-
球孢白僵菌Beauveria bassiana是一种可寄生在多种昆虫上具较强致病性的真菌,已被广泛用作防治森林、农作物虫害的生物农药[1-2];它产生大量如非核糖体多肽、聚酮类等次生代谢产物,可抑制多种腐生或寄生线虫[3-4]。聚酮、非核糖体多肽及其杂合化合物如聚酮中的红霉素[5]、四环素[6];非核糖体多肽中的青霉素、头孢霉素[7-8];聚酮和非核糖体多肽的杂合化合物如他克莫司[9]、雷帕霉素[10]、博来霉素[11]、埃博霉素[12]等具有免疫抑制剂或抗肿瘤等生理活性[13]。迄今为止,球孢白僵菌中的聚酮和非核糖体多肽杂合型化合物仅有卵孢白僵菌素经基因敲除和异源表达鉴定的报道[3]。聚酮类和非核糖体肽是2类典型的小分子天然次生代谢化合物,分别由聚酮合酶(polyketide synthases, PKS)、非核糖体肽合成酶(nonribosomal peptide synthetases, NRPS)催化简单单体脂肪酸或氨基酸缩合形成[13-14]。真菌聚酮合酶主要属于Ⅰ型PKS[13, 15],其结构域通常包括酮体合成酶(ketosynthase, KS),酰基转移酶(acyltransferase, AT),脱水酶(dehydratase, DH),甲基转移酶(methyltransferase, MT),烯酯酰还原酶(enoylreductase, ER),酮体还原酶(ketoreductase, KR),酰基载体蛋白(acyl carier pretein, ACP)和末端释放酶等[13],短链醇脱氢酶(short chain dehydrogenase/reductase,SDR)家族在PKS中属末端释放酶[16],PKS中的SDR结构域对底物选择具一定的特异性[17];NRPS是一种由多模块组成的多酶复合体,由腺苷酰化结构域(adenylation domain, A),肽酰载体蛋白结构域(phosphopantetheine attachment site, PP)和缩合结构域(condensation domain, C)等3个核心结构域按照特定的时空顺序排列组成[18]。真菌PKS/NRPS酶蛋白复合物含有N端的PKS组件和C端的NRPS组件[19],可催化生成含有酰基和氨酰基构件的聚酮和氨基酸或肽类型化合物的生物合成[9, 20];同时聚酮和氨基酸或肽类化合物能够通过它们化学属性的混合,扩展其产物的生物活性,埃博霉素(epothilone)、雷帕霉素(rapamycin)和博来霉素(bleomycin)是这类混合分子中重要代表,编码这些化合物的PKS/NRPS基因大小均约10 kb[21]。随着高通量测序技术的发展,真菌基因组数据不断增加,基因组数据显示目前已通过结构鉴定、表型筛选和生物活性测定等传统方法分离的天然次生代谢产物仅为冰山一角,真菌基因组中尚有大量的天然次生代谢产物生物合成基因存在;该类基因大多数条件下处于沉默状态,其参与合成的化合物大多尚未被鉴定[22];应用基因组挖掘技术可将这些沉默的生物合成基因分离出来,再通过改变培养基配方和条件或进行遗传学修饰等方式诱导以激发它们全部的次生代谢潜力[23-24]。为了发掘球孢白僵菌中聚酮和非核糖体多肽杂合类化合物的生物合成基因,本研究采用基因组挖掘的方式从其基因组数据中获得1条PKS/NRPS基因(命名Bbpks2),通过生物信息学分析预测其潜在功能,尝试寻找该基因的表达条件,为最终确定其天然产物及对其异源表达奠定基础。
-
昆虫致病真菌球孢白僵菌(菌株号:YH02)来源于云南云百草生物技术有限公司,将YH02菌株接种在MY固体培养基(malt/yeast extract medium)上,置于25 ℃恒温条件下培养,每隔2周转接1次;用于基因组DNA和总RNA提取的真菌接种于液体MY培养基中,置于120 r·min-1和25 ℃条件下培养5~7 d后,收获约0.5 g菌丝体,用吸水纸彻底吸干后在液氮环境下用研钵研磨成粉末用于基因组DNA和总RNA的提取。MY培养基的具体配方麦芽糖6.0 g·L-1,酵母提取物4.0 g·L-1,固体培养基配置时加20.0 g·L-1琼脂。
-
试验涉及试剂包括:真菌基因组DNA提取试剂盒(北京康为世纪生物科技有限公司),高保真聚合酶、真菌总RNA提取试剂盒(大连宝生物工程公司),反转录试剂盒(赛默飞世尔科技有限公司),无缝克隆试剂盒(Biomiga)。主要使用仪器为NanoDropTM 2000紫外分光光度计(赛默飞世尔科技有限公司)、Azure C300凝胶成像系统(Azure Biosystems)、隔水式恒温培养箱(上海博迅实业有限公司医疗设备厂)、多功能组合摇床(江苏太仓实验设备厂)。
-
首先以曲霉中已报道过的PKS基因为模板,利用本地BLAST(basic local alignment search tool)对球孢白僵菌基因组进行扫描,获得可能含有PKS基因的重叠群(contigs),然后利用antiSMASH(https://antismash.secondarymetabolites.org/)软件进行验证,并利用GENEFISH对重叠群进行开放阅读框扫描,通过生物信息学分析获得白僵菌基因组中Bbpks2基因的DNA序列。
根据DNA序列设计2对特异引物(Bbpks25F:5′-ATGTCTTCGCACCAAAACGA-3′;Bbpks2MR:5′-AACAGCACTGTCACTGTCCAC-3′;Bbpks2MF:5′-AACGCTGGCTATAATGCTCTT-3′;Bbpks23R:5′-GGCTTCCTGGAGCGGTAA-3′)用于Bbpks2基因全长cDNA的克隆。以球孢白僵菌的cDNA为模板,用HiFi高保真酶分别扩增出Bbpks2基因的5′片段和3′片段,经DNA纯化试剂盒将这2个片段纯化后,按照无缝克隆试剂盒说明书中的具体步骤将Bbpks2基因的5′片段和3′片段连接起来,获取全长cDNA。将该基因片段连接到克隆载体Peasy-T3上,转化到Trans-T1感受态细胞中,进行蓝白斑筛选后,通过菌落PCR筛选含有插入片段的阳性克隆送到上海生工进行测序。
-
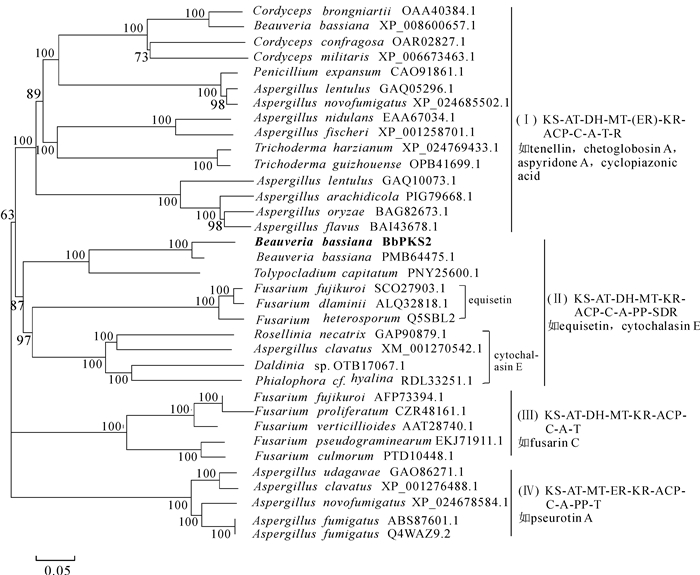
从美国生物技术信息中心(NCBI)上选择功能已知且鉴定过化合物的PKS/NRPS蛋白序列,包括黄曲霉Aspergillus flavus,米曲霉A. oryzae中编码环匹阿尼酸(cyclopiazonic acid)生物合成的CpaA基因(BAI43678,BAG82673),扩展青霉Penicillium expansum中编码球毛壳甲素(chetoglobosin A)生物合成的CheA基因(CAO91861),棒曲霉A. clavatus中编码细胞松弛素E(cytochalasin E)生物合成的CcsA基因XM_001270542,异孢镰刀菌Fusarium heterosporum中编码伊快霉素(equisetin)的EqiS基因(Q5SBL2),水稻恶苗病菌Fusarium fujikuroi,串珠赤霉菌Gibberella moniliformis及假禾谷镰孢菌Fusarium pseudograminearum等真菌中编码镰刀菌素C(fusarin C)生物合成的FusA(AFP73394),FusS(AAT28740),pks10(EKJ71911)等基因,烟曲霉Aspergillus fumigatus中编码假散囊菌素A(pseurotin A)的PsoA基因(ABS87601),球孢白僵菌中编码卵孢白僵菌素(tenellin)的TenS基因(XP_008600657)等蛋白序列用于聚类分析,采用MEGA 7.0中的Clustal W程序对所选择的PKS蛋白序列和球孢白僵菌中的BbPKS1蛋白序列进行比对后,采用邻位相接法(neighbor-joining),自检举1 000次,其余采用默认参数构建系统进化树。依据聚类结果,从基因起源与功能分化的角度预测该基因功能,并进一步推测该基因产生的聚酮化合物。
-
根据分离得到的Bbpks2基因的DNA序列,设计特异检测引物(TBbpks2F:TCTCCTTGGCAAGGTTACTG;TBbpks2R:ACCACGCTCCATCATGTACT)。然后将球孢白僵菌培养在添加不同碳源、氮源的培养基上。不同碳源实验的基础培养基为6.0 g·L-1麦芽浸粉,3.0 g·L-1酵母提取物,分别添加4.0 g·L-1乳糖、甘露醇、麦芽糖、葡萄糖、山梨醇、果糖、肌醇;不同氮源实验的基础培养基为1.8 g·L-1麦芽糖,6.0 g·L-1葡萄糖,分别添加4.0 g·L-1牛肉浸粉、酪蛋白胨、胰蛋白胨、马铃薯。培养14 d后,从每种培养基上收获0.5 g菌丝体,采用真菌总RNA提取试剂盒按照说明书步骤提取各自的总RNA,并选择浓度合适的RNA采用反转录试剂盒将其反转录成cDNA,再用特异引物TBbpks2F和TBbpks2R检测该基因的表达情况。
在琼脂糖凝胶电泳检测过程中,各PCR产物和1 kb DNA marker的点样量均为2.5 μL,电泳结束后,利用Azure C300凝胶成像系统自带的图像分析软件分析各条带的积分光密度,比较各PCR产物条带的积分光密度值,并以该值与1 kb marker的条带进行比较作为相对表达量绘制柱状图。
-
经无缝克隆获得Bbpks2基因的全长cDNA。分析显示:Bbpks2基因全长12 051 bp,编码4 016个氨基酸;经过序列相似性及保守结构域的分析显示,该基因编码蛋白的结构域组织顺序为KS-AT-DH-MT-KR-ACP-C-A-PP-SDR;该蛋白同时含有PKS和NRPS酶蛋白的结构域,由此推断该蛋白是一种PKS/NRPS酶蛋白复合体。
-
球孢白僵菌中BbPKS2蛋白与其他真菌PKS/NRPS蛋白的分子系统进化树显示(图 1),该聚类树可分为4个较大的亚类(分支Ⅰ,Ⅱ,Ⅲ,Ⅳ),各个亚类间的结构域组织顺序存在差异,至少存在1个结构域的不同;镰刀菌属Fusarium真菌中编码伊快霉素、不同真菌中参与编码细胞松弛素E的酶蛋白聚到同一分支上,且二者亲缘关系明显较近。球孢白僵菌的BbPKS2蛋白与这2类酶蛋白被归类到分支Ⅱ中,该亚类的结构域组织顺序为KS-AT-DH-MT-KR-ACP-C-A-PP-SDR;BbPKS2与球孢白僵菌JEF007菌株(PMB64475.1)、头状虫草Tolypocladium capitatum(PNY25600.1)聚在一个亚分支中;推测BbPKS2蛋白所参与合成的天然次生代谢产物与伊快霉素、细胞松弛素E存在一定的结构相似性。
-
在含有不同碳氮源添加物的培养基上,Bbpks2基因表达差异极其显著。当添加不同碳源添加物时(图 2),在乳糖培养基上,Bbpks2基因的表达量最高,其表达量显著高于其他培养基;而在麦芽糖培养基上,Bbpks2基因不表达;Bbpks2基因表达量从高到低依次为乳糖,葡萄糖,山梨醇,肌醇,甘露糖,果糖,其中以乳糖为碳源添加物的培养基上其表达量最低至少为其他添加物的3.4倍以上。当添加不同氮源添加物时(图 3),Bbpks2基因均有明显表达,在含牛肉浸粉培养基上,Bbpks2基因的表达量最高,其表达量最低为其他的1.3倍以上;而在含酪蛋白胨的培养基上,该基因表达量最低。可见该基因受碳源中乳糖、氮源中牛肉浸粉的影响较大;而受到碳源中麦芽糖、氮源中酪蛋白胨的影响则可以忽略。
-
球孢白僵菌主要是在机械压力和酶共同作用下侵染昆虫寄主[25],在昆虫体内通过次生代谢途径中相关酶类所产生的毒素致死寄主[1]。但该真菌次生代谢途径中所产生的毒素除少数外,尚有大量的次生代谢产物生物合成基因未与相关化合物关联。结合基因组挖掘技术和一菌多产物策略(one strain many compounds,OSMAC)、异源表达等操作技术可促进该问题的解决,从真菌基因组数据中可大量挖掘得到PKS,NRPS等生物合成基因,通过序列相似性、结构域、基因片段的大小及聚类分析等初步推断该基因的功能,初步了解该酶所参与合成化合物的某些结构[15, 26]。如编码2-吡啶酮卵孢白僵菌素(2-pyridone tenellin)酶蛋白的PKS/NRPS基因约12.9 kb[27],编码镰刀菌素C酶蛋白的fusA基因(PKS/NRPS)约11.9 kb[28];BALI等[17]在大肠埃希菌Escherichia coli中过量表达其SDR等末端释放结构域,发现该结构域对结构中含有环己酮的底物具较高活性。同时因本研究中的Bbpks2基因尚未发现有较高同源性基因存在,通过结构域分析推测该基因所编码的酶蛋白属于PKS/NRPS,参与聚酮/非核糖体多肽的生物合成,且该化合物与伊快霉素、细胞松弛素E存在某些共同的结构;通过比较分析伊快霉素、细胞松弛素E,综合分析该化合物可能含吡咯烷酮结构。
基因挖掘技术为迅速发现隐藏在基因组中的次生代谢产物生物合成基因和新生物合成机制提供了一种强力有效的方法。挖掘获得聚酮、非核糖体多肽等天然次生代谢产物的生物合成基因后,通过系统变换培养条件,如通过改变培养基配方、培养方式、渗透压及一菌多产物(OSMAC)等方式可选择性地上调转录因子,诱导激发它们全部的产生次生代谢产物的潜力,将基因组中沉默的PKS和NRPS等基因诱导表达出来[22-23]。本研究通过分析Bbpks2基因在含不同碳氮源添加物培养基上的具体表达情况,发现该基因在以乳糖为碳源、以牛肉浸粉为氮源的培养基上能够获得较大量的表达;在含有不同培养基成分的培养基上,该基因的表达情况差异显著;在以麦芽糖为碳源的培养基上,该基因不表达,在以酪蛋白胨为氮源的培养基上,该基因表达量显著低于在以牛肉浸粉、马铃薯浸粉和胰蛋白胨等作为氮源的培养基的表达量。这种同一基因在含不同成分培养基上出现表达差异的现象存在于许多真菌中,如长松萝Usnea longissima中UsNRPS基因可被蔗糖和果糖强烈诱导,而被天冬氨酸抑制[29];通过OSMAC策略从邬氏黄丝曲霉Talaromyces wortmannii中获得3种新型的聚酮[23];从三线镰刀菌Fusarium tricinctum在不同培养基上培养筛选获得了2个新型的聚酮:fusarielin K和fusarielin L[30]。这种基因在不同培养条件下的表达研究有助于找出不同次生代谢基因的最适表达培养基类型,对通过真菌菌丝体获得新颖的次生代谢化合物,及将相应化合物和具体基因关联起来具有指导意义。
本研究通过基因组挖掘技术从球孢白僵菌基因组中发掘得到了一个PKS/NRPS基因Bbpks2,克隆得到其全长cDNA,用聚类分析的方式对其终产物结构进行预测,并通过添加不同碳源和氮源筛选能够诱导该基因表达的最佳培养基条件,为球孢白僵菌中PKS/NRPS基因的异源表达及基因功能鉴定,发掘新颖或具潜在生物活性的聚酮和非核糖体多肽杂合化合物奠定了前期基础。
Gene cloning of a PKS/NRPS gene with an SDR domain
-
摘要: 聚酮和非核糖体多肽的复合化合物具有独特的生理活性,它们由聚酮合酶/非核糖体肽合成酶(PKS/NRPS)催化合成。目前球孢白僵菌Beauveria bassiana中含SDR结构域的PKS/NRPS酶的生物合成机制尚不清楚,采用基因挖掘技术从球孢白僵菌基因组中分离得到1个PKS/NRPS基因(命名Bbpks2),利用生物信息学分析对其功能进行预测并检测该基因在以6.0 g·L-1麦芽提取物和3.0 g·L-1酵母提取物为基本氮源培养基,7种碳源添加物和以1.8 g·L-1麦芽糖和6.0 g·L-1葡萄糖为基本碳源培养基,4种氮源添加物培养基上的具体表达情况,其中每种添加物含量为4.0 g·L-1。结果显示:Bbpks2基因长度为12 051 bp,编码4 016个氨基酸;其结构域组织顺序为KS-AT-DH-MT-KR-ACP-C-A-PP-SDR,是一种含有SDR结构域的PKS/NRPS;系统进化分析发现,BbPKS2与球孢白僵菌JEF007菌株(PMB64475.1)、头状虫草Tolypocladium capitatum(PNY25600.1)等的PKS/NRPS蛋白聚在同个分支中,可能参与一种聚酮/非核糖体多肽类化合物的生物合成;比较不同氮源、碳源添加物对Bbpks2基因表达的影响,发现该基因在添加了乳糖的培养基上的表达量是其他碳源添加物的3.4倍以上,添加了牛肉浸粉的培养基上表达量是其他氮源添加物的1.3倍以上。该研究为下一步通过异源表达鉴定Bbpks2基因的具体功能,及其调控机理研究和基因资源利用奠定基础。
-
关键词:
- 森林保护学 /
- 球孢白僵菌 /
- 杂合PKS/NRPS /
- 生物信息学分析
Abstract: The compound of a hybrid polyketide/nonribosomal peptide, which usually has some particular bioactive activity, is catalyzed by an enzyme of hybrid PKS/NRPS. A hybrid PKS/NRPS containing a SDR domain from Beauveria bassiana has not yet clearly elucidated. To inveastigate this type of gene, the present study obtained a hybrid PKS/NRPS gene (named Bbpks2) from B. bassiana using genome-mining, predicted its gene function with a bioinformatic analysis, and detected the expression level of Bbpks2 in the medium by adding 7 carbon additives(adding 4.0 g·L-1 of every carbon additives) using 6.0 g·L-1 malt extract and 3.0 g·L-1 yeast extract as basic medium, or 4 nitrogen additives(adding 4.0 g·L-1 of every carbon additives) using 1.8 g·L-1 maltose and 6.0 g·L-1 as basic medium, 3 replications every treatment. Results showed that the Bbpks2 gene had 12 051 bp, encoded 4 016 amino acids, and had a sequential domain organization of KS-AT-DH-MT-KR-ACP-C-A-PP-SDR. The proteins of BbPKS2 could belong to a hybrid PKS/NRPS, which was comprised of a short-chain dehydrogenase reductase (SDR) domain. BbPKS2, PMB64475.1 from the strain of B. bassiana JEF007, and PNY25600.1 from Tolypocladium capitatum were three hybrid PKS/NRPS proteins that clustered in the same clade meaning that BbPKS2 could take part in the biosynthesis of a compound belonging to the hybrid polyketides/nonribosomal peptides. Comparing the expression level of the Bbpks2 gene when appending different C or N additives, the expression level of the Bbpks2 gene with lactose added to the basic medium was found to be at least 3.4 times higher than the other C additives, and beef extract 1.3 times higher than the other N additives. The present study could provide some help in further identifying the specific function of the Bbpks2 gene through heterologous expression and could provide a basis for explaining the molecular regulation mechanism when utilizing the gene resource of B. bassiana.-
Key words:
- forest protection /
- Beauveria bassiana /
- hybrid PKS/NRPS /
- bioinformatics analysis
-
-
[1] 宋晓兵, 彭埃天, 程保平, 等.一株侵染柑橘木虱的球孢白僵菌的分离及鉴定[J].植物保护, 2017, 43(4):139-144. SONG Xiaobing, PENG Aitian, CHENG Baoping, et al. Isolation and identification of a Beauveria bassiana strain infecting Diaphorina citri[J]. Plant Prot, 2017, 43(4):139-144. [2] 王月, 文斌, 马军, 等.球孢白僵菌(Beauveria bassiana)对寒地温室4种害虫防控研究[J].生物灾害科学, 2017, 40(4):284-287. WANG Yue, WEN Bin, MA Jun, et al. Control effects of Beauveria bassiana on 4 conservatory pests in frigid regions[J]. Biol Disaster Sci, 2017, 40(4):284-287. [3] XIAO Guohua, YING Shenghua, ZHENG Peng, et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana[J]. Scic Rep, 2012, 2:483. doi:10.1038/srep00483. [4] 刘丹丹, 段玉玺, 陈立杰. Snf 907-76发酵液杀线虫活性测定及活性物质分离纯化[J].农药, 2007, 46(9):644-646. LIU Dandan, DUAN Yuxi, CHEN Lijie. Determine nematicidal of ferment liquid of Snf 907-76 and isolation and purification nematocide[J]. Agrochemicals, 2007, 46(9):644-646. [5] DONADIO S, STAVER M J, MCALPINE J B, et al. Modular organization of genes required for complex polyketide biosynthesis[J]. Science, 1991, 252(5006):675-679. [6] KIM E S, BIBB M J, BUTLER M J, et al. Sequences of the oxytetracycline polyketide synthase-encoding otc genes from Streptomyces rimosus[J]. Gene, 1994, 141(1):141-142. [7] AHARONOWITZ Y, COHEN G, MARTIN J F. Penicillin and cephalosporin biosynthetic genes:structure, organization, regulation, and evolution[J]. Ann Rev Microbiol, 1992, 46:461-495. [8] MARTIN F J. New aspects of genes and enzymes for beta-lactam antibiotic biosynthesis[J]. Appl Microbiol Biotechnol, 1998, 50(1):1-15. [9] MOTAMEDI H, SHAFIEE A. The biosynthetic gene cluster for the macrolactone ring of the immunosuppressant FK506[J]. Eur J Biochem, 2010, 256(3):528-534. [10] SCHWECKE T, APARICIO J F, MOLNAR I, et al. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin[J]. Proc Natl Acad Sci USA, 1995, 92(17):7839-7843. [11] DU Liangcheng, SÁNCHEZ C, CHEN Mei, et al. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase[J]. Chem Biol, 2000, 7(8):623-642. [12] TANG Li, SHAH S, CHUNG L, et al. Cloning and heterologous expression of the epothilone gene cluster[J]. Science, 2000, 287(5453):640-642. [13] COX R J. Polyketides, proteins and genes in fungi:programmed nano-machines begin to reveal their secrets[J]. Org Biomol Chem, 2007, 5(13):2010-2026. [14] SESHIME Y, JUVVADI P R, KITAMOTO K, et al. Aspergillus oryzae type Ⅲ polyketide synthase CsyA is involved in the biosynthesis of 3, 5-dihydroxybenzoic acid[J]. Bioorg Med Chem Lett, 2010, 20(16):4785-4788. [15] KROKEN S, GLASS N L, TAYLOR J W, et al. Phylogenomic analysis of type Ⅰ polyketide synthase genes in pathogenic and saprobic ascomycetes[J]. Proc Natl Acad Sci USA, 2003, 100(26):15670-15675. [16] SISKOS A P, BAERGA-ORTIZ A, BALI S, et al. Molecular basis of Celmer's rules:stereochemistry of catalysis by isolated ketoreductase domains from modular polyketide synthases[J]. Chem Biol, 2005, 12(10):1145-1153. [17] BALI S, O'HARE H M, WEISSMAN K J. Broad substrate specificity of ketoreductases derived from modular polyketide synthases[J]. Org Biomol Chem, 2006, 7(3):478-484. [18] CHALLIS G L, RAVEL J, TOWNSEND C A. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains[J]. Chem Biol, 2000, 7(3):211-224. [19] 黎志凤, NGUIMBI E, 李越中, 等.埃博霉素(Epothilones)的PKS/NRPS杂合基因簇[J].生物工程学报, 2003, 19(5):511-515. LI Zhifeng, NGUIMBI E, LI Yuezhogn, et al. The PKS/NRPS hetero-gene cluster of epothilones[J]. Chin J Biotechnol, 2003, 19(5):511-515. [20] BOETTGER D, HERTWECK C. Molecular diversity sculpted by fungal PKS-NRPS hybrids[J]. Chem Biol Chem, 2013, 14(1):28-42. [21] MIYANAGA A, KUDO F, EGUCHI T. Protein-protein interactions in polyketide synthase-nonribosomal peptide synthetase hybrid assembly lines[J]. Nat Prod Rep, 2018, 14(1):28-42. [22] BOK J W, HOFFMEISTER D, MAGGIOHALL L A, et al. Genomic mining for Aspergillus natural products[J]. Chem Biol, 2006, 13(1):31-37. [23] LIU Wencai, YANG Fan, ZHANG Ran, et al. Production of polyketides with anthelmintic activity by the fungus Talaromyces wortmannii using one strain-many compounds (OSMAC) method[J]. Phytochem Lett, 2016, 18:157-161. [24] YUAN Chao, GUO Yuhua, WANG Haiying, et al. Allelopathic polyketides from an endolichenic fungus Myxotrichum sp. by using OSMAC strategy[J]. Sci Rep, 2016, 6:19350. [25] 季香云, 杨长举.白僵菌的致病性与应用[J].中国生物防治学报, 2003, 19(2):82-85. JI Xiangyun, YANG Changju. Infection and application of Beauveria spp.[J]. Chin J Biol Control, 2003, 19(2):82-85. [26] WEBER T, BLIN K, DUDDELA S, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters[J]. Nucleic Acids Res, 2015, 43(W1):W237-W243. [27] ELEY K L, HALO L M, SONG Z, et al. Biosynthesis of the 2-pyridone tenellin in the insect pathogenic fungus Beauveria bassiana[J]. Chem Biol Chem, 2007, 8(3):289-297. [28] SONG Z, COX R J, LAZARUS C M, et al. Fusarin C biosynthesis in Fusarium moniliforme and Fusarium venenatum[J]. Chem Biol Chem, 2004, 5(9):1196-1203. [29] 原晓龙, 陈剑, 张传光, 等.长松萝中非核糖体肽合成酶NRPS基因克隆与鉴定[J].西部林业科学, 2016, 45(1):14-20. YUAN Xiaolong, CHEN Jian, ZHANG Chuanguang, et al. The genetic clone and identification of non-ribosomal peptides synthetases (NRPS) from Usnea longissima[J]. J West China For Sci, 2016, 45(1):14-20. [30] HEMPHILL C F, SUREECHATCHAIYAN P, KASSACK M U, et al. OSMAC approach leads to new fusarielin metabolites from Fusarium tricinctum[J]. J Antibiot, 2017, 70(6):726-732. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.2019.06.024







 下载:
下载: