-
香榧Torreya grandis ‘Merrillii’是红豆杉科Taxaceae榧树Torreya grandis的优良变异类型,经人工选育后嫁接繁殖栽培的优良品种,也是目前榧树中唯一进行人工规模栽培的品种[1-2]。它集果用、油用、药用、材用和绿化观赏为一体,经济价值非常高[3],老产区在浙江会稽山区的诸暨、柯桥、嵊州、东阳和磐安等5县(市、区),分布范围狭窄,产量极其有限,产品供不应求。全国10余个省(市、自治区)均在引种栽植[4]。近年来,随着香榧产业的发展,香榧绿藻病害日趋严重,给榧农造成重大经济损失,尤其是人工林中,其对香榧叶片的光合作用产生极大影响并造成落花落果,甚至导致香榧树死亡,逐渐引起了榧农的重视。吾中良等[4]首次报道了香榧绿藻病害。香榧绿藻Chlorella sp.为小球藻属Chlorella的变异类型,是附着在榧树枝和叶上形成表面粗糙的灰绿色苔状物[5],因病原归属于绿藻门Chlorophyta,且在香榧枝叶上分布较多,生产上一般称之为香榧绿藻,但也有研究者认为其是青苔(moss)和地衣(lichens)[6]。专家学者对其防治方法进行了研究[5, 7],但研究仍处于起步阶段,其危害机理尚不明确,病原种类也尚无定论。本研究结合传统形态学方法和分子生物学技术,通过对香榧绿藻的培养观察及18S rDNA和内转录间隔区(ITS)基因序列分析,探索了香榧绿藻的生活习性,初步鉴定了香榧绿藻的种属类别,为香榧绿藻的综合防治和香榧产业的健康发展提供新的思路。
HTML
-
2017年7月中旬,在浙江省杭州市临安区、安徽省黄山市和浙江省诸暨市3个地区分别采集患有香榧绿藻病的香榧叶片,密封带回实验室,用灭菌后的小刀片从患病叶片上刮取香榧绿藻混合物用于离体培养。
-
绿藻的培养采用SE液/固体培养基[8],其中液体培养基配方主要成分如下:硝酸钠250 mg·L-1,三水磷酸氢二钾75 mg·L-1,七水硫酸镁75 mg·L-1,二水氯化钙25 mg·L-1,磷酸二氢钾175 mg·L-1,氯化钠25 mg·L-1,六水合三氯化铁5 mg·L-1,A5处理液1.000 g·L-1(主要包括硼酸2.860 g·L-1,四水氯化锰1.810 g·L-1,七水硫酸锌0.220 g·L-1,五水硫酸铜0.079 g·L-1,四水合钼氨酸0.390 g·L-1)和Fe-EDTA处理液1.000 g·L-1(包括乙二胺四乙酸钠10.000 g·L-1和六水合三氯化铁0.810 g·L-1)[9]。配制好的液体培养基自然pH值为(6.70±0.05),在1 L液体培养基中加入1.500 g琼脂糖,经高温灭菌(121 ℃,30 min)冷凝后形成SE固体培养基。
-
取少量香榧绿藻混合物,置于盛有100 mL SE液体培养基的三角瓶中静置培养,温度(23±2)℃,光照强度为6 000 lx(120 μmol·m-2·s-1),24 h持续光照,培养2周[10]。
-
采用平板划线分离法将混合培养后的香榧绿藻接种在SE固体培养基上,继续培养1周后,根据藻体的形态特征将不同形态的绿藻用接种针取出,分别培养于SE液体培养基中,并进行放大培养进一步纯化,直至各组培养液中只有一种形态的绿藻,获得A和B组2组香榧绿藻,定期观察藻液,并用荧光显微镜拍照记录。
-
从A和B组香榧绿藻培养液中取1~2滴处于对数增长期的藻液,分别置于盛有100 mL灭菌SE液体培养基的三角瓶中,在温度为(23±2)℃,光照强度为6 000 lx(120 μmol·m-2·s-1),24 h持续光照的培养室中,静置培养1个月,做3个重复。每天早晚将培养瓶震动1次,防止藻细胞贴壁生长,且每隔2 d在紫外分光光度计680 nm[11]下测定光密度。
-
从A和B组香榧绿藻培养液中分别取1~2滴对数增长期的藻液,加入pH值为6.0,7.0,8.0,9.0,10.0,11.0(用0.1 mol·L-1盐酸和0.1 mol·L-1氢氧化钠调节)的SE液体培养基中,原培养基(自然pH值)为对照组,培养3周,做3个重复,且每隔2 d在紫外分光光度计680 nm下测定吸光度[10]。
-
① 香榧绿藻DNA的提取。取100 mL对数生长期的藻液,分装至50 mL离心管中,6 000 r·min-1离心10 min,弃上清,收集的藻体研磨后用TaKaRa MiniBEST Plant Genomic DNA Extraction Kit(编码9768)试剂盒提取基因组DNA。②引物合成。用于真核绿藻18S rRNA基因的上游引物MA1(5′-CGGGATCCGTAGTCATATGCTTGTCTC-3′)和下游引物MA2(5′-CGGAATTCCTTCTGCAGGTTCACC-3′),由上海生工生物技术有限公司进行合成。③PCR扩增。以香榧绿藻样品的基因组DNA为模板,对香榧绿藻18S rDNA进行PCR扩增。PCR反应体系为50 μL,包括:双蒸水21 μL,上下引物各1 μL,DNA模板1 μL,2×Gflex PCR缓冲液(镁离子, dNTP plus)25 μL,Tks Gflex DNA Polymerase(1.25×16.67 nkat·L-1)1 μL。PCR反应条件:94 ℃预变性1 min;98 ℃变性10 s,55 ℃退火15 s,68 ℃延伸60 s,30个循环。PCR产物取5 μL经10 g·L-1(1×TAE)琼脂糖凝胶电泳检测后,观察并成像[12]。扩增产物由大连宝生物工程有限公司(TaKaRa)进行测序。
-
以香榧绿藻样品的基因组DNA为模板,分别以正反向引物对香榧绿藻ITS区进行PCR扩增,并对PCR产物进行测序。香榧绿藻ITS正反向引物序列[13-14]:1800f: ACCTGCGGAAGGATCATT, LR 1850: CCTCACGGTACTTGTTC由大连宝生物工程有限公司(TaKaRa)进行合成。PCR反应体系和反应条件与18S rDNA序列获取条件一致。
-
测序结果与美国国立生物技术信息中心(NCBI)已知的数据库进行比较,并用Genetyx软件进行多重序列对齐排列,通过MEGA 7.0软件邻接(NJ)法绘制系统发育树,基于Kimura2-Prameter方法计算遗传距离值,计算Bootstrap值,重复1 000次[15]。
1.1. 试验材料
1.1.1. 香榧绿藻
1.1.2. 供试培养基
1.2. 试验方法
1.2.1. 绿藻的混合培养
1.2.2. 绿藻的分离纯化培养
1.2.3. 香榧绿藻生长曲线的绘制
1.2.4. pH值对香榧绿藻生长的影响
1.2.5. 18S rDNA序列获取
1.2.6. ITS序列获取
1.2.7. 系统发育树分析
-
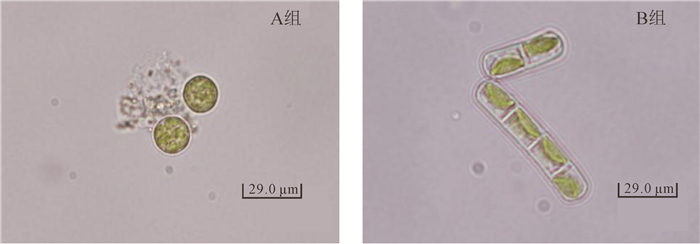
对3个不同地点采样的香榧绿藻培养观察发现:香榧绿藻主要由2种形态的绿藻构成,按形状、大小等细胞特性将分离纯化的香榧绿藻分为A组和B组,其形态结构见图 1。临安和黄山地区采集的香榧绿藻均由A组和B组不同形态的绿藻组成,而诸暨地区采集的香榧绿藻只含A组形态的绿藻。表明不同地区香榧绿藻的形态不尽相同,但主要以A组形态绿藻为主,这可能与当地土壤中的藻类分布有关,因为绿藻主要通过空气中扬尘进行传播[16],同时地理位置的差异也会导致藻类形态的多样性[10]。从生活习性上来看,香榧绿藻属于气生藻Luftalgen aerophtische类植物的树生藻,与小球藻Chlorella sp.不同的是其主要在树干以及枝叶表面生长,它们所需要的水分可以从降雨和蒸汽中得到[17]。
藻类的鉴定方法包括传统形态学方法、生物化学法和分子生物学法。传统形态学方法是通过对藻类的培养观察,根据藻体的形状、大小等形态学特征鉴定藻的种类。本研究通过荧光显微镜观察结果显示:A组香榧绿藻为单细胞圆形,直径10.36~22.79 μm,叶绿体布满整个细胞,细胞壁薄且光滑;非油性,淡绿色单胞,无鞭毛。B组香榧绿藻为单细胞椭圆形或长椭圆形,两端钝圆,长21.07~25.21 μm,宽10.36~12.43 μm,叶绿体多位于细胞的一侧;油性,淡绿色,细胞间以单链相连,无分支,基部有一着生细胞,与李润霞等[18]鉴定的黄瓜绿藻病病原丝藻Ulothrix sp.的形态特征基本一致。2组香榧绿藻的形态结构与前人从沙漠和海洋分离纯化的藻种形态不一致[19-21],应该是树生藻为适应寄生环境形成的特殊形态结构。
-
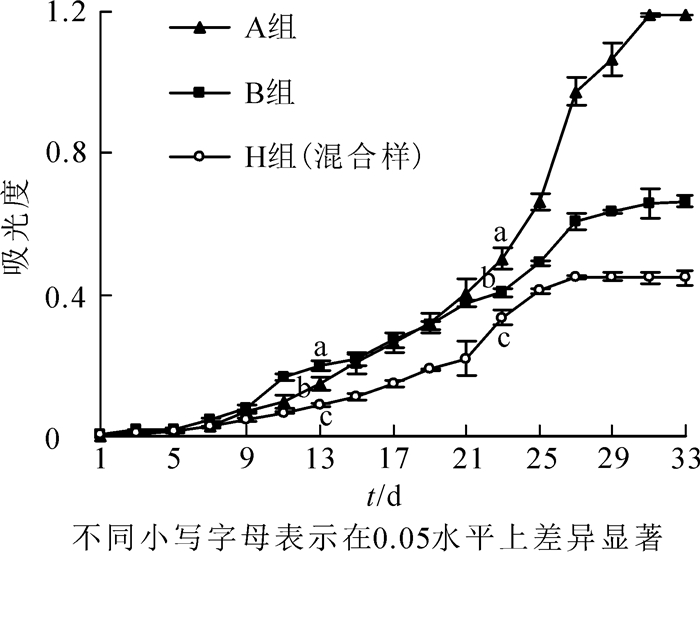
香榧绿藻生长特性与大部分藻类相似,在培养初期(适应期)藻体生长缓慢,进入对数生长期后,生长速率迅速增加,藻液也由浅绿色变为暗绿色,直到饱和停止增长,进入衰亡期后,藻体逐渐死亡并下沉,藻液颜色开始变黄,最后至无色、腐烂[10]。由图 2可知:A组香榧绿藻的适应期较长,前4 d是适应期,第5天进入对数生长期,第31天进入稳定期;B组香榧绿藻的适应期短,第2天即进入对数生长期,第29天进入稳定期;2组香榧绿藻的混合样H适应期也较短,第3天进入对数生长期,第27天进入稳定期。
H组为A和B组香榧绿藻的混合样。比较发现:香榧绿藻培养前期生长速率从大到小依次为B组、A组、H组,第9~13天差异显著(P<0.05);香榧绿藻培养19 d后生长速率从大到小依次为A组、B组、H组,培养23 d时达显著水平(P<0.05),表明2组香榧绿藻生长期不一致,B组香榧绿藻在生长前期占有优势,而A组香榧绿藻在生长后期占优势,且2组香榧绿藻存在竞争关系,导致2组香榧绿藻混合后生长速率最慢(图 2)。通过生长曲线结果可知:2组香榧绿藻均表现出较长的生长周期,1个月内均没有进入衰亡期,因此香榧绿藻在香榧叶上能长期生存下来。
-
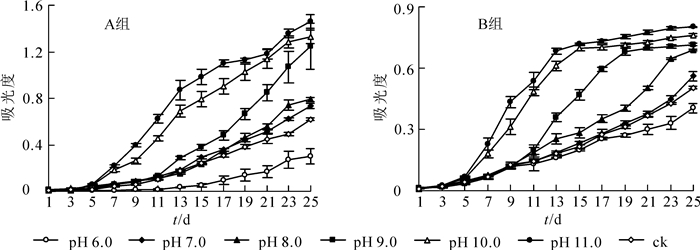
由图 3可知:A组香榧绿藻在pH 10.0和pH 11.0的培养基上生长最好,在度过适应期(前5 d)后出现爆发式增长,长势明显优于其他各组,pH 9.0培养基中的A组香榧绿藻在第11天增长速度加快,长势逐渐接近pH 10.0和pH 11.0的香榧绿藻,且pH 6.0的培养基会明显抑制A组香榧绿藻的生长。香榧绿藻的生长速率从大到小依次为pH 11.0、pH 10.0、pH 9.0、pH 8.0、pH 7.0、对照组、pH 6.0。得出A组香榧绿藻最适生长的pH值为11.0,说明A组香榧绿藻适合在碱性环境中生长,酸性环境会抑制A组香榧绿藻的生长。
由图 3可知:B组香榧绿藻同样在pH 10.0和pH 11.0的培养基上生长最好,在度过适应期(前3 d)后出现爆发式增长,长势明显优于其他各组,分别在第15天和第13天进入稳定期;pH 8.0和pH 9.0培养基中的B组香榧绿藻在第9天增长速度加快,长势逐渐接近pH 10.0和pH 11.0,并分别在第23天和第19天进入稳定期;且pH 6.0的培养基会明显抑制B组香榧绿藻的生长。香榧绿藻生长速率从大到小依次为pH 11.0、pH 10.0、pH 9.0、pH 8.0、pH 7.0、对照组、pH 6.0。得出B组香榧绿藻最适生长的pH值为11.0,说明B组香榧绿藻也适合在碱性环境中生长,酸性环境同样会抑制B组香榧绿藻的生长。相比于A组香榧绿藻,B组香榧绿藻更早进入稳定期,且光密度值明显低于A组香榧绿藻。这可能与细胞的排列结构有关,链状结构限制了B组香榧绿藻的增长。
培养基中pH值是藻类生长代谢等许多生理过程的重要影响因子,pH值会影响培养基中藻细胞对元素的吸收和利用以及对代谢产物的利用,从而影响藻类的光合作用和呼吸作用[22-23]。每一种藻类都有它自身适合的pH值范围,改变pH值会影响藻类的生长和光合作用[24-25]。赵娜等[26]研究表明:小球藻的最适生长pH值为7.0,而斜生栅藻Scenedesmus obliquus的最适生长pH值为9.0。本研究显示:香榧绿藻在pH 11.0环境下生长最好,说明其具有较好的耐碱能力,这与斜生栅藻对pH值的反应规律相似[26],表明香榧绿藻的生活习性与斜生栅藻更为接近。在碱性环境中,细胞可维持较好的分裂速度,这也与袁丽娜等[27]的研究结果一致:低pH值不利于藻生长,在一定范围内较高的pH值能促进藻细胞增殖。因此,香榧绿藻适合生长在碱性环境中,酸性环境可有效抑制香榧绿藻的生长,这也为香榧绿藻的防治提供了新思路。
-
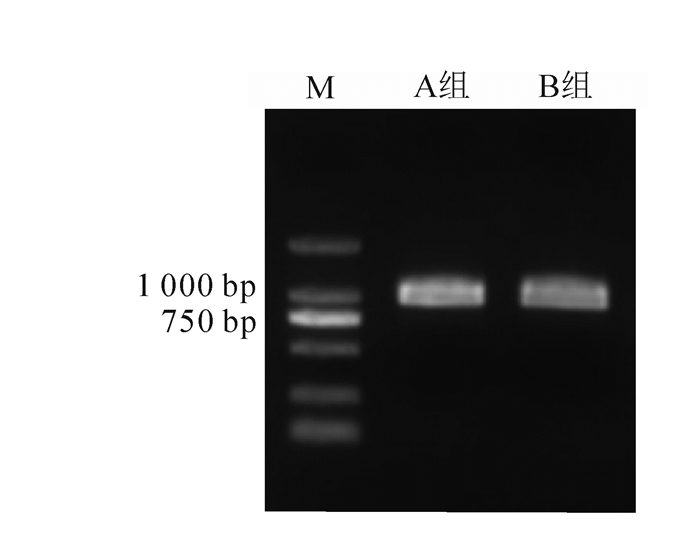
对具有代表性的2组香榧绿藻CTG1003-XQ(A组)和CTG1003-SZ(B组)进行18S rDNA序列分析,扩增出长度均为1 678 bp的基因片段(图 4)。
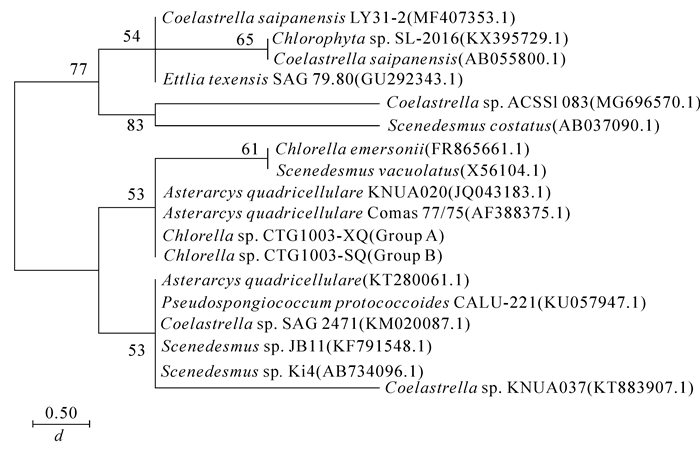
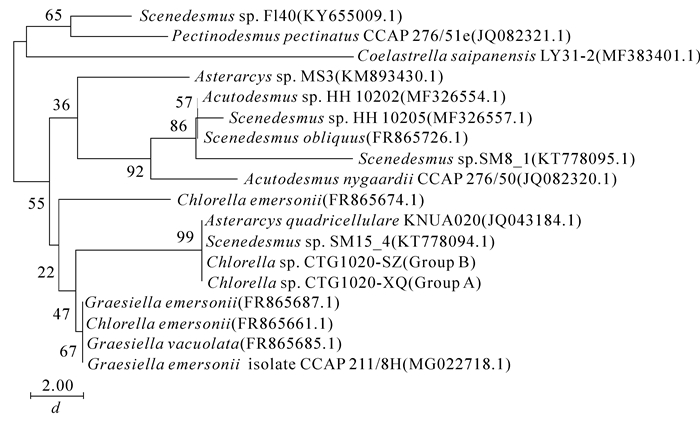
NCBI中BLAST结果表明:香榧绿藻A组和B组的18S rDNA碱基序列完全相同,且与GenBank上属于栅藻科Scenedesmace的Asterarcys quadricellulare KNUA020(JQ043183.1)和Asterarcys quadricellulare Comas 77/75(AF388375.1)的同源性高,相似率均达100%。由图 5可以看出:A组和B组香榧绿藻跟栅藻科聚簇成一支,跟Asterarcys quadricellulare KNUA020和Asterarcys quadricellulare Comas77/75的节点支持率为53,遗传距离为0.00,说明这2组香榧绿藻很有可能属于Asterarcys quadricellulare。
传统形态学方法存在一定局限性,藻类形态差异小、鉴定周期长、形态不稳定[28]等因素导致其物种鉴定面临着不少难题。对此分子鉴定技术应运而生,它是根据物种的基因差异来定性分类物种,可以快速稳定鉴定藻类[29]。近年来,国内外学者通过对18S rRNA基因片段的差异分析鉴定了多种藻类[19, 30-31],但由于其片段保守性,仅适用于分析高阶元如属间或目间的物种[32-33]。本研究通过对香榧绿藻18S rDNA序列分析发现:香榧绿藻基因序列与栅藻科的Asterarcys quadricellulare亲缘关系较近,印证了本研究中香榧绿藻对pH值的反应规律与斜生栅藻Scenedesmus obliquus的结果相同,表明香榧绿藻属于栅藻科。这与本研究形态学分类的结果不一致,与早期研究者对香榧绿藻的认知不同。
-
对具有代表性的2组香榧绿藻CTG1003-XQ(A组)和CTG1003-SZ(B组)的ITS区基因片段进行序列分析,扩增出总长度均为874 bp的基因片段(图 6),且2组香榧绿藻的碱基序列完全一致。根据FAGGIAN等[34]的划分方法,香榧绿藻ITS1区长297 bp,5.8S rDNA长287 bp,ITS2区长290 bp。
NCBI中BLAST结果表明:香榧绿藻A组和B组的ITS区基因序列与GenBank上Asterarcys quadricellulare和Scenedesmus sp. SM15_4(KT778094.1)的同源性高,相似率均达100%。由图 7可以看出:A组和B组香榧绿藻跟栅藻科聚簇成一支,跟Asterarcys quadricellulare和Scenedesmus sp.的节点支持率为99,遗传距离为0.00,表明2组香榧绿藻均与栅藻科的Asterarcys quadricellulare以及Scenedesmus sp.的ITS区基因序列一致。本结果进一步证实了香榧绿藻与栅藻科的亲缘关系,与本研究中香榧绿藻18S rDNA序列比对的结果基本一致。
香榧绿藻18S rDNA序列和ITS区基因序列均与Asterarcys quadricellulare相似率达100%,表明香榧绿藻与Asterarcys quadricellulare属于同一物种,与丝藻和小球藻亲缘关系较远。香榧绿藻ITS区基因序列与栅藻科的物种Scenedesmus sp.的ITS区基因序列相似率也达100%,但两者的18S rDNA序列相差较大,表明栅藻ITS区基因序列种间保守性较强,不利于种间的区分。这与前人对其他物种的研究结果相反[35-36]。一般认为,ITS区基因序列变异性更大,说明不同物种间基因片段的保守性不尽相同。
结合形态分析发现:A组香榧绿藻的形态特征也与Asterarcys quadricellulare相近[30],说明A组香榧绿藻属于Asterarcys quadricellulare,B组香榧绿藻形态与Asterarcys quadricellulare形态差异较大,可能为其变种。HONG等[30]发现:Asterarcys quadricellulare来自于韩国庆北国立大学(Kyungpook National University)花园土壤中的一种微型藻类,含有大量的长链多不饱和脂肪酸和脂肪醇,说明香榧绿藻也有产生长链多不饱和脂肪酸和脂肪醇的潜力,具有一定的科研价值。而GenBank上Scenedesmus sp.的ITS区基因序列样品来源于中国浙江[37],与香榧绿藻亲缘关系较近,对香榧绿藻的鉴定具有重要意义。
2.1. 香榧绿藻正置荧光显微镜观察结果
2.2. 2组香榧绿藻的生长曲线
2.3. pH值对香榧绿藻生长的影响
2.4. 2组香榧绿藻18S rDNA序列分析
2.5. 2组香榧绿藻ITS序列分析
-
本研究表明:香榧绿藻由A组和B组2种不同形态的藻类组成,并主要以A组形态绿藻为主,2种形态的香榧绿藻间存在竞争关系。根据形态学鉴定表明:A组香榧绿藻与引起番茄绿藻病的椭圆小球藻形态相近,B组香榧绿藻与引起黄瓜绿藻病的丝藻形态相近;但香榧绿藻对pH值的反应规律与斜生栅藻相近。序列比对也发现:2种形态的香榧绿藻的18S rDNA和ITS区基因序列均与栅藻科的Asterarcys quadricellulare的同源性高,相似率达100%。因此,本研究将A组香榧绿藻初步鉴定为栅藻科的Asterarcys quadricellulare,B组香榧绿藻为其变种。且2组香榧绿藻的ITS区基因序列与GenBank上的栅藻科物种Scenedesmus sp.相同。pH值会显著影响香榧绿藻的生长,酸性环境可有效抑制香榧绿藻的生长,这也为香榧绿藻的防治工作提供了新思路。
















 DownLoad:
DownLoad: