-
乙烯响应因子(ERF)是AP2/ERF(apetala2/ethylene response factor)大家族中的1个亚族,最早从烟草Nicotiana tabacum中分离发现,都含有1个AP2结构域。ERF受乙烯诱导表达,并且具有与生物胁迫抗性基因启动子结合的能力[1]。随着研究深入,ERF结构中各类基序的作用被广泛关注,其中AP2结构域作为DNA结合域更是受到深入研究。在各类植物生长代谢与非生物胁迫响应等方面,ERF也受到农林研究者的重视。除乙烯外,其他信号分子及表达调控机制也影响着ERF的行使功能。本研究以ERF的结构特征、生物学功能以及有关的调控机制为主题,对近年有关ERF的研究进行综述,以期为ERF表达调控和功能研究提供思路。
HTML
-
ERF含有1个AP2结构域,AP2结构域是ERF的DNA结合域,是其行使转录调控的关键。NAKANO等[2]根据系统发育关系、外显子-内含子结构和蛋白质基序分别将拟南芥Arabidopsis thaliana和水稻Oryza sativa中的ERF分为12组和15组。这种分类方式适用多种植物,如苜蓿Medicago sativa[3]、二穗短柄草Brachypodium distachyon[4]、人参Panax ginseng[5]等。同组的ERF因为有着相似的结构和保守基序,通常具有相似的性质[2]。
AP2结构域最早发现于拟南芥AP2基因中,由大约60个氨基酸组成[1, 6]。AP2结构域的N端存在1个碱性亲水区,含有3个反平行的β-折叠,这3个β-折叠在识别顺式作用元件中具有重要作用,C端有1个两亲性的α-螺旋,可能参与其他转录因子或DNA的相互作用[7]。ERF根据AP2结构域上特定位置的不同,氨基酸残基可分为ERF与DREB,由于残基的不同,ERF和DREB对启动子的亲和性和识别特异性会有差异[8]。ERF和DREB通常结合的顺式元件分别是GCC(核心序列为GCCGCC)和DRE/CRT(dehydration-responsive element/C-repeat,核心序列为CCGAC)[9]。除这两者外,ERF还可以结合其他的基序,例如刚毛柽柳Tamarix hispida的ThCRF1可与GCC、DRE、TTG1和TTG2基序结合[10]。ERF与基序结合的特异性似乎受到某种机制的调控,根据接收到的信号改变对基序的结合倾向,例如拟南芥中ERF1在茉莉酸诱导下与茉莉酸响应基因启动子上的GCC基序结合,但受到非生物胁迫时,ERF1优先与胁迫抗性基因启动子上的DRE基序结合[11]。
-
ERF在AP2域外含有一些保守的氨基酸基序。有些基序在ERF参与转录调控时起到重要作用,当基序缺失或突变时会导致功能的改变或缺失。比如EAR(ERF-associated amphiphilic repression)类基序,这类基序有2种保守序列,分别为LxLxLx和DLNxxP[12]。EAR基序可直接影响ERF转录调控的功能,例如拟南芥中ERF4有2种亚型,其中ERF4-R(含有EAR基序)能抑制基因表达,而ERF4-A(不含EAR基序)能上调基因表达[13]。有的基序具有激活结构域的功能,比如EDLL基序具有激活转录的功能,因保守的谷氨酸(E)、天冬氨酸(D)和亮氨酸(L)残基而得名,属于酸性的激活结构域[14]。基序还有其他功能,如拟南芥ESR1中的ESR基序是激活结构域,当用VP16替换ESR基序时,ESR1丧失了促进芽再生的能力[15]。ERF功能基序可以通过基因编辑的方式添加至目标ERF基因中,修饰后ERF的功能根据基序的类别发生改变,例如将以EAR基序为基础改造的SRDX序列添加至ERF中,使修饰后的ERF具有转录抑制的功能[16]。
1.1. ERF结构概述
1.2. 功能基序
-
ERF的生物学功能涉及范围非常广,胁迫和生长发育都有它的参与。ERF的功能主要由直接调控目标基因实现,也有一些是通过调控其他转录因子实现(表1)。
植物 ERFs 目的基因及调控方式 目的基因说明 效果 拟南芥 ERF4 CAT3 过氧化氢酶,ROS清除酶系统之一 根据ERF4形态不同,调控的效果不同[13] 拟南芥 ERF96 上调PDF1.2a、PR-3/4和ORA59 生物胁迫抵御基因,ORA59为转录
因子增强植物对病原体抗性[17] 拟南芥 ERF74 上调RbohD 呼吸暴发氧化酶,增加活性氧的产量 启动胁迫初期活性氧暴发,增强信号传导[18] 拟南芥 Ⅸb组ERF 上调CYP81F2 杀菌剂吲哚类硫苷合成关键酶 增加吲哚类硫苷合成,减少植物间真菌传播[19] 拟南芥 WRI4 上调LACS1、KCR1、PAS2、ECR和WSD 参与表皮蜡质合成 增加蜡质合成,保护植物受逆境伤害[20] 拟南芥 RAP2.2和RAP2.12 上调LBD41和PCO1 低氧响应蛋白质 受低氧胁迫诱导,保持低氧应答基因在低氧环境下的稳定[21] 拟南芥 AtERF72 下调IRT1、HA2和上调CLH1 IRT1、HA2铁吸收,CLH1叶绿素降解 受缺铁环境诱导,起到负调控效果[22] 苹果 MdERF2 上调MdACS3a 果实成熟过程中乙烯合成关键酶基因 进一步促进乙烯生成[23] 苹果 MdERF4 下调MdERF3 转录因子,响应盐胁迫并提高盐胁迫抗性 削弱植物对盐胁迫抗性[24] 水稻 OsERF71 上调OsXIP 抑制微生物来源的木聚糖酶 增强植物抗性[25] 芜菁 BrERF72 上调BrLOX4、BrAOC3和BrOPR3 参与茉莉酸合成 加速叶片衰老[26] 番茄 JRE4 上调DWF5和GAME4 甾醇还原酶和配糖生物碱代谢相关 促进糖苷生物碱的合成[27] 番茄 ERF68 上调COPA、Sw-5a、AOS和
下调CAB参与细胞程序性死亡 促进细胞程序性死亡,防止病原体扩散[28] 月季 RhERF1和RhERF4 下调RhBGLA1 β-半乳糖苷酶,加速花瓣脱落 防止花瓣脱落[29] 罂粟 PsAP2 上调AOX1a 抗氧化酶之一 增强植株的ROS清除能力[30] 碧冬茄 PhERF2 上调ADH1-2 乙醇脱氢酶 提高植物对水淹抗性[31] 山葡萄 VaERF092 上调VaWRKY33 转录因子,调控胁迫相关基因 增强植物对低温的抗性[32] 小果野蕉 MaERF10 下调MaLOX7/8、MaAOC3和MaOPR4 参与茉莉酸合成 抑制茉莉酸信号,增强冷害[33] 小果野蕉 MaDEAR1 下调MaEXP1/3、MaPG1、MaXTH10、MaPL3和MaPME3 修饰细胞壁,与果实软化有关的基因 防止果实过早软化[34] 甜橙 CitERF13 上调CitPPH和CitNYC 参与叶绿素降解 加速果实褪绿[35] 甜橙和椪柑 CitERF6 上调CitPPH 参与叶绿素降解 加速果实褪绿[35] 桃子 PpeERF2 下调PpeNCED2/3和PpePG1 ABA合成与细胞壁降解 防止果实过快软化[36] 番木瓜 CpERF9 下调CpPME1/2和CpPG5 细胞壁降解 防止果实过早软化[37] 枇杷 EjERF39 Ej4CL1 木质素合成基因 促进果实木质化[38] 说明:苹果Malus domestica、芜菁Brassica rapa、月季Rosa chinensis、罂粟Papaver somniferum、碧冬茄Petunia×hybrida、山葡萄 Vitis amurensis、小果野蕉Musa acuminata、甜橙Citrus sinensis、椪柑Citrus reticulata、桃Amygdalus persica、番木瓜Carica papaya、枇杷Eriobotrya japonica Table 1. Function of ERF
-
ERF在植物受到胁迫时通过调控胁迫响应的相关基因以应对各种胁迫。生物胁迫中,ERF抵御真菌病原体最直接的方式是上调有关防卫基因,如拟南芥中ERF96上调PDF和PR这类病理相关基因[17];拟南芥中Ⅸb组ERF可以上调CYP81F2促进杀菌剂吲哚类硫苷合成[19]。还有通过促进程序性死亡以防止病原体的扩散,如番茄Solanum lycopersicum中ERF68可以上调有关细胞程序性死亡的基因促进细胞死亡[28]。非生物胁迫下ERF通过调控各种非生物胁迫抗性相关基因表达,提高植物抗逆性。碧冬茄中PhERF2在植株受水淹的情况下上调ADH1-2乙醇脱氢酶的表达,提高植株对水淹的抗性[31]。枇杷的EjERF39受低温诱导,上调木质素合成基因Ej4CL1,促进果实木质化,减轻冷害[38]。ERF可以通过调控转录因子来增强植物抗性,例如山葡萄中VaERF092受低温胁迫诱导并通过增强VaWRKY33表达,从而间接增强植株对低温的抗性[32]。ERF还可以通过调控渗透调节物质以增强植物的抗性,例如小豆Vigna angularis中VaERF3在盐碱胁迫中及水稻OsERF71在干旱胁迫中都可以诱导脯氨酸的积累,分别提高相应的胁迫抗性[39-40]。
ERF可能在逆境条件下负调控植物抗性,这会加剧植物所受的伤害。ERF可以通过直接抑制相关抗性基因的表达降低植物的抗性。拟南芥AtERF72会受缺铁环境诱导并抑制参与铁吸收的IRT1和HA2的表达,进一步抑制铁吸收[22]。白桦Betula platyphylla中BpERF11受盐胁迫和干旱胁迫诱导表达,抑制LEA和脱水蛋白基因的表达,并加剧干旱胁迫导致的伤害[41]。ERF还能通过抑制相关转录因子的表达以起到间接负调控。苹果中MdERF4抑制具有盐胁迫抗性的MdERF3表达,从而削弱了植物对盐胁迫的抗性[24]。还有ERF通过抑制植物激素或信号分子以负调控胁迫的响应基因。小果野蕉中MaERF10受低温胁迫诱导,与TIFY蛋白MaJAZ3互作通过抑制茉莉酸合成基因的表达以抑制茉莉酸信号途径,从而使果实表现出明显的冷害[33]。CaDRAT1在辣椒Capsicum annuum受到干旱胁迫时,通过抑制脱落酸(abscisic acid,ABA)合成基因的表达从而抑制了依靠ABA信号的干旱响应基因的表达[42]。
ERF负调控胁迫下的植物抗性或许是为维持其他系统的稳定。番茄SlERF84可以增强干旱和盐胁迫抗性,但会削弱生物胁迫抗性,原因可能是SlERF84通过增强活性氧(reactive oxygen species,ROS)清除能力以减缓细胞死亡,从而削弱植物对病原体的抗性[43]。苹果中MdERF4对MdERF3的调控可能是盐胁迫下维持乙烯在植物体内稳态的反馈调节机制[24]。番薯Ipomoea batatas中IbERF4抑制非生物胁迫响应基因表达,这或许是因为IbERF4主要参与调控开花和叶片衰老而导致的[44]。
-
果实成熟的标志包括褪绿和变软。ERF可以通过调控相关基因促进叶绿素的降解,比如甜橙和椪柑中CitERF6上调果实中参与叶绿素降解的CitPPH,从而促进果实褪绿[45];甜橙CitERF13可以直接上调CitPPH和CitNYC加速果实褪绿[35]。一般果实在成熟后会发生软化的现象,软化会影响果实的货架期寿命以及运输,ERF能通过调控相关基因调节软化的过程,比如桃子中PpeERF2通过抑制PpeNCED2、PpeNCED3(参与ABA合成)和PpePG1(参与细胞壁降解)的表达,以防止果实过早软化[36];同样还有番木瓜中CpERF9和小果野蕉MaDEAR1,都是通过抑制相关细胞壁降解的基因表达,防止果实过早软化[34, 37]。
-
花或叶的衰老和脱落是植物的生理现象,但衰老和脱落并非为一种过程。衰老背后的本质是细胞的程序式死亡。以拟南芥为例,拟南芥WRKY53被视为叶片衰老的核心转录因子,ESP/ESR负调控WRKY53,AtERF4和AtERF8可以直接抑制ESP/ESR的表达从而间接加速叶的衰老[46]。拟南芥中过氧化氢(H2O2)正向增强WRKY53的表达,ERF4的2种亚型直接调控CAT3的表达以控制H2O2的量从而间接调控衰老进程[13]。ERF还可以通过调节其他信号分子及其信号转导来调节花或叶的衰老。芜菁中BrERF72由茉莉酮酸甲酯诱导并上调茉莉酸合成基因表达从而通过茉莉酸信号途径加速叶片衰老[26]。KHASKHELI等[47]在月季花中筛选出可以上调细胞分裂素含量的基因RhERF113,在沉默其表达后,花加速衰老,而在补充外源细胞分裂素后可以使衰老速度恢复到正常水平。脱落是另一种过程,脱落器官在基部的离区细胞受乙烯等激素信号的诱导完成脱落。在乙烯信号传导过程中有一类名为EDF(ethylene response DNA-binding factors)的蛋白质位于EIN3的下游[48],EDF在植物开花期间促进花的衰老和脱落。一种名为FUF1的ERF可抑制EDF的表达从而达到延长花期的效果[49]。此外,月季的β-半乳糖苷酶基因RhBGLA1会加速花瓣的脱落,而RhERF1和RhERF4可以抑制此基因的表达,防止花瓣脱落[29]。
2.1. ERF在胁迫响应时的功能
2.2. ERF在果实成熟中的功能
2.3. ERF在花叶衰老与脱落中的功能
-
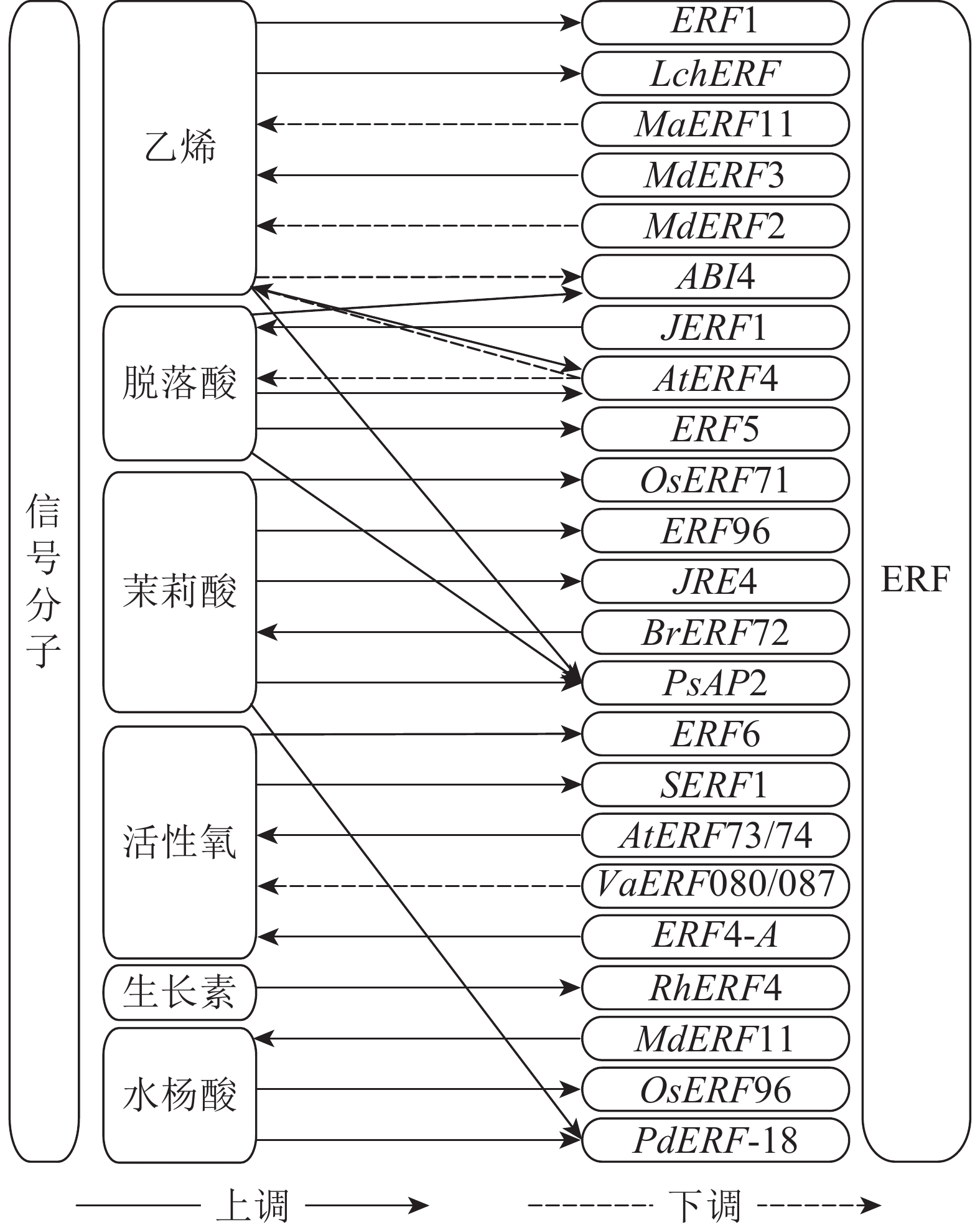
ERF受各类信号分子的诱导从而表达并行使相应的功能(图1),这些信号分子通过在植物体内的信号转导,逐级将体内和体外的环境信息传递,最终完成对ERF的表达调控。
作为ERF的命名来源,乙烯对ERF的调控关系最先被发现[1]。乙烯经过信号转导,诱导ERF在植物体内调控胁迫防御及生长发育等生理过程。以拟南芥为例,乙烯与位于内质网膜上的各类乙烯受体结合,使与受体关联的激酶CTR1失活,无法磷酸化EIN2的C端[50]。未被磷酸化的EIN2从而进入细胞核,使转录因子EIN3能稳定存在并发挥作用[51],随即EIN3激活包括ERF1等转录因子的转录,这些转录因子调控其他的乙烯响应基因以完成乙烯信号响应。除ERF1外还有许多受乙烯信号诱导的ERF,如枸杞Lycium chinense中的LchERF受乙烯诱导增强植株盐胁迫抗性[52],拟南芥中AtERF4受乙烯诱导降低了对乙烯的敏感性[53]。
ABA在植物对抗非生物胁迫时能通过诱导相关的ERF增强抗性。比如番茄ERF5基因的转录受外源ABA诱导并能增强植物对干旱和盐胁迫的抗性[54]。ABA与乙烯2种信号在部分情况下也存在拮抗作用,例如拟南芥中ABA信号能激活下游ERF基因ABI4,而乙烯信号通过EIN3能够抑制ABI4的表达,两者的拮抗作用影响抗坏血酸与ROS的含量[55]。ERF也可以反过来调控ABA信号传导。拟南芥AtERF4受ABA诱导并抑制ABA的信号传导,ABA响应基因明显下调[53]。
ERF和其他植物激素也有调控关系,这其中发现较多的是茉莉酸信号与ERF之间的调控,茉莉酸信号对ERF的调控主要表现在抗逆性和生长代谢等方面。例如在抵御胁迫时水稻OsERF71和拟南芥ERF96都可由茉莉酸信号诱导表达,提高植物对生物胁迫的抗性[17, 25]。在植物生长代谢方面,茉莉酸信号可由ERF进一步促进茉莉酸的信号传导,如芜菁BrERF72可由茉莉酮酸甲酯诱导并上调参与茉莉酸合成相关基因从而加速叶片衰老[26]。茉莉酸信号通过ERF还能调控其他代谢相关的产物生成,例如番茄JRE4受茉莉酸诱导上调,JRE4直接上调甾醇还原酶和配糖生物碱代谢相关基因促进糖苷生物碱的合成[27]。生长素信号诱导ERF参与了脱落有关的调控,月季花瓣离层RhERF4受生长素诱导且抑制下游RhBGLA1的转录,从而抑制了离层果胶的降解,延缓花瓣脱落[29]。水杨酸是重要的植物防御激素,它本身能诱导ERF的表达,例如水稻的OsERF96和山海关杨Populus deltoides‘Shanhaiguan’的PdERF-18[56-57]。
ROS在ERF接收胁迫信号过程中具有重要作用。胁迫初期位于细胞膜上的蛋白质感应到胁迫信息,需要次级信使将胁迫信息传至内部。ROS作为次级信使之一能够将细胞内的信号快速传递至细胞核,激活一系列蛋白激酶(protein kinases,PKs)或蛋白磷酸酶(protein phosphatases,PPs),而PKs和PPs可以触发对转录因子磷酸化或脱磷酸化级联反应[58-59]。例如拟南芥中ROS会触发MPK6,随后MPK6磷酸化ERF6,最后ERF6调控下游相关ROS响应基因[60]。除此之外,还有水稻中受ROS诱导的SERF1,通过MAPK5磷酸化后激活,提高水稻的盐胁迫抗性[61]。
-
在ERF基因转录后,目前有选择性剪接和miRNA为主的2种调控方式。选择性剪接在参与非生物胁迫的DREB中较为常见,如水稻的OsDREB2A/2B[62]和玉米Zea mays的ZmDREB2A[63],只有在胁迫下才会产生具有功能的有效转录本。选择性剪接还可导致功能转变,例如拟南芥中,ERF4-A和ERF4-R的存在比率由RNA结合蛋白FPA控制,当叶片开始衰老时,ERF4-R的存在比率比未衰老时高,从而抑制抗氧化酶的表达以促进叶片的衰老[13]。miRNA作为一类非编码RNA,主要通过降解mRNA或是沉默目标基因的翻译以抑制目标基因的表达。如miRNA172通过抑制AP2/ERF蛋白的翻译从而对调控生殖器官的发育以及抑制干细胞增殖起到了关键的作用[64]。选择性剪接和miRNA都是针对mRNA的修饰方式,选择性剪接使ERF可以产生多种转录本,丰富了ERF功能的多样性。miRNA对mRNA进行直接修饰可以特异性负调控ERF,明晰miRNA对ERF的表达调控机制可以促进miRNA技术在ERF功能上的应用。
有一些调控机制可以降解或激活已翻译成蛋白质的ERF。泛素化是降解ERF的调控方式之一,例如拟南芥中,响应干旱胁迫的AtERF53在非胁迫情况下会被带有RING结构域的E3泛素连接酶RGLG2泛素化,使其被26s蛋白酶分解[65]。Ⅶ组的ERF被独特的N端规则所调节,这些ERF的N端在正常生长环境下会被植物半胱氨酸氧化酶Pco1/2修饰,随后被N端识别蛋白ATE1/2和PRT6识别并降解。其中半胱氨酸受氧气(O2)与一氧化氮(NO)氧化,而非生物胁迫能抑制NO的积累。在植物处于缺氧状态或是胁迫的情况下,Ⅶ组的ERF才得以稳定存在,从而激活胁迫相关的应答基因[66-67]。不同于前两者,磷酸化是一种激活ERF的机制,对ERF是重要的调控途径。不同激酶磷酸化通常会导致ERF的功能发生变化,包括活性的变化和细胞器的定位等。WANG等[60]研究发现:拟南芥ERF6上的磷酸化位点可以被MPK6特异性识别并进行磷酸化,磷酸化后的ERF6转录因子对其下游基因具有更强的激活活性。尽管野大豆Glycine soja的GsERF7本身含有核定位信号基序,但只有被GsSnRK1磷酸化之后才会从细胞质被重新定位至细胞核,从而发挥转录调控功能[68]。
-
ERF有些功能是通过调控植物中的信号分子完成的,比如ERF通过调控植物激素合成途径中的关键酶基因以控制植物激素信号通路。ERF通过调控乙烯合成关键基因的表达,从而调节乙烯的生物合成和信号传导,例如苹果果实中的MdERF3受乙烯诱导且能上调MdACS1的表达,从而加速乙烯的合成[23]。ERF还可以抑制乙烯合成关键酶基因的表达,以控制乙烯的生成,例如HAN等[69]在小果野蕉果实中发现MaERF11能够有效抑制MaACO1的转录从而抑制乙烯的合成。除此之外,ERF也可以参与果实成熟过程中乙烯合成模式的转换。例如苹果果实成熟初期,MdERF2通过抑制酶活性较高的MdACS1的表达,并上调酶活性较低MdACS3a的表达,来限制乙烯在果实中的积累以放慢苹果的成熟速度[23, 70]。
ERF也可以通过调控ABA合成相关基因从源头调控ABA的信号,例如烟草中JERF1直接上调ABA合成相关基因NtSDR以增加ABA的含量,增强烟草对盐胁迫和低温胁迫的抗性[71]。ERF还可抑制ABA信号,例如拟南芥AtERF4过表达株系的种子经ABA处理后,相比于野生型种子,ABA信号转导下游基因表达量明显下降[53],这说明AtERF4是通过抑制ABA的信号传导调控了ABA的信号。桃子PpeERF2可以直接抑制ABA合成相关基因PpeNCED2、PpeNCED3的表达以抑制ABA的合成[72]。其他植物激素如茉莉酸和水杨酸等也受到ERF的调控,例如芜菁BrERF72上调参与茉莉酸合成相关基因从而加速叶片衰老[26],苹果MdERF11通过促进水杨酸的合成以增强植株对生物胁迫的抗性[73]。
ERF可以通过调控呼吸暴发氧化酶同系物(respiratory burst oxidase homolog,Rboh)家族基因,加速ROS生成,从而促进胁迫信号传递,完成早期胁迫响应。比如拟南芥中的AtERF73/74在胁迫初期能调控Rboh家族基因,Rboh催化H2O2生成,H2O2将胁迫信号继续传递[18, 74]。信号传递后过量的ROS会对植物产生氧化伤害,ERF通过加强ROS清除系统以清除过量的ROS。例如SUN等[75]将山葡萄中受低温胁迫诱导的VaERF080/087转入拟南芥,转基因植株较野生型在低温胁迫下活性氧清除酶具有更高活性,也表现出更好的低温抗性。拟南芥ERF4-A可以上调CAT3的表达从而减缓细胞死亡[13]。植物激素可以通过ERF调节抗氧化系统以控制ROS,例如罂粟中PsAP2可由乙烯、茉莉酸和ABA诱导并通过上调AOX1a增强植株的抗氧化能力[30]。
-
ERF与蛋白质协作互作可以介入到多种反应。通过与相应的蛋白质协作能大幅提高转录激活能力。比如GmERF5与GmbHLH、GmEIF互作,可提高大豆Glycine max对病原体的抗性[76],OsERF3和WOX11互作调控细胞分裂素响应基因RR2以调节水稻的根冠发育[77],EjERF39和EjMYB8互作调控木质素合成基因以增强枇杷果实低温抗性[38]。具有转录抑制功能的ERF通过招募辅抑制因子以起到加强抑制的作用,例如在小果野蕉果实中,MaERF11可以抑制果实成熟相关基因的表达并可招募组蛋白去乙酰化酶MaHDA1增强抑制效果[69]。ERF通过与其他转录因子之间发生互作以竞争与目标启动子结合的机会,例如苹果MdERF2通过其N端与MdERF3的DNA结合域结合,从而使MdERF3无法与目标基因启动子结合[23]。
3.1. 植物对ERF的调控机制
3.1.1. 信号分子对ERF的调控
3.1.2. 其他因子对ERF的调控
3.2. ERF对下游基因的调控机制
3.2.1. ERF对信号分子的调控
3.2.2. 蛋白质互作
-
ERF是植物特有的转录因子,其AP2结构域作为DNA结合域受到大量关注,但在AP2结构域外尚有很多基序是未知的,且这些基序能否应用在基因工程中也需要进一步研究。ERF的功能受到很多农林领域研究者的关注,但对于ERF这个大家族来说,得到明确功能注释的仅有少数,且主要出自少数模式作物,后期研究应对其他植物中ERF进行大量的功能验证。园艺领域研究者对ERF调控果实成熟和花叶的衰老与脱落的关注较多,未来可以对园艺植物在切花或果实保鲜和花果期调控等课题深入研究。ERF既受信号分子调控,又调控信号分子的生成和传导。目前有大量关于ERF与各类信号分子之间调控的研究,但大多数并未涉及到信号交叉或完整的调控网络,或许未来研究者可以关注在信号调控网络中ERF所发挥的节点作用。植物中一些因子对ERF调控使ERF适宜地发挥作用,这些调控机制有助于理解ERF发挥功能的过程,但很少得到具体应用。目前来看,应用miRNA具有一定的可行性,或许未来应更深入miRNA调控ERF的研究。ERF与蛋白质相互作用类型广泛,具有结构域的特异性,但这方面的机理研究还有很多空缺,未来有关ERF-蛋白质特异性结合的机理应该得到深入研究。










 DownLoad:
DownLoad: