, SAITO T, OHSHIMA T, et al. Terminal RELP analysis to determine the oral microbiota with hyposalivation [J]. Archives of Microbiology, 2014, 196(7): 489 − 496. [30] PARKES R J, CALDER A G. The cellular fatty-acids of 3 strains of Desulfobulbus, a propionate-utilizing sulfate reducing bacterium [J]. FEMS Microbiology Ecology, 1985, 31: 361 − 363. [31] van GYLSWYK N O. Succiniclasticum ruminis gen. nov., sp. nov., a ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism [J]. International Journal of Systematic Bacteriology, 1995, 45(2): 297 − 300. [32] KLAENHAMMER T R, KLEEREBEZEM M, KOPP M V, et al. The impact of probiotics and prebiotics on the immune system [J]. Nature Reviews Immunology, 2012, 12(10): 728 − 734. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.20220482






 下载:
下载: