-
土壤盐碱化是全球范围内非常严峻的环境问题,目前已有超过13.81亿hm2的土地受到过量盐分的影响[1]。过高的盐分会在植物体内引发渗透胁迫、离子胁迫和氧化胁迫,导致水分吸收受阻,离子平衡及细胞稳态破坏,从而抑制植物的生长与发育,甚至造成植株死亡[2−3]。植物通常通过渗透调节、离子稳态调节和活性氧清除等途径应对盐胁迫[2−3],而植物激素在这些过程中发挥着重要调控作用[4−5]。已有研究表明:褪黑素(melatonin)[6]、茉莉酸甲酯(methyl jasmonate,MeJA)[7]、水杨酸(salicylic acid,SA)[8]、细胞分裂素(cytokinin,CK)[9]、乙烯(ethylene)[10]以及脱落酸(abscisic acid,ABA)[11]等多种植物激素参与盐胁迫的响应。
脱落酸被认为是植物应对逆境,尤其是盐胁迫的核心信号因子[11]。外源施加脱落酸可有效缓解盐胁迫对植物生长的抑制[12];施加脱落酸合成抑制剂钨酸钠(Na2WO4),则会削弱植物的渗透调节能力,阻碍气孔关闭并加剧活性氧积累,最终导致植株生长受抑[13−16]。在盐胁迫条件下,植物体内脱落酸会迅速升高,从而启动一系列防御机制。首先,脱落酸能够被受体PYRABACTIN RESISTANCE/PYRABACTIN RESISTANCE-LIKE(PYR/PYL)感知,受体结合脱落酸后可以抑制蛋白磷酸酶(protein phosphatase 2C,PP2C)的活性,进而解除对Ⅲ类SnRK2蛋白激酶的抑制,并使其下游靶蛋白发生磷酸化[17]。活化的SnRK2激酶进一步调控多种生理过程,包括离子转运和气孔关闭等[18],并通过调控多种转录因子介导渗透物质的生物合成[19−22]。
芙蓉菊Crossostephium chinense是菊花Chrysanthemum × morifolium的近缘属物种,能够在700 mmol·L−1 氯化钠(NaCl)的胁迫条件下维持正常的形态和生理功能。甘菊C. lavandulifolium是菊花同属植物,在500 mmol·L−1 氯化钠胁迫下即整株死亡,对盐敏感,耐盐性差[23]。已有研究表明:将芙蓉菊作为母本,甘菊为父本进行属间远缘杂交能够获得杂交后代[24]。也有研究对父母本甘菊、芙蓉菊与杂交后代统一进行了耐盐评价,发现后代耐盐性介于两亲本之间,其中FG26后代相对较为耐盐[23]。这表明芙蓉菊可通过属间杂交将耐盐等优良性状导入栽培菊花,获得杂交后代。尽管前期研究表明芙蓉菊具有强耐盐性,但其耐盐相关的及与后代遗传有关的生理及分子机制,特别是脱落酸在芙蓉菊盐胁迫过程中所起的作用和影响尚不明确。基于此,本研究以耐盐种质芙蓉菊,盐敏感种质甘菊及较耐盐的杂交后代FG26为研究材料,系统评估盐胁迫下植株内源脱落酸的动态变化;分析外源施加脱落酸及脱落酸合成抑制剂钨酸钠处理对耐盐性相关关键生理指标的影响,包括脯氨酸(proline,Pro)、超氧化物歧化酶(superoxide dismutase,SOD)和丙二醛(malondialdehyde,MDA),并结合转录组数据[25],解析脱落酸在芙蓉菊及杂交后代盐胁迫适应过程中的生理分子层面的调控机制,以期为耐盐菊花新品种的创制提供理论依据和基因资源。
-
选用耐盐型芙蓉菊、盐敏感型甘菊及以芙蓉菊作为母本、甘菊为父本获得的较耐盐杂交后代FG26为研究材料[23−24]。试验材料种植于国家花卉工程研究中心研发温室。3种材料的扦插苗在蛭石∶珍珠岩为1∶1(体积比)的基质中生根,随后移栽至草炭∶珍珠岩∶松针土为6∶3∶1(体积比)的基质中,在人工气候室中培养1.5个月。培养条件为光周期12 h (光照)/12 h (黑暗)、温度25 ℃和相对湿度60%。
-
选择生长状态良好且一致的植株进行试验,每个处理设置3个生物学重复。参考盐胁迫转录组的研究[25],采用700 mmol·L−1氯化钠溶液处理植株,并分别于处理0、0.5、1.0、3.0、6.0、12.0和24.0 h在中段叶位采集叶片样品0.5 g。样品迅速用铝箔纸包裹,经液氮速冻后转移至−80 ℃超低温冰箱保存,用于内源激素质量分数测定,所用样品均为鲜质量。脱落酸质量分数的测定采用酶联免疫吸附法(ELISA)[26]。
-
为筛选出适宜的处理浓度,预试验以清水处理为对照,将3种材料的扦插苗分别用不同浓度的氯化钠(0、100、300 mmol·L−1)结合不同浓度的脱落酸(0、100、150、200 μmol·L−1)或钨酸钠(0、1、2 mmol·L−1)处理7 d。结果表明,300 mmol·L−1 氯化钠结合200 μmol·L−1 脱落酸或1 mmol·L−1钨酸钠处理,生理指标变化最显著(P<0.05),3种材料间差异最大,且植株均未死亡。因此,正式试验选用300 mmol·L−1 氯化钠、200 μmol·L−1 脱落酸及1 mmol·L−1钨酸钠作为处理浓度。
选择生长状态良好且一致的植株进行试验,每个处理设置5个生物学重复,胁迫时间为7 d。以0 mmol·L−1 氯化钠处理并施加清水为对照组(ck),以300 mmol·L−1 氯化钠溶液处理并施加清水(T1)、300 mmol·L−1 氯化钠溶液处理并施加200 μmol·L−1 脱落酸(T2),以及300 mmol·L−1 氯化钠溶液处理并施加1 mmol·L−1 钨酸钠溶液(T3)为试验组。试验在人工气候室中进行,培养条件为光周期12 h (光照)/12 h (黑暗)、温度25 ℃和相对湿度60%。处理结束后在中段叶位采集叶片样品0.5 g,经铝箔纸包裹、液氮速冻后,保存于−80 ℃冰箱,用于内源脱落酸、脯氨酸、超氧化物歧化酶活性及丙二醛测定。所用样品均为鲜质量。
-
脯氨酸采用可见分光光度法测定[27],超氧化物歧化酶活性采用WST-1法测定[28],丙二醛采用硫代巴比妥酸法(TBA)测定[29]。所用试剂盒均购自南京建成生物科技有限公司。
-
根据已发表转录组数据[25],提取芙蓉菊中与盐胁迫响应有关的PYRs/PYLs、PP2Cs和SnRK2s基因的TPM(transcripts per million)值,并利用TBtools绘制基因表达热图[30]。
以拟南芥Arabidopsis thaliana脱落酸信号通路中盐胁迫响应有关的PYRs/PYLs、PP2Cs和SnRK2s基因的蛋白质序列为查询序列[2−3],使用 BLASTP v2.5.0+在芙蓉菊转录组及其他18个被子植物的基因组中搜索对应的同源基因,E的阈值设为1.0×10−5[31]。为进行系统发育分析,各基因的氨基酸序列使用 MAFFT v7.505 进行比对[32],并通过 PAL2NAL v14 生成对应的基于密码子的核苷酸比对[33]。随后使用 IQ-TREE 2.1.4-beta 构建最大似然系统发育树,参数设置为“-bb
1000 -nt AUTO”[34],并利用MEGA11进行可视化[35]。本研究用到的转录组和基因组数据均下载于已发表的研究论文(表1)。表 1 系统发育分析所用的基因组或转录组数据
Table 1. Genomic and transcriptomic data used for phylogenetic analyses
科名 种名 缩写 参考文献 科名 种名 缩写 参考文献 菊科 Asteraceae 芙蓉菊Crossostephium chinense Crch [25] 茄科 Solanaceae 番茄Solanum lycopersicum Soly [46] 野菊Chrysanthemum indicum Chin [36] 十字花科 Brassicaceae 拟南芥Arabidopsis thaliana Arth [47] 菊花Chrysanthemum × morifolium Chmo [37] 杨柳科 Salicaceae 欧洲山杨Populus trichocarpa Potr [48] 菊花脑Chrysanthemum nankingense Chna [38] 葡萄科 Vitaceae 葡萄Vitis vinifera Vivi [49] 甘菊Chrysanthemum lavandulifolium Chla [39] 毛茛科 Ranunculaceae 蓝花耧斗菜Aquilegia coerulea Aqco [50] 生菜Lactuca sativa Lasa [40] 禾本科 Poaceae 玉米Zea mays Zema [51] 向日葵Helianthus annuus Hean [41] 水稻Oryza sativa Orsa [52] 牛蒡Arctium lappa Alap [42] 木兰科Magnoliaceae 鹅掌楸Liriodendron chinense Lich [53] 艾Artemisia argyi Arar [43] 无油樟科Amborellaceae 无油樟Amborella trichopoda Amtr [54] 黄花蒿Artemisia annua Aran [44] 三齿蒿Artemisia tridentata Artr [45] -
采用Excel 2021进行数据统计和处理,用SPSS 26软件进行单因素方差分析(one-way ANOVA),采用Duncan’s多重比较检验(P<0.05)分析不同处理间的差异显著性。
-
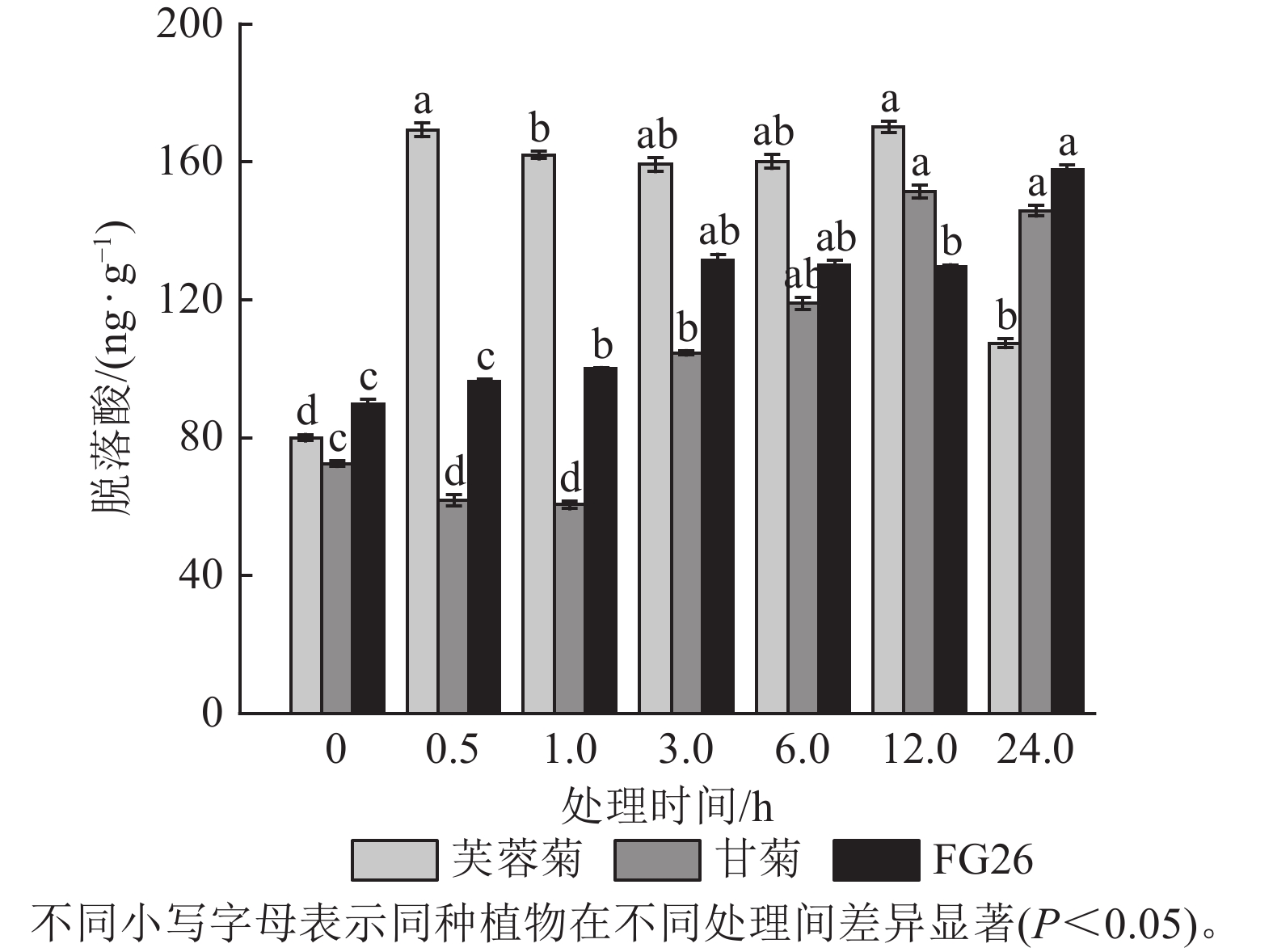
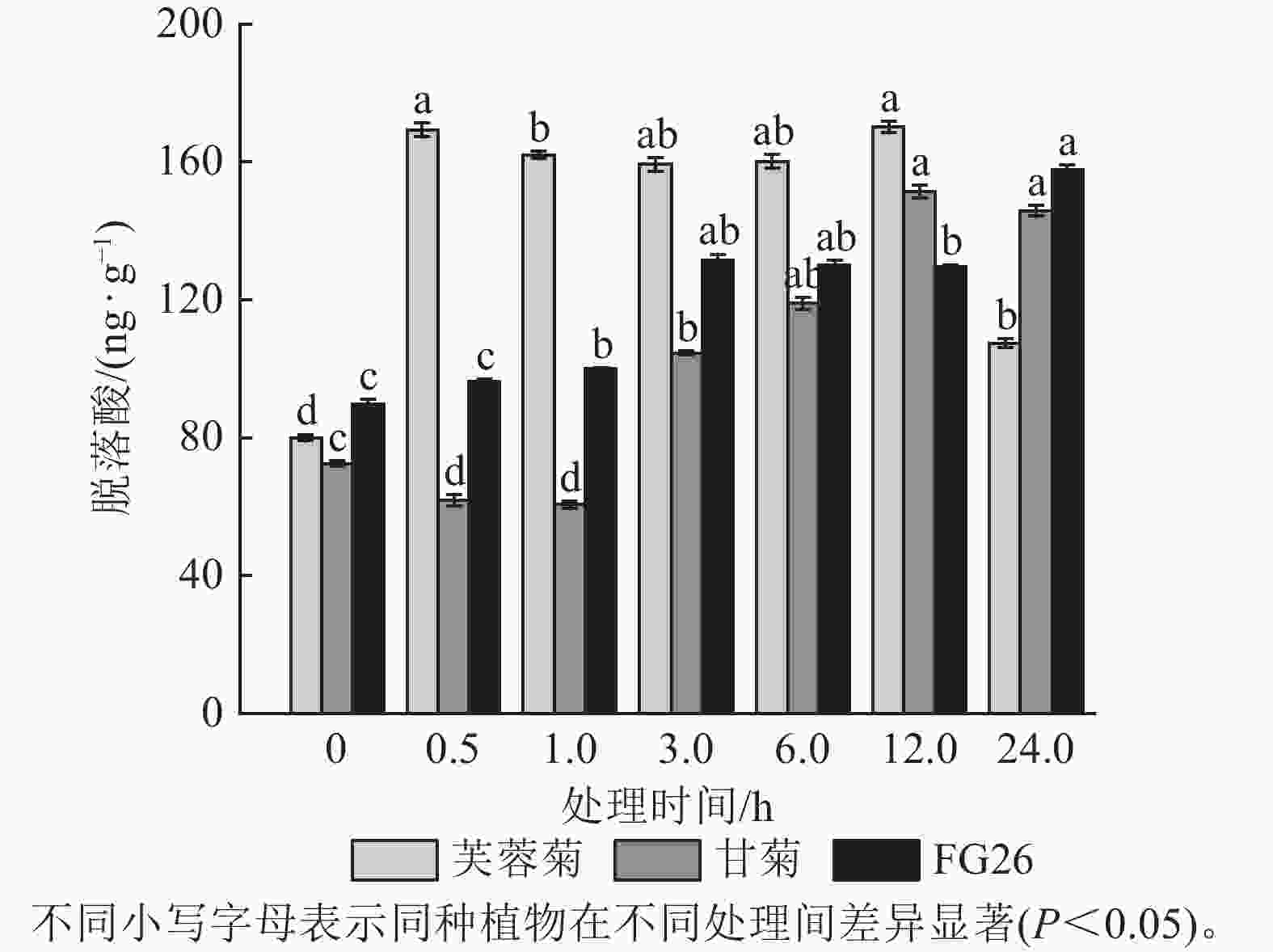
盐胁迫会影响植物代谢,进而对植物内源激素产生影响,所以内源激素水平的变化可以反映植物对盐胁迫的响应。由图1可知:盐胁迫显著改变了3种植物叶片的内源脱落酸质量分数(P<0.05)。芙蓉菊的脱落酸质量分数在盐处理后0.5 h即快速响应,显著升高至169.37 ng·g−1(P<0.05),较处理0 h增加 111.71%。随后较长时间内(1~12 h),脱落酸质量分数持续保持较高的水平,直至24 h显著下降(P<0.05),恢复至接近处理前的水平。甘菊的脱落酸质量分数呈现先下降后上升再下降的趋势,响应过程较为缓慢,在12 h才达到峰值(151.40 ng·g−1)。后代FG26的脱落酸质量分数则一直呈现上升趋势,在24 h时仍呈现出上升趋势(157.78 ng·g−1)。这些结果表明:脱落酸在3种植物盐胁迫响应过程中均发挥作用,但芙蓉菊中脱落酸对盐胁迫的响应速度更加迅速,在1~12 h内都保持着较高质量分数。FG26的脱落酸一直保持着上升趋势,而甘菊的脱落酸调控可能存在延迟,响应速度较慢,且在24 h内甘菊与FG26的脱落酸质量分数都低于芙蓉菊在0.5 h的脱落酸质量分数,甘菊的峰值质量分数也小于FG26的最终质量分数。
-
由图2可知:T1处理显著提升3种植物脱落酸质量分数(P<0.05),较ck分别增加3.23、1.53及5.61倍。T2处理进一步提高了三者脱落酸质量分数,较T1分别增加2.03、2.14及2.99倍,且T1、T2处理中芙蓉菊的脱落酸质量分数均最高,FG26的脱落酸上升倍数最高。相反,在T3处理时,降低了脱落酸质量分数,芙蓉菊脱落酸质量分数较T1下降了6.31%,甘菊下降幅度最大达12.72%,FG26下降幅度最小达4.00%。
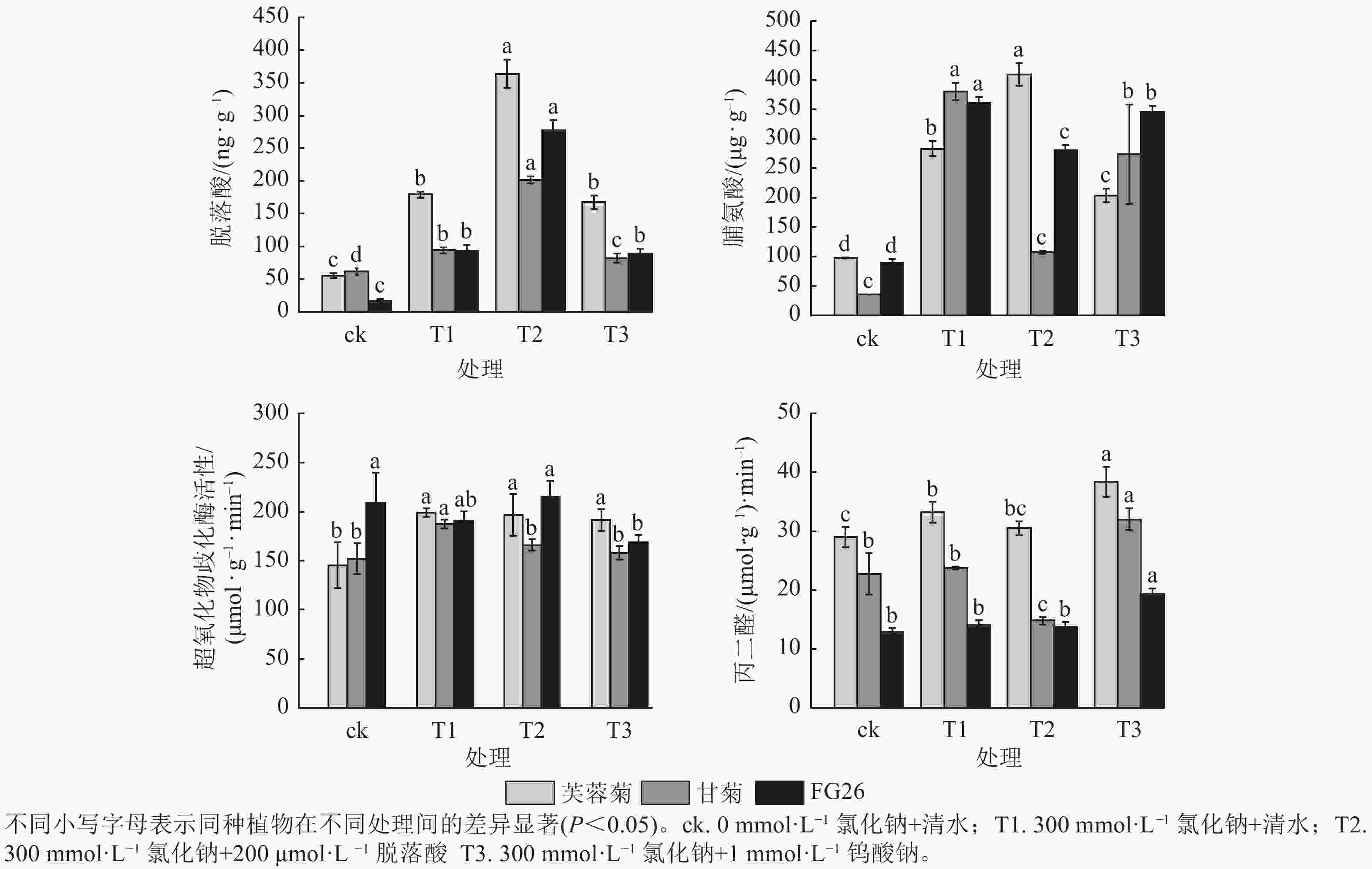
图 2 外源施加脱落酸和钨酸钠对盐胁迫下芙蓉菊、甘菊及其杂交后代FG26生理指标的影响
Figure 2. Effects of exogenous ABA and Na2WO4 on physiological indicators of Crossostephium chinense, Chysanthemum lavandulifolium and their hybrid FG26 under salt-stress condition
T1处理使3种植物脯氨酸质量分数显著上升(P<0.05),较ck分别增加2.90、10.69及4.03倍,三者脯氨酸质量分数及上升倍数和耐盐力强弱排序相反,盐敏感型甘菊的脯氨酸质量分数上升倍数最高,可能是因为甘菊作为盐敏感物种,遭受的胁迫更严重,促使渗透调节物质被动积累,但其细胞结构或保护系统无法有效利用这些物质,最终仍死亡。T2处理促进芙蓉菊中脯氨酸质量分数较盐处理组上调44.42%,而在甘菊和FG26中,脯氨酸质量分数较T1分别降低71.74%和22.27%,可能暗示着脯氨酸代谢途径对高浓度脱落酸的响应存在物种特异性。在T3处理中,3种植物的脯氨酸质量分数较T1分别下降了28.00%、28.00%及4.45%,其中FG26的下降幅度远小于芙蓉菊与甘菊,进一步证明了脱落酸在渗透调节中的积极作用。
T1处理芙蓉菊的超氧化物歧化酶活性最高,FG26次之,甘菊最低;与ck相比,芙蓉菊、甘菊中超氧化物歧化酶活性显著升高(P<0.05),分别增加1.37和1.23倍,而FG26中变化不显著(P<0.05)。在T2和T3处理中,芙蓉菊和FG26较T1变化均不显著,可能由于其活性氧清除系统在盐胁迫下较为稳定,受到干扰较小;甘菊的超氧化物歧化酶活性较T1分别下降11.45%与15.72%,说明其抗氧化系统较为脆弱,易受胁迫扰动。
T1处理导致3种植物丙二醛质量摩尔浓度上升,较ck分别增加14.62%、4.58%和9.47%,芙蓉菊上升幅度最大,但其基础丙二醛质量摩尔浓度较甘菊高,耐盐性却最强,推测其较甘菊来说具备更强膜修复与抗氧化能力,或细胞膜结构存在差异。在T2处理中,甘菊较T1处理丙二醛质量摩尔浓度显著下降37.63% (P<0.05),而芙蓉菊和FG26变化不显著,可能暗示着脱落酸能有效缓解盐胁迫对盐敏感物种的膜损伤,对耐盐性强的物种作用不显著。在T3处理中,3种植物较T1组丙二醛质量摩尔浓度均有显著上调(P<0.05),分别上调了15.43%、34.72%及36.95%,进一步验证内源脱落酸在维持细胞膜稳定性中的关键作用。
-
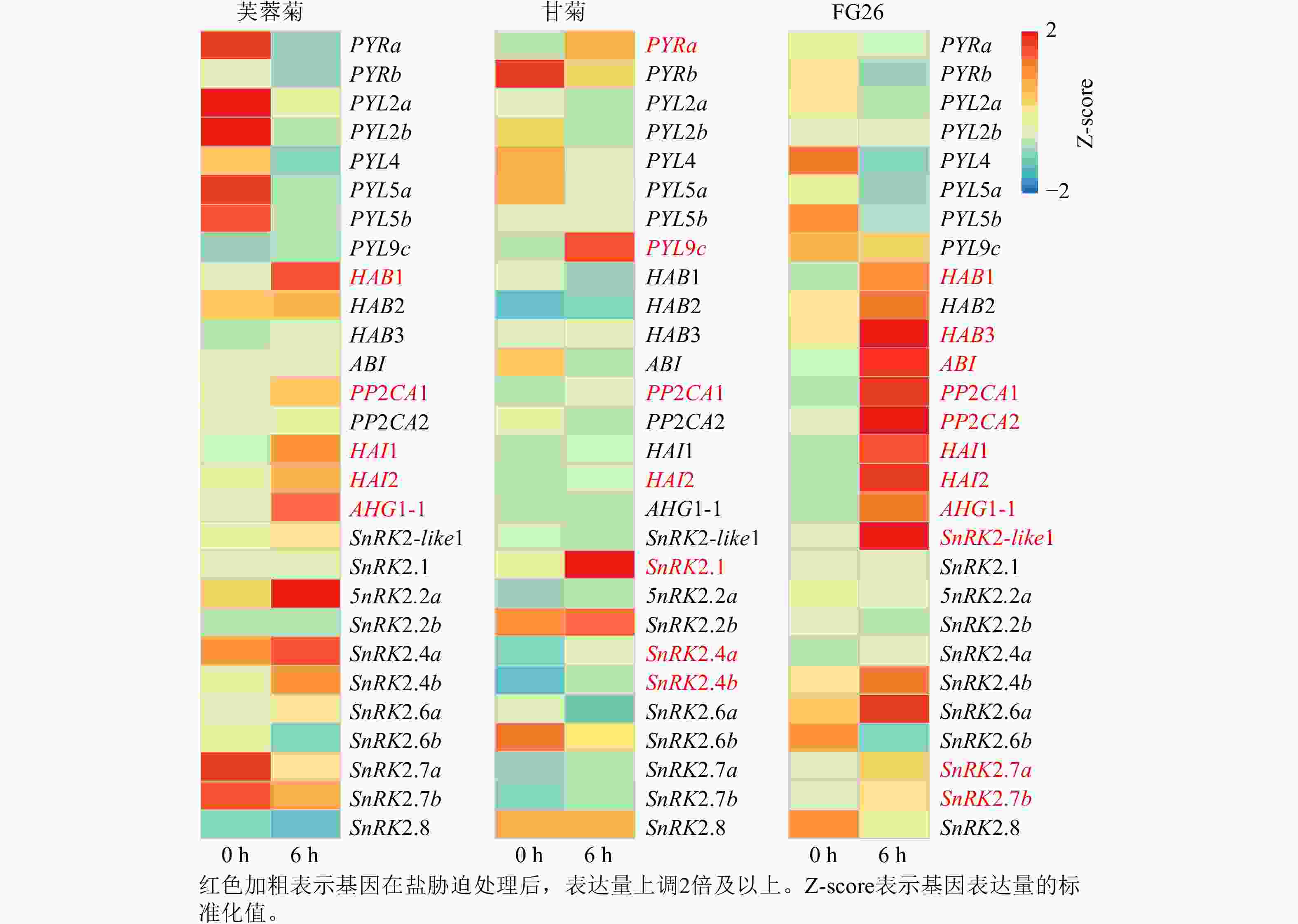
基于前期已发表的芙蓉菊转录组数据[25],共鉴定到脱落酸信号通路中响应盐胁迫关键基因28个,其中包括8个PYRs/PYLs基因,9个PP2Cs基因和11个Ⅲ类SnRK2s基因(图3)。在上述28个候选基因中,芙蓉菊中有5个PP2Cs基因在盐胁迫处理6 h后表达显著上调。值得注意的是,PP2CA1和HAI2在盐胁迫处理6 h后3种植物中的表达水平均显著提高,暗示它们在盐胁迫响应中的调控作用可能具有保守性。相反,另外3个基因(HAB1、HAI1和AHG1-1)则仅在盐胁迫处理6 h后的芙蓉菊和FG26中表达上调,表明PP2C类基因,尤其是HAB1、HAI1和AHG1-1,可能对芙蓉菊超强耐盐特性的获得具有重要作用,并且能够通过杂交遗传给后代,增强后代的耐盐性。
-
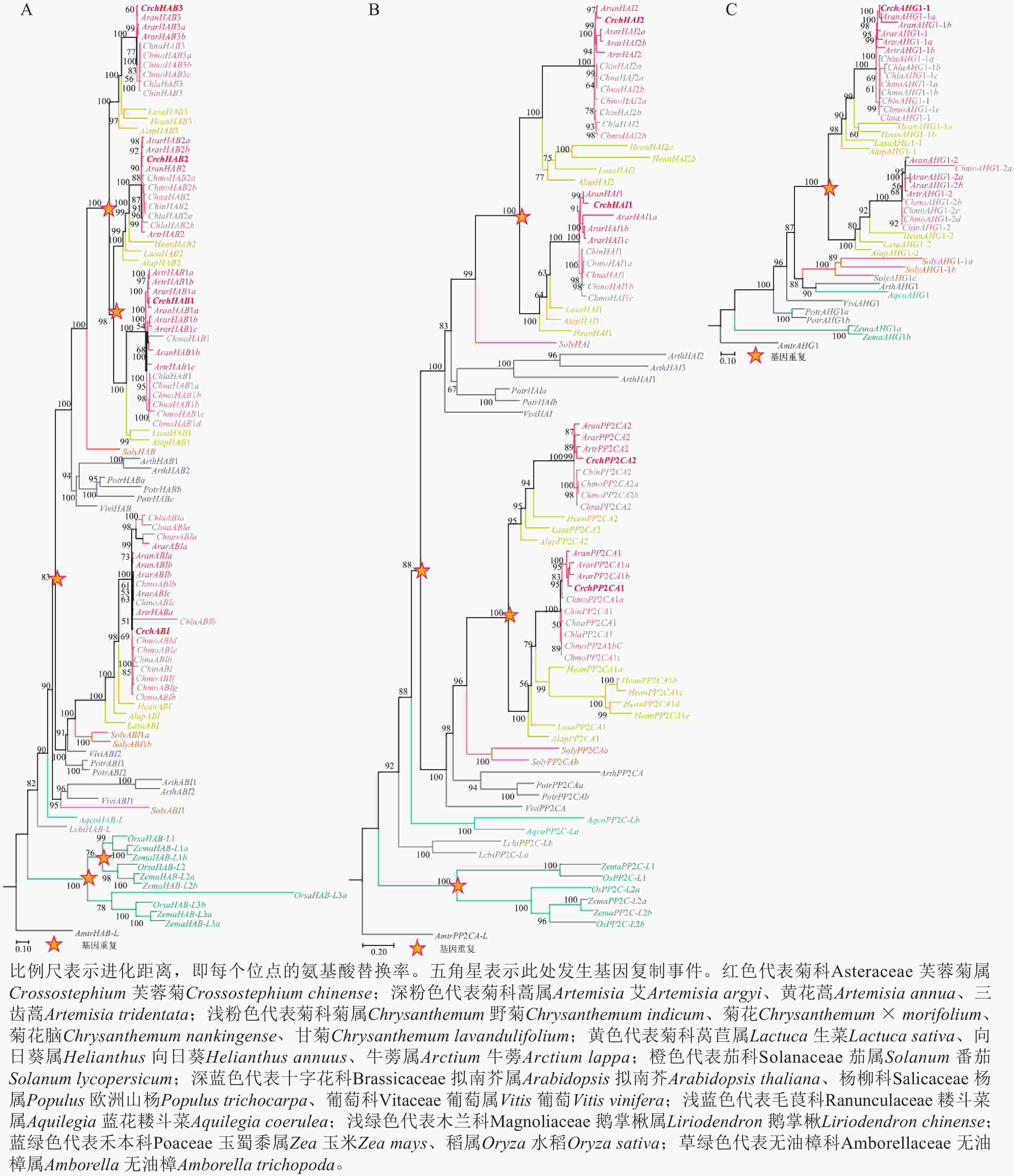
对芙蓉菊耐盐关键候选基因PP2C家族基因的进化历史进行了研究。由图4A可知:HAB类基因在芙蓉菊的进化过程中发生了3次基因重复,形成了CrchHAB1、CrchHAB2、CrchHAB3和CrchABI等4个拷贝。其中1次重复事件发生于核心真双子叶最近共同祖先中,其余2次发生于菊科Asteraceae植物的最近共同祖先中。类似地,由图4B可知:PP2CA类基因同样经历了3次重复事件。其中1次重复事件发生在核心真双子叶最近共同祖先中,形成PP2CA和HAI等2个分支。随后,这2个分支分别在菊科最近共同祖先中各发生1次重复,形成CrchPP2CA1、CrchPP2CA2、CrchHAI1和CrchHAI2等4个拷贝。由图4C可知:AHG1基因在菊科最近共同祖先中发生过1次重复,但在芙蓉菊中丢失了其中1个拷贝,仅保留CrchAHG1-1。上述结果表明:芙蓉菊PP2C家族基因经历了与菊科多数植物相似的基因重复事件,暗示基因重复后的表达分化可能在芙蓉菊耐盐特性形成中发挥了重要作用。
-
植物激素在植物应对非生物胁迫的过程中发挥着关键作用[55]。本研究结果表明:盐胁迫导致芙蓉菊、甘菊及其杂交后代FG26的内源脱落酸质量分数均显著升高。这与盐胁迫下燕麦Avena sativa[56]、向日葵Helianthus annuus[57]的脱落酸变化趋势相似,表明脱落酸在盐胁迫响应中具有重要作用。芙蓉菊表现出更快速强烈的脱落酸响应,其峰值最高,FG26次之,甘菊最低。这可能与3种植物的耐盐性差异有关。
T2处理提高了3种植物的内源脱落酸质量分数,其中芙蓉菊内源脱落酸质量分数最高,FG26的上升倍数最高。这与盐胁迫下外源脱落酸对楸树Catalpa bungei内源脱落酸质量分数的影响结果相符[58]。T3处理下,甘菊的脱落酸质量分数下降幅度最大,其次是FG26,芙蓉菊最小。表明芙蓉菊具有更强的脱落酸合成及代谢调控能力。渗透调节方面,T1处理的甘菊脯氨酸质量分数和增幅变化表明:在盐胁迫下可能是因为甘菊作为盐敏感物种,遭受的胁迫更严重,促使渗透调节物质被动积累,但其细胞结构或保护系统无法有效利用这些物质,最终仍死亡。T2处理可显著提高芙蓉菊的脯氨酸质量分数,这与刺槐Robinia pseudoacacia[59]、鹰嘴紫云英Astragalus cicer[60]在盐胁迫时外施脱落酸的脯氨酸变化一致。甘菊与FG26的脯氨酸质量分数下降,可能暗示着脯氨酸代谢途径对高质量分数脱落酸的响应存在物种特异性。
T1处理下芙蓉菊和甘菊的超氧化物歧化酶活性上升。这与望春玉兰Magnolia biondii[61],八棱海棠Malus × robusta[62]在盐胁迫时外施脱落酸的超氧化物歧化酶的变化一致。T2及T3处理组对芙蓉菊和FG26的超氧化物歧化酶质量分数均无显著影响,表明其活性氧清除系统具备较强的稳定性。甘菊的超氧化物歧化酶质量分数易受干扰,说明其抗氧化系统较为脆弱。T1处理导致3种植物的丙二醛均有上升,这与望春玉兰[61]、红豆草Onobrychis cyri[63]在盐胁迫下的丙二醛质量摩尔浓度变化一致。芙蓉菊丙二醛质量摩尔浓度相对增幅较大,表明芙蓉菊可能具备更强的细胞膜修复能力或抗氧化能力。T2处理能显著降低甘菊的丙二醛质量摩尔浓度,这与银边吊兰Chlorophyt comosum var. variegatum[64]在盐胁迫下外施脱落酸的丙二醛变化一致。但T2处理对芙蓉菊和FG26的丙二醛影响不显著,说明脱落酸可能对盐敏感物种的丙二醛作用更强。T3处理则显著提高3种植物丙二醛质量摩尔浓度,反面证实了内源脱落酸在维持细胞膜稳定性中的作用。
在分子水平上,基于转录组数据[25],芙蓉菊脱落酸信号通路关键基因中PP2Cs家族基因在盐胁迫下表现出显著差异表达。PP2CA1和HAI2在3种植物中均呈显著上调,显示其盐胁迫响应的保守性;而HAB1、HAI1和AHG1-1则仅在芙蓉菊及FG26中表达上调,表明它们可能是芙蓉菊强耐盐性形成及其向子代传递的关键分子基础。在拟南芥中过表达AtHAIs能提高植物抗逆性[65],也有研究证实AtHAB3参与脱落酸响应过程[66]。同时,已证实天女木兰Magnolia sieboldii中MsAHG1能改变脱落酸敏感性[67],并且EsHABs、EsHAIs、EsAHG1在耐盐植物盐芥Eutrema salsugineum中可以增强耐盐性[68]。系统发育分析表明:PP2Cs基因经历了与菊科多数植物类似的基因重复事件,重复后基因的表达分化也可能为芙蓉菊耐盐特性的形成提供了分子基础。这些发现与盐胁迫下芙蓉菊脱落酸的迅速响应及其他生理变化形成了生理与分子协同的耐盐机制。
-
盐胁迫下芙蓉菊、甘菊及其杂交后代FG26的内源脱落酸质量分数均显著升高,表明脱落酸在盐胁迫响应中发挥关键作用。脱落酸可调控耐盐性,但调控效应具有物种特异性,主要通过促进芙蓉菊渗透调节和减轻甘菊细胞损伤参与植物盐胁迫响应。芙蓉菊较强的耐盐性源于脱落酸的快速响应能力、脱落酸代谢稳定性以及高效的渗透调节与较稳定的抗氧化与膜保护系统。杂交后代FG26在多数生理指标上介于两亲本之间,但更倾向于耐盐亲本,表现出一定的杂交优势。通过转录组数据和系统发育分析,发现了仅在芙蓉菊及FG26中表达上调的PP2C类基因HAB1、HAI1和AHG1-1。它们可能在芙蓉菊超强耐盐性的形成中发挥核心作用,并且能够通过杂交遗传给后代。本研究不仅揭示了脱落酸在盐胁迫中的作用及在不同耐盐性菊科植物中的调控机制,还分析了芙蓉菊及其杂交后代响应盐胁迫的生理与分子层面上的变化,为耐盐菊花新品种创制提供了可参考的理论依据。
Physiological characteristics and molecular mechanisms of salt tolerance regulated by abscisic acid in Crossostephium chinense and its hybrid progeny
-
摘要:
目的 研究盐胁迫下脱落酸(ABA)对于芙蓉菊Crossostephium chinense及杂交后代耐盐性作用机制,为菊花耐盐种质培育创新奠定基础。 方法 以耐盐种质芙蓉菊、盐敏感种质甘菊Chrysanthemum lavandulifolium及两者杂交后代中较耐盐的FG26为研究材料,分析700 mmol·L−1氯化钠(NaCl)处理下内源脱落酸动态变化;以0 mmol·L−1 氯化钠处理并施加清水为对照组(ck),研究300 mmol·L−1 氯化钠溶液与清水(T1)、T1结合200 μmol·L−1 脱落酸(T2),以及T1结合1 mmol·L−1 钨酸钠溶液(T3)3种不同处理下的生理指标变化,揭示3种不同耐盐性植物的生理差异,解析脱落酸在3种植物盐胁迫中调控机制差异;利用转录组与系统发育分析,解析脱落酸信号通路关键基因表达模式及进化历史。 结果 ①700 mmol·L−1 氯化钠溶液处理下,植物内源脱落酸质量分数均显著提高(P<0.05),芙蓉菊响应速度与峰值高于FG26和甘菊。②T2处理,3种植物的内源脱落酸质量分数较T1分别增加2.03,2.14及2.99倍,均呈上升趋势。同时芙蓉菊的脯氨酸质量分数较T1上调44.42%,甘菊和FG26的脯氨酸质量分数较T1分别降低71.74%和22.27%。芙蓉菊和FG26超氧化物歧化酶活性和丙二醛质量摩尔浓度较T1变化均不显著,甘菊的超氧化物歧化酶活性较T1下降11.45%,丙二醛质量摩尔浓度较T1显著下降37.63% (P<0.05)。③盐胁迫后,脱落酸信号通路PP2C类基因HAB1、HAI1和AHG1-1仅在芙蓉菊和FG26中特异性上调,在甘菊中变化不明显。 结论 盐胁迫响应中内源脱落酸发挥重要调控作用,外施脱落酸能特异性调控不同植物生理代谢,改善抗逆能力。芙蓉菊较强的耐盐性与高效脱落酸响应、渗透调节及稳定抗氧化与膜保护系统有关。耐盐相关性状及关键基因HAB1、HAI1和AHG1-1可通过杂交部分遗传给后代FG26。证实了芙蓉菊具有作为菊花耐盐新品种培育优良种质资源的巨大潜力。图4表1参68 Abstract:Objective This study aims to investigate the role of abscisic acid (ABA) in the salt tolerance of Crossostephium chinense and its hybrid progeny under salt stress, so as to provide a basis for breeding and innovation of salt-tolerant chrysanthemum germplasm. Method Salt-tolerant C. chinense, salt-sensitive Chrysanthemum lavandulifolium, and their relatively salt-tolerant hybrid progeny FG26 were used as materials. The dynamic changes of endogenous ABA under 700 mmol·L−1 NaCl treatment were analyzed. With 0 mmol·L−1 sodium chloride treatment and application of clear water as the control group (ck), the changes in physiological indicators under three different treatments, namely 300 mmol·L−1 NaCl stress (T1), T1 combined with 200 μmol·L−1 exogenous ABA (T2), and T1 combined with 1 mmol·L−1 ABA synthesis inhibitor Na2WO4 (T3) were examined to reveal the physiological differences among the three materials with varying salt tolerance and to clarify the differential regulatory mechanisms of ABA in their responses to salt stress. The expression patterns and evolutionary history of key genes in the ABA signaling pathway were explored by transcriptomic and phylogenetic analysis. Result (1)Under 700 mmol·L−1 NaCl treatment, the endogenous ABA mass fraction in the plants increased significantly (P<0.05). The response speed and peak value of C. chinense were higher than those of FG26 and C. lavandulifolium. (2) Compared to T1, the endogenous abscisic acid mass fractions in the three plants after T2 treatment increased by 2.03, 2.14, and 2.99 times, respectively, all showing an upward trend. Meanwhile, the proline mass fraction in C. chinense increased by 44.42% compared to T1, while that in C. lavandulifolium and FG26 decreased by 71.74% and 22.27%, respectively. Compared to T1, the superoxide dismutase activity and mass molar concentration of malondialdehyde in C. chinense and FG26 showed no significant changes, while in C. lavandulifolium, the superoxide dismutase activity decreased by 11.45%, and the mass molar concentration of malondialdehyde decreased significantly by 37.63% (P<0.05). (3) After salt stress, the expression of PP2C family genes HAB1, HAI1, and AHG1-1 in the ABA signaling pathway was specifically up-regulated only in C. chinense and FG26, while no significant change was observed in C. lavandulifolium. Conclusion Endogenous ABA plays an important regulatory role in salt stress response, and exogenous application of ABA can specifically regulate physiological metabolism in different plants to enhance stress resistance. The strong salt tolerance of C. chinense may be attributed to its efficient ABA response, osmotic adjustment, and stable antioxidant and membrane protection system. Salt tolerance related traits and key genes such as HAB1, HAI1, and AHG1-1 can be partially inherited by the progeny FG26 through hybridization. It has been confirmed that C. chinense possesses great potential as a new salt-tolerant variety for breeding. [Ch, 4 fig. 1 tab. 68 ref.] -
Key words:
- salt stress /
- Crossostephium chinense /
- ABA /
- PP2C gene family
-
表 1 系统发育分析所用的基因组或转录组数据
Table 1. Genomic and transcriptomic data used for phylogenetic analyses
科名 种名 缩写 参考文献 科名 种名 缩写 参考文献 菊科 Asteraceae 芙蓉菊Crossostephium chinense Crch [25] 茄科 Solanaceae 番茄Solanum lycopersicum Soly [46] 野菊Chrysanthemum indicum Chin [36] 十字花科 Brassicaceae 拟南芥Arabidopsis thaliana Arth [47] 菊花Chrysanthemum × morifolium Chmo [37] 杨柳科 Salicaceae 欧洲山杨Populus trichocarpa Potr [48] 菊花脑Chrysanthemum nankingense Chna [38] 葡萄科 Vitaceae 葡萄Vitis vinifera Vivi [49] 甘菊Chrysanthemum lavandulifolium Chla [39] 毛茛科 Ranunculaceae 蓝花耧斗菜Aquilegia coerulea Aqco [50] 生菜Lactuca sativa Lasa [40] 禾本科 Poaceae 玉米Zea mays Zema [51] 向日葵Helianthus annuus Hean [41] 水稻Oryza sativa Orsa [52] 牛蒡Arctium lappa Alap [42] 木兰科Magnoliaceae 鹅掌楸Liriodendron chinense Lich [53] 艾Artemisia argyi Arar [43] 无油樟科Amborellaceae 无油樟Amborella trichopoda Amtr [54] 黄花蒿Artemisia annua Aran [44] 三齿蒿Artemisia tridentata Artr [45] -
[1] Food and Agriculture Organization of the United Nations. Global Status of Salt-affected Soils-Main Report [R/OL]. Rome: Food and Agriculture Organization of the United Nations, 2024[2025-08-18]. DOI:10.4060/cd3044en. [2] van ZELM E, ZHANG Yanxia, TESTERINK C. Salt tolerance mechanisms of plants [J]. Annual Review of Plant Biology, 2020, 71: 403−433. [3] ZHOU Huapeng, SHI Haifan, YANG Yongqing, et al. Insights into plant salt stress signaling and tolerance [J]. Journal of Genetics and Genomics, 2024, 51(1): 16−34. [4] 焦明翠, 蔡立格, 魏健, 等. 盐胁迫的生理危害与植物的适应机制研究进展[J]. 长春师范大学学报, 2023, 42(6): 125−132. JIAO Mingcui, CAI Lige, WEI Jian, et al. Research progress on the physiological harms of salt stress and the adaptation mechanism of plants [J]. Journal of Changchun Normal University, 2023, 42(6): 125−132. [5] ZHU Zhihui, ZHOU Yuqing, LIU Xiuyue, et al. Integrated transcriptomic and metabolomic analyses uncover the key pathways of Limonium bicolor in response to salt stress [J]. Plant Biotechnology Journal, 2025, 23(3): 715−730. [6] MAURYA V, SINGH N, SHARMA I, et al. Effect of melatonin in regulating salt stress responses in plants[M]// SHARMA A, AHAMMED G J. Melatonin in Plants: Role in Plant Growth, Development, and Stress Response. Singapore: Springer Nature Singapore, 2024: 109−139. [7] KOLUPAEV Y E, YASTREB T O. Role of jasmonate and jasmonate signaling components in plant adaptation to salt stress[M]//YASTREB T O. Regulation of Adaptive Responses in Plants. New York: Nova Science Publishers, 2024: 161−207. [8] MIR R A, ARYENDU A, SOMASUNDARAM R. Salicylic acid and salt stress tolerance in plants: a review [J]. Journal of Stress Physiology and Biochemistry, 2021, 17(3): 32−50. [9] GHASSEMI-GOLEZANI K, SAMEA-ANDABJADID S. Cytokinin signaling in plants under salt stress[M]// AFTAB T. Auxins, Cytokinins and Gibberellins Signaling in Plants. Cham: Springer International Publishing, 2022: 189−212. [10] SONG Xin, ZHANG Miao, WANG Tingting, et al. Polyploidization leads to salt stress resilience via ethylene signaling in Citrus plants [J]. New Phytologist, 2025, 246(1): 176−191. [11] BASHARAT S, SAEED W, LIU Pingwu, et al. Abscisic acid mediated salinity stress tolerance in crops[J/OL]. Plant Hormones, 2025, 1(1): e015[2025-07-30]. DOI: 10.48130/ph-0025-0014. [12] YANG Xinhui, LIU Zisheng, CHEN Jun, et al. PP2C-mediated ABA signaling pathway underlies exogenous abscisic acid-induced enhancement of saline-alkaline tolerance in potato (Solanum tuberosum L. )[J/OL]. Plants, 2025, 14(13): 1921[2025-07-30]. DOI: 10.3390/plants14131921. [13] 张云霞, 石勇, 王瑞刚, 等. 初始盐胁迫下ABA与CaM对胡杨叶片气体交换的调控[J]. 林业科学, 2008, 44(1): 57−64. ZHANG Yunxia, SHI Yong, WANG Ruigang, et al. Effects of ABA and CaM on leaf gas exchange of Populus euphratica in the process of initial salinity [J]. Scientia Silvae Sinicae, 2008, 44(1): 57−64. [14] 邓昌哲, 安飞飞, 李开绵, 等. 外源ABA及其抑制剂钨酸钠对木薯块根类胡萝卜素相关基因和蛋白的影响[J]. 生物技术通报, 2017, 33(11): 76−83. DENG Changzhe, AN Feifei, LI Kaimian, et al. Effects of ABA and its synthesis inhibitor sodium tungstate on carotenoid associated genes and enzymes of cassava tuber root [J]. Biotechnology Bulletin, 2017, 33(11): 76−83. [15] ZHANG Qitong, ZHANG Lili, GENG Biao, et al. Interactive effects of abscisic acid and nitric oxide on chilling resistance and active oxygen metabolism in peach fruit during cold storage [J]. Journal of the Science of Food and Agriculture, 2019, 99(7): 3367−3380. [16] LI Wenfang, MAO Juan, SU Jing, et al. Exogenous ABA and its inhibitor regulate flower bud induction of apple cv. ‘Nagafu No. 2’ grafted on different rootstocks [J]. Trees, 2021, 35(2): 609−620. [17] SAINI L K, SINGH N, PANDEY G K. Plant protein phosphatase 2C: critical negative regulator of ABA signaling[M]//PANDEY G K. Protein Phosphatases and Stress Management in Plants. Cham: Springer International Publishing, 2020: 83−102. [18] HEWAGE K A H, YANG Jingfang, WANG Di, et al. Chemical manipulation of abscisic acid signaling: a new approach to abiotic and biotic stress management in agriculture[J/OL]. Advanced Science, 2020, 7(18): 2001265[2025-07-30]. DOI: 10.1002/advs.202001265. [19] HOANG X L T, NHI D N H, THU N B A, et al. Transcription factors and their roles in signal transduction in plants under abiotic stresses [J]. Current Genomics, 2017, 18(6): 483−497. [20] ZHENG Yuan, CHEN Zhaojin, MA Liang, et al. The ubiquitin E3 ligase RHA2b promotes degradation of MYB30 in abscisic acid signaling [J]. Plant Physiology, 2018, 178(1): 428−440. [21] NIE Kaili, ZHAO Hongyun, WANG Xiaopei, et al. The MIEL1-ABI5/MYB30 regulatory module fine tunes abscisic acid signaling during seed germination [J]. Journal of Integrative Plant Biology, 2022, 64(4): 930−941. [22] ZHAN Qidi, SHEN Jialu, NIE Kaili, et al. MIW1 participates in ABA signaling through the regulation of MYB30 in Arabidopsis[J/OL]. Plant Science, 2023, 332: 111717[2025-07-30]. DOI: 10.1016/j.plantsci.2023.111717. [23] 刘淼. 基于转录组学的芙蓉菊与甘菊杂交后代耐盐机制研究[D]. 北京: 北京林业大学, 2022. LIU Miao. Study of the Salt Tolerance Mechanism in the Hybrids of Crossostephium chinense and Chrysanthemum lavandulifolium Based on Transcriptomic Analyses [D]. Beijing: Beijing Forestry University, 2022. [24] 陈俊通. 广义菊属远缘杂交障碍及耐盐种质创制的研究[D]. 北京: 北京林业大学, 2019. CHEN Juntong. Exploration on Reproductive Barriers in Distant Hybridization and Creation of Salt-tolerant Germplasms within Chrysanthemum in Broad Sense [D]. Beijing: Beijing Forestry University, 2019. [25] WANG Yuxin, LIU Miao, GUO Ziyu, et al. Comparative physiological and transcriptome analysis of Crossostephium chinense reveals its molecular mechanisms of salt tolerance[J/OL]. International Journal of Molecular Sciences, 2023, 24(23): 16812[2025-07-30]. DOI: 10.3390/ijms242316812. [26] ZHAO Jing, LI Gang, YI Guoxiang, et al. Comparison between conventional indirect competitive enzyme-linked immunosorbent assay (icELISA) and simplified icELISA for small molecules [J]. Analytica Chimica Acta, 2006, 571(1): 79−85. [27] ÁBRAHÁM E, HOURTON-CABASSA C, ERDEI L, et al. Methods for determination of proline in plants[M]// SUNKAR R. Plant Stress Tolerance. Totowa: Humana Press, 2010: 317−331. [28] STEWART R R, BEWLEY J D. Lipid peroxidation associated with accelerated aging of soybean axes [J]. Plant Physiology, 1980, 65(2): 245−248. [29] 赵世杰, 许长成, 邹琦, 等. 植物组织中丙二醛测定方法的改进[J]. 植物生理学通讯, 1994, 30(3): 207−210. ZHAO Shijie, XU Changcheng, ZOU Qi, et al. Improvement of determination method of malondialdehyde in plant tissues [J]. Plant Physiology Communications, 1994, 30(3): 207−210. [30] CHEN Chengjie, CHEN Hao, ZHANG Yi, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Molecular Plant, 2020, 13(8): 1194−1202. [31] CAMACHO C, COULOURIS G, AVAGYAN V, et al. BLAST+: architecture and applications[J/OL]. BMC Bioinformatics, 2009, 10: 421[2025-07-30]. DOI: 10.1186/1471-2105-10-421. [32] KATOH K, STANDLEY D M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability [J]. Molecular Biology and Evolution, 2013, 30(4): 772−780. [33] SUYAMA M, TORRENTS D, BORK P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding Codon alignments[J/OL]. Nucleic Acids Research, 2006, 34(suppl 2): W609-W612[2025-07-30]. DOI: 10.1093/nar/gkl315. [34] MINH B Q, SCHMIDT H A, CHERNOMOR O, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era [J]. Molecular Biology and Evolution, 2020, 37(5): 1530−1534. [35] TAMURA K, STECHER G, KUMAR S. MEGA11: molecular evolutionary genetics analysis version 11 [J]. Molecular Biology and Evolution, 2021, 38(7): 3022−3027. [36] DENG Yin’ai, YANG Peng, ZHANG Qianle, et al. Genomic insights into the evolution of flavonoid biosynthesis and O-methyltransferase and glucosyltransferase in Chrysanthemum indicum[J/OL]. Cell Reports, 2024, 43(2): 113725[2025-07-30]. DOI: 10.1016/j.celrep.2024.113725. [37] SONG Aiping, SU Jiangshuo, WANG Haibin, et al. Analyses of a chromosome-scale genome assembly reveal the origin and evolution of cultivated Chrysanthemum[J/OL]. Nature Communications, 2023, 14(1): 2021[2025-07-30]. DOI: 10.1038/s41467-023-37730-3. [38] SONG Chi, LIU Yifei, SONG Aiping, et al. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of Chrysanthemum flowers and medicinal traits [J]. Molecular Plant, 2018, 11(12): 1482−1491. [39] WEN Xiaohui, LI Junzhuo, WANG Lili, et al. The Chrysanthemum lavandulifolium genome and the molecular mechanism underlying diverse Capitulum types[J/OL]. Horticulture Research, 2022, 9: uhab022[2025-07-30]. DOI: 10.1093/hr/uhab022. [40] SHEN Fei, QIN Yajuan, WANG Rui, et al. Comparative genomics reveals a unique nitrogen-carbon balance system in Asteraceae[J/OL]. Nature Communications, 2023, 14(1): 4334[2025-07-30]. DOI: 10.1038/s41467-023-40002-9. [41] BADOUIN H, GOUZY J, GRASSA C J, et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution [J]. Nature, 2017, 546(7656): 148−152. [42] FAN Wei, WANG Sen, WANG Hengchao, et al. The genomes of chicory, endive, great burdock and yacon provide insights into Asteraceae Palaeo-polyploidization history and plant inulin production [J]. Molecular Ecology Resources, 2022, 22(8): 3124−3140. [43] CHEN Hongyu, GUO Miaoxian, DONG Shuting, et al. A chromosome-scale genome assembly of Artemisia argyi reveals unbiased subgenome evolution and key contributions of gene duplication to volatile terpenoid diversity[J/O]. Plant Communications, 2023, 4(3): 100516[2025-07-30]. DOI: 10.1016/j.xplc.2023.100516. [44] SHEN Qian, ZHANG Lida, LIAO Zhihua, et al. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis [J]. Molecular Plant, 2018, 11(6): 776−788. [45] MELTON A E, CHILD A W, BEARD R S, et al. A haploid pseudo-chromosome genome assembly for a keystone sagebrush species of western North American rangelands[J/OL]. G3, 2022, 12(7): jkac122[2025-07-30]. DOI: 10.1093/g3journal/jkac122. [46] ZHOU Yao, ZHANG Zhiyang, BAO Zhigui, et al. Graph pangenome captures missing heritability and empowers tomato breeding [J]. Nature, 2022, 606(7914): 527−534. [47] LAMESCH P, BERARDINI T Z, LI Donghui, et al. The Arabidopsis information resource (TAIR): improved gene annotation and new tools[J/OL]. Nucleic Acids Research, 2012, 40: D1202-D1210[2025-07-30]. DOI: 10.1093/nar/gkr1090. [48] TUSKAN G A, DIFAZIO S, JANSSON S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) [J]. Science, 2006, 313(5793): 1596−1604. [49] JAILLON O, AURY J M, NOEL B, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm Phyla [J]. Nature, 2007, 449(7161): 463−467. [50] FILIAULT D L, BALLERINI E S, MANDÁKOVÁ T, et al. The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history[J/OL]. eLife, 2018, 7: e36426[2025-07-30]. DOI: 10.7554/eLife.36426. [51] HUFFORD M B, SEETHARAM A S, WOODHOUSE M R, et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes[J]. Science, 2021, 373(6555): 655−662. [52] OUYANG Shu, ZHU Wei, HAMILTON J, et al. The TIGR rice genome annotation resource: improvements and new features[J/OL]. Nucleic Acids Research, 2007, 35: D883-D887[2025-07-30]. DOI: 10.1093/nar/gkl976. [53] CHEN Jinhui, HAO Zhaodong, GUANG Xuanmin, et al. Author correction: Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation[J/OL]. Nature Plants, 2019, 5(3): 328[2025-07-30]. DOI: 10.1038/s41477-018-0323-6. [54] CAREY S B, AKÖZBEK L, LOVELL J T, et al. ZW sex chromosome structure in Amborella trichopoda [J]. Nature Plants, 2024, 10(12): 1944−1954. [55] VEGA A, O’BRIEN J A, GUTIÉRREZ R A. Nitrate and hormonal signaling crosstalk for plant growth and development [J]. Current Opinion in Plant Biology, 2019, 52: 155−163. [56] 马祥, 李中兴, 杨荣尘, 等. 盐胁迫对不同耐盐性燕麦糖类及内源激素含量变化的影响[J/OL]. 草业学报, 2025-09-11[2025-07-30]. http://kns.cnki.net/kcms/detail/62.1105.S.20250910.1258.004.html. MA Xiang, LI Zhongxing, YANG Rongchen, et al. The effect of salt stress on the changes of sugar and endogenous hormone content in oats with different salt tolerance [J]. Acta Prataculturae Sinica, 2025-09-11[2025-07-30]. http://kns.cnki.net/kcms/detail/62.1105.S.20250910.1258.004.html. [57] 李海洋, 李爱学, 王成, 等. 盐胁迫对苗期向日葵内源激素含量的影响[J]. 干旱地区农业研究, 2018, 36(6): 92−97. LI Haiyang, LI Aixue, WANG Cheng, et al. Effects of salt stress on endogenous hormone contents in sunflower seedlings [J]. Agricultural Research in the Arid Areas, 2018, 36(6): 92−97. [58] 张钰, 陈慧, 王改萍. 外源ABA对楸树幼苗NaCl胁迫的缓解效应及其生长生理响应特征[J]. 西北植物学报, 2023, 43(6): 996−1005. ZHANG Yu, CHEN Hui, WANG Gaiping. Alleviating effects of exogenous ABA on Catalpa bungei seedlings under NaCl stress and growth physiological response characteristics [J]. Acta Botanica Boreali-Occidentalia Sinica, 2023, 43(6): 996−1005. [59] 沈惠娟, 李梅枝, 梁成喜. 盐胁迫下ABA对刺槐幼苗体内腐胺、脯氨酸和保护酶系统的影响[J]. 浙江林学院学报, 1992, 9(3): 57−63. SHEN Huijuan, LI Meizhi, LIANG Chengxi. Effects of ABA on putrescine, proline and protective enzyme system in Robinia pseudoacacia seedlings under salt stress [J]. Journal of Zhejiang Forestry College, 1992, 9(3): 57−63. [60] 马福钦, 王彦, 郑晓琳, 等. 盐胁迫下外源脱落酸对鹰嘴紫云英种子萌发及幼苗生理特性的影响[J]. 核农学报, 2025, 39(8): 1797−1806. MA Fuqin, WANG Yan, ZHENG Xiaolin, et al. Effects of exogenous abscisic acid on seed germination and seedling physiological characteristics of Astragalus cicer seedlings under salt stress [J]. Journal of Nuclear Agricultural Sciences, 2025, 39(8): 1797−1806. [61] 沈徐悦, 张浪, 陈蓉蓉, 等. 盐胁迫对望春玉兰幼苗形态和相关生理指标的影响[J]. 浙江农林大学学报, 2021, 38(2): 289−295. SHEN Xuyue, ZHANG Lang, CHEN Rongrong, et al. Effects of salt stress on morphology and related physiological indices of Magnolia biondii seedlings [J]. Journal of Zhejiang A&F University, 2021, 38(2): 289−295. [62] 王亚丽. 外源ABA对盐胁迫下八棱海棠苗木生长及生理特性的影响[J]. 山西林业科技, 2024, 53(2): 25−28. WANG Yali. Effect of exogenous ABA on the growth and physiological characteristics of Malus robusta seedlings under salt stress [J]. Shanxi Forestry Science and Technology, 2024, 53(2): 25−28. [63] 田戈, 南丽丽, 王利群, 等. 盐胁迫下外源ABA对红豆草幼苗生长与生理特性的影响[J]. 草业学报, 2025, 34(10): 95−106. TIAN Ge, NAN Lili, WANG Liqun, et al. Effects of exogenous ABA on growth and physiological characteristics of Onobrychis cyri seedlings under salt stress [J]. Acta Prataculturae Sinica, 2025, 34(10): 95−106. [64] 宁朋, 王菲, 肖雨, 等. 外源ABA与盐胁迫对银边吊兰生长及生理特性的影响[J]. 江西农业大学学报, 2021, 43(2): 287−295. NING Peng, WANG Fei, XIAO Yu, et al. Effects of ABA and salt stress on the growth and physiological characteristics of Chlorophytum comosum var. variegatum [J]. Acta Agriculturae Universitatis Jiangxiensis, 2021, 43(2): 287−295. [65] ZHANG Jihong, LI Xiushan, HE Zhimin, et al. Molecular character of a phosphatase 2C (PP2C) gene relation to stress tolerance in Arabidopsis thaliana [J]. Molecular Biology Reports, 2013, 40(3): 2633−2644. [66] 王博雅. 拟南芥中NSOS1与HAB3基因调控逆境应答的分子机理研究[D]. 杨凌: 西北农林科技大学, 2014. WANG Boya. Study of the Molecular Mechanisms of AtNSOS1 and AtHAB3 Genes Regulating Stress Response in Arabidopsis thaliana [D]. Yangling: Northwest agriculture and forestry university of science and technology, 2014. [67] 孙宏涛. 天女木兰A类PP2C基因克隆及其在种子休眠解除中的功能验证[D]. 沈阳: 沈阳农业大学, 2022. SUN Hongtao. Cloning of Group A PP2C Gene in Magnolia sieboldii and Its Functional Verification in the Release of Seed Dormancy [D]. Shenyang: Shenyang Agricultural University, 2022. [68] 张恒阳. 盐芥PP2C基因应对非生物胁迫分子机制及功能研究[D]. 济南: 山东师范大学, 2024. ZHANG Hengyang. Research on Molecular Mechanisms and Function of Eutrema salsugineum PP2C Gene in Response to Abiotic Stress [D]. Ji’nan: Shandong Normal University, 2024. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.20250469






 下载:
下载: