-
山核桃Carya cathayensis属胡桃科Juglandaceae山核桃属Carya乔木树种,是浙江区域优势明显的特色经济干果,其坚果种仁具有较高的营养价值和经济效益[1]。近年来向贵州石漠化地区推广山核桃种植,生长良好,并已陆续挂果[2]。尽管生产上已解决了无性嫁接繁殖的问题[3],但山核桃长期处于野生、半野生状态[4],种下无品种,以实生繁殖。与山核桃同属的薄壳山核桃Carya illinoensis则品种极其繁多,2个物种均含油率高,且具互补的优势。如山核桃种子小,壳硬,风味独特,而薄壳山核桃则种子大,壳薄,也有其独特的风味。因此,自20世纪60年代开始,2个物种的杂交试验发现:山核桃与薄壳山核桃正反交均可育,杂种后代在生长性状上具有杂种优势,但不管正交还是反交,其子代在表型上均与母本相似[5-6]。尽管山核桃为二倍体物种[7-8],但近年的研究发现:山核桃存在起源于珠心细胞且属于不定胚生殖范畴的无融合生殖现象及假配合现象[9]。随机扩增多态DNA(random amplified polymorphic DNAs,RAPD),扩增片段长度多态(amplified fragment length polymorphism,AFLP),简单序列重复间隔区(inter-simple sequence repeat,ISSR),序列相关扩增多态(sequence-related amplified polymorphism,SRAP),简单序列重复(simple sequence repeat,SSR)等分子标记的分析发现:山核桃多态性不佳,尽管存在一些多态的位点,但比例较低[10-12],且利用有限的标记在山核桃中发现存在印迹数量性状位点(imprinted quantitative trait loci, iQTLs)[13]。基因组印迹与DNA甲基化有关[14],而DNA甲基化与染色质结构修饰、小分子RNA调控及核小体重塑[15]、组蛋白共价修饰、基因沉默和RNA编辑等一起在生物体内进行表观遗传调控基因的表达,其中DNA甲基化是研究最为透彻的表观遗传现象之一[16]。甲基化敏感扩增多态(methylation sensitive amplification polymorphism, MSAP)标记是在AFLP技术基础上衍生的基于PCR扩增的DNA甲基化检测方法,它将AFLP中Mse Ⅰ替换成2个对甲基化位点敏感程度存在差异的同裂酶Hpa Ⅱ和Msp Ⅰ,可检测全基因组范围内CCGG位点的甲基化状况,同时不仅可以获得非甲基化的遗传位点(1, 1),还可以获得包括半甲基化位点(1, 0),内部甲基化位点(0, 1),超甲基化或突变位点(0, 0)在内的共4类标记位点。MSAP在不同生物中具有通用性,被广泛应用于动植物资源鉴定、生物发育调控研究、杂种优势分子机理研究[17]等,并已应用于巴西橡胶树Hevea brasiliensis[18],杉木Cunninghamia lanceolata[19],叶籽银杏Ginkgo biloba var. epiphylla[20],地中海柏木Cupressus sempervirens[21],落叶松Larix[22]和毛白杨Populus tomentosa[23]等林木的研究中。为了从标记方面对山核桃开展深入研究,本研究借鉴其他物种的MSAP体系,建立适用于山核桃的MSAP析体系,并将其应用于天然群体的山核桃,旨在为山核桃的遗传学研究提供依据。
HTML
-
从浙江省杭州市临安区太湖源镇指南村(7株,30°21′N,119°34′E),临安区清凉锋镇林竹村(17株,30°06′N,118°58′E),安徽省宁国市仙霞镇潘张村(6株,30°24′N,119°17′E)3个山核桃天然居群中分别采集盛果期成年单株的叶片,用于DNA的提取。采样单株彼此间相距至少50 m。
薄壳山核桃叶片样品采自浙江省杭州市余杭区长乐林场中国林业科学研究院亚热带林业研究所种质资源收集圃盛果期成年单株的叶片(4株,30°18′N,119°51′E)。
-
采用郭金剑[24]改良的十六烷基三甲基溴化铵(CTAB)法提取山核桃和薄壳山核桃基因组DNA,用核酸蛋白质分析仪(ND-1000,NanoDrop,美国)测定波长260和280 nm时的光密度值[D(260),D(280)]及反映基因组DNA质量的D(260)/D(280)与DNA浓度;用质量分数为1%的琼脂糖凝胶电泳检测DNA的优劣。
-
基于洪舟等[19]用于杉木的MSAP分析体系,在山核桃中就酶切连接时间、预扩增产物稀释倍数、选择性扩增的模板DNA用量及PCR退火温度(第1轮)与扩增循环数(第2轮)进行优化,建立适用于山核桃MSAP分析体系。DNA限制性酶切与接头连接采用1步法。在此基础上对酶切连接时间(4,6和8 h)进行实验。因为预扩增与选择性扩增的目的与程序有差异,所以有必要就具体的物种或研究对象,对实验进行一定的优化[25]。在预扩增阶段,参考已有的杉木MSAP分子标记体系[19]以及苹果Malus pumila[26],脐橙Citrus sinensis[27]等其他经济物种的MSAP反应体系,测试3种不同的预扩增产物稀释倍数(10,20和40倍),同时试验选择性扩增的模板DNA用量(2和4 μL)。借鉴三叶木通Akebia trifoliata的MSAP反应体系[28],对选择性扩增程序(94 ℃变性2 min:第1轮PCR 94 ℃ 30 s, 65 ℃ 30 s,每个循环递减0.7 ℃,72 ℃ 1 min,共13个循环;第2轮PCR 94 ℃ 30 s,56 ℃ 30 s,72 ℃ 1 min;最后72 ℃延伸10 min)中第2轮PCR的循环数(20,25和30次)及第1轮PCR中退火温度的递减幅度(每循环分别降低0.7,1.0和1.3 ℃)进行优化。以上试验内容按序进行,待前1个参数优化后便固定下来进行后1个参数的优化。选扩产物用质量分数为1%琼脂糖凝胶电泳检测。选扩产物电泳检测有扩增产物,且条带清晰,则进行96对引物组合(表 1)的筛选,用体积分数为6%变性聚丙烯酰胺凝胶电泳分离,并对胶进行常规银染。
EcoRⅠ(E)引物 序列(5′→3′) HpaⅡ/MspⅠ(HM)引物 序列(5′→3′) E1 GACTGCGTACCAATTCCGT HM-1 GATGAGTCTAGAACGGTAG E2 GACTGCGTACCAATTCCAC HM-2 GATGAGTCTAGAACGGTAC E3 GACTGCGTACCAATTCAAC HM-3 GATGAGTCTAGAACGGTTG E4 GACTGCGTACCAATTCAGA HM-4 GATGAGTCTAGAACGGTTC E5 GACTGCGTACCAATTCAAA HM-5 GATGAGTCTAGAACGGTGT E6 GACTGCGTACCAATTCAAT HM-6 GATGAGTCTAGAACGGTGC E7 GACTGCGTACCAATTCAGC HM-7 GATGAGTCTAGAACGGTCT E8 GACTGCGTACCAATTCCTG HM-8 GATGAGTCTAGAACGGTCG E9 GACTGCGTACCAATTCCAC E10 GACTGCGTACCAATTCCTG E11 GACTGCGTACCAATTCCAT E12 GACTGCGTACCAATTCACG Table 1. Sequences of primers for selective amplification in MSAP
-
用建立的适用于山核桃的MSAP分析体系对天然居群山核桃样品进行分析时,HpaⅡ/MspⅠ选择性扩增引物在序列末端加荧光5′-FAM(羟基荧光素)标记(北京睿博兴科生物科技有限公司)。选择性扩增产物送北京睿博兴科生物科技有限公司进行毛细管电泳检测(ABI 3730XL)。
-
建立山核桃MSAP分析体系对薄壳山核桃DNA进行分析,了解山核桃的MSAP分析体系是否适用于薄壳山核桃。
-
在山核桃MSAP分析结果中提取非甲基化遗传位点信息,并将其与RAPD,AFLP,ISSR,SRAP,SSR分子标记的结果进行比较。
-
毛细管电泳检测选扩产物后,甲基化与非甲基化位点由GeneMarker V2.0.0软件产生。软件默认去除小于70 bp的位点,各位点上有扩增产物时记为“1”,无扩增产物时记为“0”。使用Excel,将MSAP分析2组酶EcoRⅠ/HpaⅡ和EcoRⅠ/MspⅠ酶切产物的PCR 0或1的结果转换成MSAP分析结果(0,0),(1,0),(0,1)和(1,1)4种位点数据,用基于R语言的msap软件[30]分析数据,包括甲基化敏感位点(methylation sensitive locus,MSL)与非甲基化敏感位点(non-methylation locus,NML),即遗传位点的判断,并分析总位点数、MSL数、NML数、多态的MSL数及其比例,多态的NML数及其比例,MSL及NML各自的香农信息指数(Shannon diversity index,I)。基于Wilcoxon Rank Sum检验的遗传多样性与表观遗传多样性的差异显著性,获得各居群(0,0),(1,0),(0,1)和(1,1)4种位点各自的比例,并分别对MSL和NML进行分子方差分析(AMOVA),最后基于主坐标分析(Principal coordinates analysis,PCoA)来评估MSL和NML的分化。
1.1. 材料
1.2. 方法
1.2.1. 基因组DNA的提取
1.2.2. 山核桃MSAP体系的建立
1.2.3. 天然居群山核桃的分析
1.2.4. 山核桃MSAP分析体系对薄壳山核桃的适用性
1.2.5. 山核桃MSAP标记与其他分子标记的比较
1.2.6. 数据处理
-
提取的基因组DNA经琼脂糖电泳检测,结果条带明亮、清晰,质量较好。核酸蛋白质分析仪测定结果表明:基因组DNA D(260)/D(280)为1.80~2.10,纯度较高,可用于后续的MSAP标记分析。
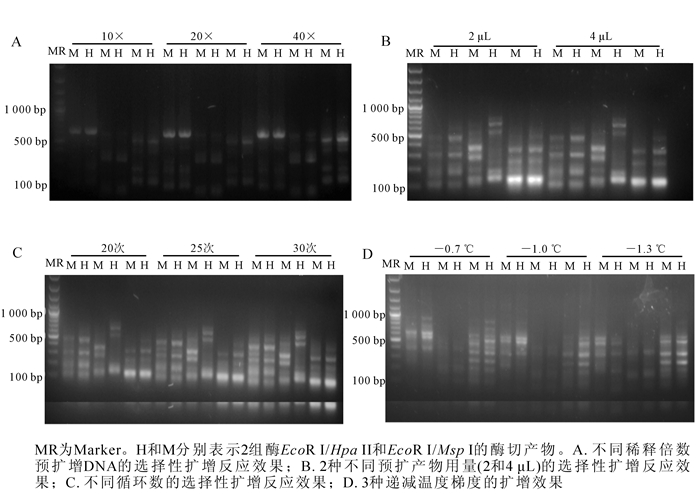
实验发现:500 ng基因组DNA在37 ℃的水浴锅中双酶切-连接反应4,6和8 h,其中,6 h效果最佳,没有大分子DNA(图 1A)。将酶切完全且带接头的DNA产物进行预扩,琼脂糖电泳检测预扩产物在100~1 000 bp呈弥散状(图 1B),说明效果较好,可用作下一步选择性扩增的模板。

Figure 1. Results of double enzyme digested DNAs in hickory (A) and different pre-amplified DNA samples(B)
在预扩增基础上,预扩产物模板3种不同稀释倍数的试验发现,预扩产物稀释40倍时,选择性扩增结果最佳(图 2A)。此外,发现预扩增产物用量为2 μL时,选择性扩增结果最佳(图 2B)。而选择性扩增第2个PCR循环数为25时选扩反应效果最佳(图 2C)。当第2个PCR循环递减温度为-0.7 ℃时,选择性扩增条带数较少;当温度变化梯度为-1.0 ℃时,清晰条带数量偏少;而每循环降低1.3 ℃为最佳温度变化梯度(图 2D)。
实验结果最佳的山核桃MSAP分析体系如下:500 ng DNA在37 ℃水浴锅中酶切连接6 h;酶切连接反应产物稀释10倍进行预扩增。选择性扩增反应体系为10×PCR缓冲液2.0 μL,dNTPs(10 mmol·L-1,生工生物工程(上海)股份有限公司)0.5 μL,25 mmol·L-1氯化镁1.5 μL,稀释40倍的预扩增产物2.0 μL,选择性扩增引物各1.5 μL,Taq酶(5×16.67 mkat·L-1,上海申能博彩生物科技有限公司)0.5 μL,补水至20.0 μL。选择性扩增的PCR程序为94 ℃预变性2 min;94 ℃变性30 s,65 ℃退火复性30 s,72 ℃延伸1 min,共13个循环,每个循环退火温度递减1.3 ℃;94 ℃变性30 s,56 ℃退火复性30 s,72 ℃延伸1 min,共25次循环;72 ℃延伸10 min,4 ℃保持。从96对选择性扩增引物组合中筛选出了能扩增清晰条带,重复性好,且适用于山核桃MSAP分析的引物5对,分别为HM-2/E4,HM-3/E2,HM-3/E11,HM-5/E8,HM-8/E8。
-
利用EcoRⅠ/HpaⅡ和EcoRⅠ/MspⅠ不同的酶切位点,MSAP分析得到4种5′-CCGG位点信息。非甲基化遗传位点(1,1)HpaⅡ和MspⅠ2种酶均能进行酶切,毛细管电泳时出现H(EcoRⅠ/HpaⅡ酶切产生)与M(EcoRⅠ/MspⅠ酶切产生)双峰;半甲基化位点(1,0)仅HpaⅡ能进行酶切;内部胞嘧啶甲基化位点(0,1)则相反,仅MspⅠ能进行酶切;而超甲基化位点(0,0)2种酶均不能酶切,电泳图上没有峰,只能通过样品间的对比来推断30个山核桃样品5对选择性扩增引物共产生389个位点,且387个位点为多态位点,多态性水平为99.48%,其中引物组合HM-3/E2扩增的位点数(191)最多,引物组合HM-3/E11扩增的多态性位点(18)最少;MSL 140个,多态位点65个,甲基化敏感多态位点比例为46.42%;NML 249个,多态位点233个,非甲基化遗传多态位点比例为93.57%。MSL和NML的香农信息指数(I)平均分别为0.554和0.173。
-
由于NML是遗传位点[29],在与山核桃其他分子标记(RAPD,ISSR,SRAP,AFLP,SSR)分析结果相比较时,采用NML的数据。结果表明:MSAP分子标记的多态性水平(93.57%)最高,其他每对引物的扩增位点数、多态引物比例、多态位点比例、每对引物扩增得到的多态位点数等各项指标也高于其他分子标记(表 2)。
标记名称 使用引物/对(个) 获得多态的引物/对(个) 扩增总位点数/个 每对(个)引物扩增位点数 扩增多态性位点总数/个 多态引物比例/% 多态位点比例/% 每对(个)引物扩增多态位点数 MSAP 5 5 249 49.80 233 100.00 93.57 46.60 RAPD 16 12 121 7.56 25 75.00 20.66 1.56 ISSR 17 9 143 8.41 20 52.94 13.99 1.18 SRAP 15 9 173 11.53 27 60.00 15.61 1.80 AFLP 11 8 32 72.72 2.91 SSR 30 0 30 1.00 0 0.00 0 0 Table 2. A comparison among the amplification results of various molecular markers in hickory
-
由于MSAP技术中所用的2种同裂酶HpaⅡ和MspⅠ均不能有效地识别非5′-CCGG位点的胞嘧啶甲基化和内外侧胞嘧啶都甲基化的5′-CCGG位点[19],且(0,0)位点在实验上是没有扩增产物的位点,也不排除位点缺失所致[29],因此MSAP检测到的甲基化位点数比实际的偏低。定义半甲基化类型与内侧胞嘧啶甲基化类型位点数之和为总甲基化位点数,并将一个村的样品视作来自一个居群。基于MSAP分析结果可以看出:山核桃甲基化相对水平变异系数在20%左右,总甲基化相对水平和非甲基化相对水平分别为25.99%±2.42%和33.38%±3.66%,非甲基化相对水平大于总甲基化相对水平,且两者间差异极显著(P<0.001);总甲基化中半甲基化相对水平和内侧胞嘧啶甲基化相对水平分别为7.89%±1.53%和18.10%±3.94%,内侧胞嘧啶甲基化相对水平远大于半甲基化相对水平,且两者间差异极显著(P<0.001)(表 3)。对3个居群山核桃之间的甲基化状态进行比较,其中林竹村总甲基化相对水平(28.00%)和内部胞嘧啶甲基化相对水平(21.33%)最高,而其半甲基化相对水平(6.66%)和非甲基化相对水平(29.16%)最低;潘张村的半甲基化相对水平(9.60%)和非甲基化水平(35.61%)最高,其总甲基化相对水平(23.31%)和内侧胞嘧啶甲基化相对水平(13.71%)最低;指南村各项相对水平居中。此研究中山核桃总甲基化相对水平和内侧胞嘧啶甲基化相对水平高,则半甲基化相对水平和非甲基化相对水平则相对较低,反之亦然(表 3)。
甲基化类型 均值±标准差/% 变异系数/% 最大值/% 最小值/% 非甲基化(1, 1) 33.38 ± 3.66 10.96 35.61 29.16 半甲基化(1, 0) 7.89 ± 1.53 19.32 9.60 6.66 内侧胞嘧啶甲基化(0, 1) 18.10 ± 3.94 21.76 21.33 13.71 总甲基化(1, 0)(0, 1) 25.99 ± 2.42 9.29 28.00 23.31 全甲基化或位点缺失(0, 0) 40.96 ± 2.95 7.19 43.84 37.96 Table 3. Relative methylation levels in hickory
-
由表 4可见:指南村山核桃遗传多样性(0.176±0.053)和表观遗传多样性(0.591±0.099)都最丰富,其次为潘张村的山核桃(0.163±0.070和0.588±0.089),林竹村山核桃遗传多样性(0.145±0.035)和表观遗传多样性(0.541±0.130)最低。变异系数遗传多样性在24.16%~42.81%,表观遗传多样性为15.12%~24.07%,表明遗传多样性高的居群,表观遗传多样性也相对较高,说明表观变异与遗传变异可能存在一定的关联,但又不完全依赖于经典遗传。3个居群的整体表观遗传多样性(0.574±0.011)远大于整体的遗传多样性(0.162±0.052),经Wilcoxon秩和检验分析(W=16 656.5,P<0.000 1)表明,两者具有极显著差异。
居群 甲基化位点 非甲基化位点 香农信息指数 变异系数/% 香农信息指数 变异系数/% 潘张村 0.588 ± 0.089 15.12 0.163 ± 0.070 42.81 指南村 0.591 ± 0.099 16.78 0.176 ± 0.053 30.15 林竹村 0.541 ± 0.130 24.07 0.145 ± 0.035 24.16 均值 0.574 ± 0.011 18.73 0.162 ± 0.052 32.39 Table 4. Genetic (NML) and epigenetic (MSL) diversities assessed by Shannon diversity index in three natural populations
-
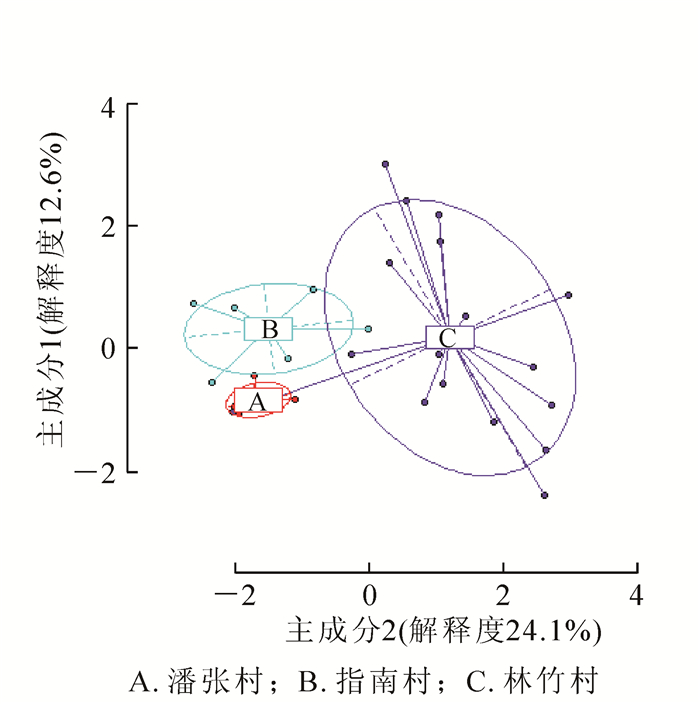
非甲基化位点(NML)的AMOVA分析表明:山核桃天然居群几乎所有的遗传变异存在于居群内(98.76%),只1.24%存在于居群间,居群间分化不显著(分化系数Φst=0.012,P=0.235,P>0.05,表 5)。甲基化敏感位点(MSL)的AMOVA分析表明:山核桃天然居群76.70%的表观遗传变异存在于居群内,而23.30%表观遗传变异存在于居群间,且居群间分化差异极显著(Φst=0.233,P<0.000 1,表 5)。通过主坐标分析:各居群间的个体可以大致汇聚在一起,说明居群间存在明显的表观遗传分化,其中林竹村居群分别与指南村居群和潘张村居群之间存在少部分交集,而指南村居群与潘张村居群间相互独立(图 3)。居群间的表观遗传分化系数(0.233)远大于居群间的遗传分化系数(0.012),这说明山核桃天然居群间表观遗传分化程度要高于遗传分化程度。Mantel检验(相关系数r=0.125,P>0.05)发现,遗传背景对表观遗传变异的影响不显著。
位点 变异来源 自由度 平方和 均方 方差 方差分量百分率/% 非甲基化遗传位点 居群间 2 22.94 11.47 0.129 4 1.24 居群内 27 279.00 10.33 10.330 0 98.76 总计 29 302.00 10.41 甲基化位点 居群间 2 78.93 39.47 3.273 0 23.30 居群内 27 290.80 10.77 10.770 0 76.70 总计 29 369.80 12.75 Table 5. Analysis of molecular variance (AMOVA) for both non-methylated (NML) and methylated loci (MSL) in three natural populations
-
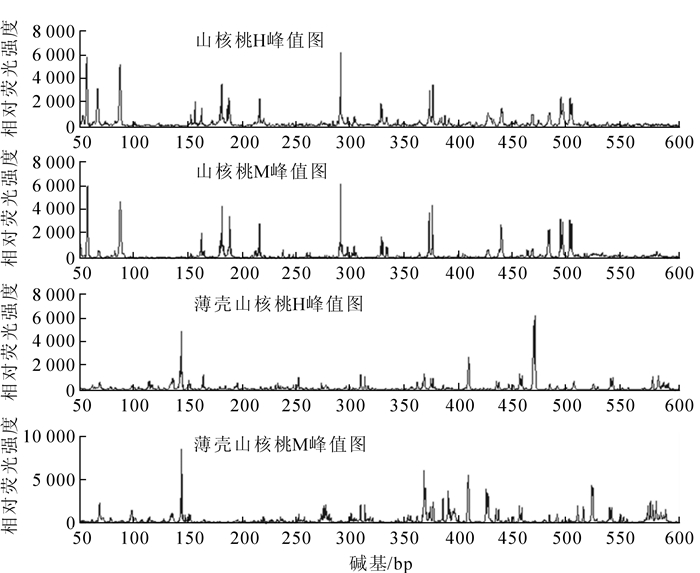
从山核桃中筛选的引物组合(HM-3/E2,HM-3/E11,HM-5/E8,HM-8/E8)对4个薄壳山核桃个体进行MSAP分析发现:只有1对引物组合HM-5/E8有扩增产物,且其位点及位点数量与山核桃存在差异(图 4),其他3对引物均没有扩增产物,说明山核桃MSAP体系也适用于薄壳山核桃,但由于不同物种之间的差异性,薄壳山核桃适用的MSAP引物还需进一步筛选。
2.1. 山核桃MSAP分析体系的建立
2.2. 天然居群山核桃的分析
2.2.1. 山核桃MSAP分析
2.2.2. 山核桃MSAP标记与其他标记分析结果的比较
2.2.3. 甲基化/非甲基化相对水平分析
2.2.4. 山核桃遗传和表观遗传多样性分析
2.2.5. 山核桃遗传与表观遗传分化及结构分析
2.3. 山核桃MSAP分析体系在薄壳山核桃上的应用
-
本研究的MSAP分析表明:山核桃中确实存在甲基化,且与山核桃中其他的分子标记比较发现,MSAP分析每对引物获取的遗传位点数、每对引物扩增的多态位点数、多态引物的比例、多态位点的比例都最高;如果不用MSAP分析,则不可能获得这些遗传位点的信息,说明甲基化对山核桃遗传位点的发现与获取有影响。常规分子标记在山核桃中多态性不佳,影响了山核桃的遗传分析。位点甲基化后使得染色质变得异常紧凑[30],且影响DNA的正常酶切,因而使通常可获得较多多态标记的AFLP分析在山核桃中的适用性也不佳。内部胞嘧啶甲基化相对水平远大于半甲基化相对水平,说明山核桃以内部胞嘧啶甲基化为主,尽管山核桃中超甲基化或突变位点的比例不低。对于超甲基化或突变位点,不同的人有不同的看法,有些研究人员将其看作是超甲基化位点[31-32]。如地中海柏木Cupressus sempervirens[21],有些研究人员将其视为无信息位点[33]。与其他遗传标记相比,这些超甲基化或突变位点在实验上是无扩增产物的位点,排除在扩增位点之外。这些位点至少不可能都是超甲基化或突变位点,有待于进一步研究。山核桃整体及各居群内的表观遗传多样性均大于遗传多样性,与很多其他植物种群的表观遗传研究结果一致,如Viola cazorlensis[34],日本的一种入侵植物[35]等。类似的研究结果同样出现在一些动物种群的表观变异研究中[36]。
3个居群非甲基化相对水平排序与遗传多相性排序间无对应关系,但遗传多样性高的居群,表观遗传多样性也相对较高,说明表观变异与遗传变异可能存在一定的关联,须进一步进行研究。此外,甲基化变异主要存在于居群内,且居群间差异显著,说明山核桃甲基化变异具有居群特异性。研究表明:甲基化在植物对环境的刺激反应中发挥着作用[37],也即环境变化会影响DNA甲基化,但DNA甲基化又在基因组印迹、发育基因调节及基因表达水平的自然变异中发挥作用[38]。山核桃幼胚甲基化比例就随着幼胚的发育逐渐升高,且不同发育时期在基因组DNA甲基化水平上存在较大差异[39]。但越来越多的证据表明:不改变DNA序列的表观遗传信息可以在世代间遗传[40]。从受精卵到成熟的生物体,DNA甲基化模式通过连续的细胞分裂将这些信息传递给子细胞,维持细胞特有的功能[41],而生物选择许多表观遗传过程是为了确保发育过程的保真度[42]。盐胁迫处理2个基因型水稻Oryza sativa的研究发现:有一定比例的DNA甲基化位点是基因型特有的,与其类似的遗传背景相一致;并在耐盐及盐敏感基因型中发现了几个具有稳定甲基化变化的位点,可作为基因型特有的表观遗传标记[43]。在大花蕙兰Cymbidium hybridium[44]和水稻[45]中发现了2类甲基化位点,其中一类甲基化状态保持不变,而另一类为不同发育阶段或品种间有变化的甲基化位点。也就是说,一定比例的甲基化位点是可遗传的。因此,从MSAP标记中获取的遗传位点信息,不仅可用于山核桃遗传分析,也可和遗传位点信息一起用于遗传图谱的构建[46],对解析QTL基因对表型的作用具有一定的意义。DNA甲基化使物种的遗传研究趋于复杂化,增加了研究的难度。
有性植物研究中越来越多支撑材料表明:无整合生殖是有性生殖的脱分化[47],是可逆地叠加在有性途径上的一种生殖方式[48]。表观遗传调控无融合生殖是一个很吸引人的假设,将是继续研究天然无融合生殖的关键焦点[48],表观遗传机制是区分有性与无融合生殖控制事件的关键[49-50],表观途径的突变导致无融合生殖样的表型[48]。无融合生殖遗传上常出现大规模的分离畸变[51],山柳菊属Hieracium物种连锁图谱有无标记或“连锁的”大片段[52],而许多无融合生殖位点交换受到抑制[48],且转录保守等位基因与无融合种子形成之间有很强的关联[53]。甲基化使得DNA结构上更加紧凑[30],难以酶切产生交换。同时杉木中树高、胸径和材积性状的杂种优势[19]及地中海柏木全同胞子代高和直径生长[21]均与甲基化有关。甲基化既与无融合生殖有关,又与杂种优势有关。因此,山核桃甲基化的初步分析为该物种的后续深入研究打下了基础。













 DownLoad:
DownLoad: