-
种子萌发是高等植物生命周期新的开始,它始于吸水止于胚根突出,经历一系列复杂的生物学过程。一般情况,种子萌发可划分为3个发育阶段:3 h内种子迅速吸水膨胀(阶段Ⅰ),随后储藏物动员(阶段Ⅱ),最后胚根突出完成萌发(阶段Ⅲ)[1]。种子萌发完成后将进入幼苗发育阶段,其中胚根向下生长逐渐发育为植物的根,而胚轴和胚芽向上生长发育为植物的茎和叶。在深层土壤中,光很少参与种子萌发的调控。因此,研究黑暗条件下种子萌发的分子调控网络,不仅有助于揭示种子暗萌发的分子机制,而且对于指导作物播种也具有重要意义。
经过长期自然选择,高等植物已进化出多种光感受器,以适应复杂多变的自然环境。在拟南芥Arabidopsis thaliana中,至今已发现3类光受体,即红光/远红光受体光敏色素、蓝光受体隐花色素以及紫外光受体。在调节种子萌发方面,以光敏色素(PHY)研究较为深入。在黑暗中,PHY失活,其靶标蛋白光敏色素互作因子(PIF1)积累,抑制赤霉素(GA)信号和激发脱落酸(ABA)信号,从而抑制种子萌发;在红光或远红光下,光敏色素B (PHYB) 或光敏色素A (PHYA)被激活促进PIF1降解,减除 PIF1 对GA信号的抑制和ABA信号的激发,从而促进种子萌发[2−4]。
不同物种光调控种子萌发的机制存在一定差异。如光促进拟南芥种子萌发,而抑制番茄Solanum lycopersicum种子萌发。在拟南芥中,PHYA 和 PHYB 分别感知远红光和红光调控种子萌发[5−6],然而,在番茄中红光或远红光下PHYA均抑制种子萌发[7]。DONG等[8]对244份烟草Nicotiana tabacum种质资源进行研究,根据萌发对光的敏感性将烟草种子划分为浅光休眠种子和深度光休眠种子,浅光休眠种子在光下萌发率>90%,暗光下萌发率>50%;而深度光休眠种子在光下萌发率>90%,暗光下萌发率<20%。本研究以浅光休眠种子类群中暗萌发率较高的代表野生型烟草Y85为材料,对萌发前后的种子样本进行转录组和蛋白组测序,通过联合分析挖掘黑暗下浅光休眠种子萌发的调控网络。
-
野生型烟草Y85种子由贵州省烟草科学研究院提供。为避免光休眠、成熟度和后熟期差异对实验结果造成影响,种子均为授粉后40 d收获,在40 ℃机械烘干36 h脱粒。
-
采用质量分数为0.5%硫酸铜溶液进行种子消毒处理;15 min后,用蒸馏水反复冲洗已消毒的种子;最后用滤纸吸附种子表面水分待用。配置质量分数为0.8%的琼脂溶液,进行高压灭菌,将灭菌液倒入直径为9 cm的培养皿中制作琼脂发芽床。每个处理设3次重复,每次重复100粒种子,以10×10模式均匀点播在琼脂床表面,然后将培养皿放置在人工气候箱中(江南仪器RXZ-36C),设置人工气候箱的温度为25 ℃,光照强度为0 lx,相对湿度为80%,进行种子萌发实验。
-
分别收集暗萌发第2天和第4天的种子作为转录组测序的样本。使用TRIzol试剂(Tiagen Biochemical)提取种子的总RNA。使用NanoDrop 2000分光光度计(Thermo Science)评估纯度和定量,同时使用Agilent 2100生物分析仪(Agilent Technologies)评估RNA的完整性。使用TruSeq Stranded mRNA LT Sample Prep Kit (Illumina)构建基因库。转录物测序和分析由APTBIO生物技术有限公司(中国上海)完成。文库在Illumina HiSeq X Ten平台上进行测序,产生150 bp的成对末端读数。使用Trimomatic[9]处理FASTQ格式的原始读数,并为每个样品生成原始读数。通过从原始数据中去除低质量的读数和poly-N的读数,获得准确的读数用于深入分析。使用HISAT2[10]将处理后的读数映射到野生型烟草Y85基因组上。使用Cufflinks[11]计算每个基因的每千碱基转录每百万映射读取的片段值[12],使用HTSeq-Coun获得每个基因的读数。差异表达分析使用DESeq (2012)R[13]进行。P<0.05和差异倍数(FC)>2.0或<0.5被用作差异表达的阈值。对差异表达基因(DEGs)进行了层次聚类分析,以探索基因表达模式。随后,应用数学模型对转录组数据进行分析。首先,筛选出在2和4 d发芽的DEGs,根据超几何分布,使用R软件包对DEGs进行了基因本体(GO)富集及京都基因和基因组百科全书(KEGG)路径富集分析。
-
将种子样品在液氮中磨成细粉,用3 mL含有1 mm 苯甲基磺酰氟的提取缓冲液[50 mmol·L−1 三羟甲基氨基甲烷盐酸盐(Tris-HCl),pH 8.0,0.1 mol·L−1 氯化钾(KCl),5 mmol·L−1 EDTA,质量分数为30%蔗糖]提取500 mg样品,持续2 h。在13 000 r·min−1下离心15 min后,将上清液转移到新的试管中。在上清液中加入5倍体积的体积分数为10%冷TCA丙酮,在20 ℃下保存2 h,然后以13 000 r·min−1离心15 min。用体积分数为90%的冷丙酮冲洗,在20 ℃下放置30 min,然后以13 000 r·min−1离心5 min。这个步骤重复3次。
冻干后,最终的颗粒在80 ℃下储存或立即进行分析。将蛋白质样品加入0.5 mL标准缓冲液(质量分数为4%十二烷基硫酸钠,1 mmol·L−1 二硫苏糖醇,150 mmol·L−1 Tris-HCl,pH 8.0),在沸水中孵育5 min,然后超声处理10次(持续时间为5 min,时间间隔为5 min)。
在13 000 r·min−1下离心40 min后,用聚氰基丙烯酸正丁酯法测定蛋白质浓度。用200 μL 尿酸缓冲液(8 mol·L−1尿素,150 mmol·L−1 Tris-HCl,pH 8.0)稀释约200 μg蛋白质样品,在14 000 r·min−1下离心30 min,然后加入200 μL UA缓冲液(50 mmol·L−1碘乙酰胺)。加入100 μL UA缓冲液,在黑暗中孵化30 min,离心20 min,重复2次。加入100 μL的溴酚蓝缓冲液(50 mmol·L−1三乙基碳酸铵,pH 8.5),离心20 min。此步骤重复2次,然后加入40 μL胰蛋白酶溶液(在40 μL DS缓冲液中加入2 μg Pro mega的胰蛋白酶)。样品在37 ℃下培养约17 h,并通过离心收集多肽。用BCA法测定多肽含量。光密度[D(280)]为1.1,表示1 g·L−1。使用iTRAQ (Applied Biosystems) 4plex试剂盒的标准方案对每个样品约100 μg多肽分别进行标记[14]。
-
使用TIANGEN RNA Prep Pure植物总RNA提取试剂盒(离心柱型)提取总RNA。使用NanoDrop-2000测定RNA的浓度和纯度。使用TaKaRaPrimeScript™ II First Strand cDNA Synthesis Kit将提取的RNA进行逆转录。使用TaKaRaTBGreen®Premix Ex Taq™ II (TliRNaseH Plus), Batch Fluorescence Quantification Kit 20 µL反应系统进行RT-PCR:10 µL TB Green Premix Ex Taq(2×) (TliRNaseH Plus),0.4 µL ROX参考染料(50×),2 µL稀释至40 mg·L−1的cDNA,0.8 µL上游和下游引物,其余用水补足。RT-PCR反应的条件是在95 ℃下预变性0.5 min,95 ℃循环5 s,58 ℃循环30 s。使用Step One Plus实时PCR仪测定每个基因的相对表达水平,每个样品重复3次。根据LIVAK等[15]的方法计算相对表达水平[15]。
-
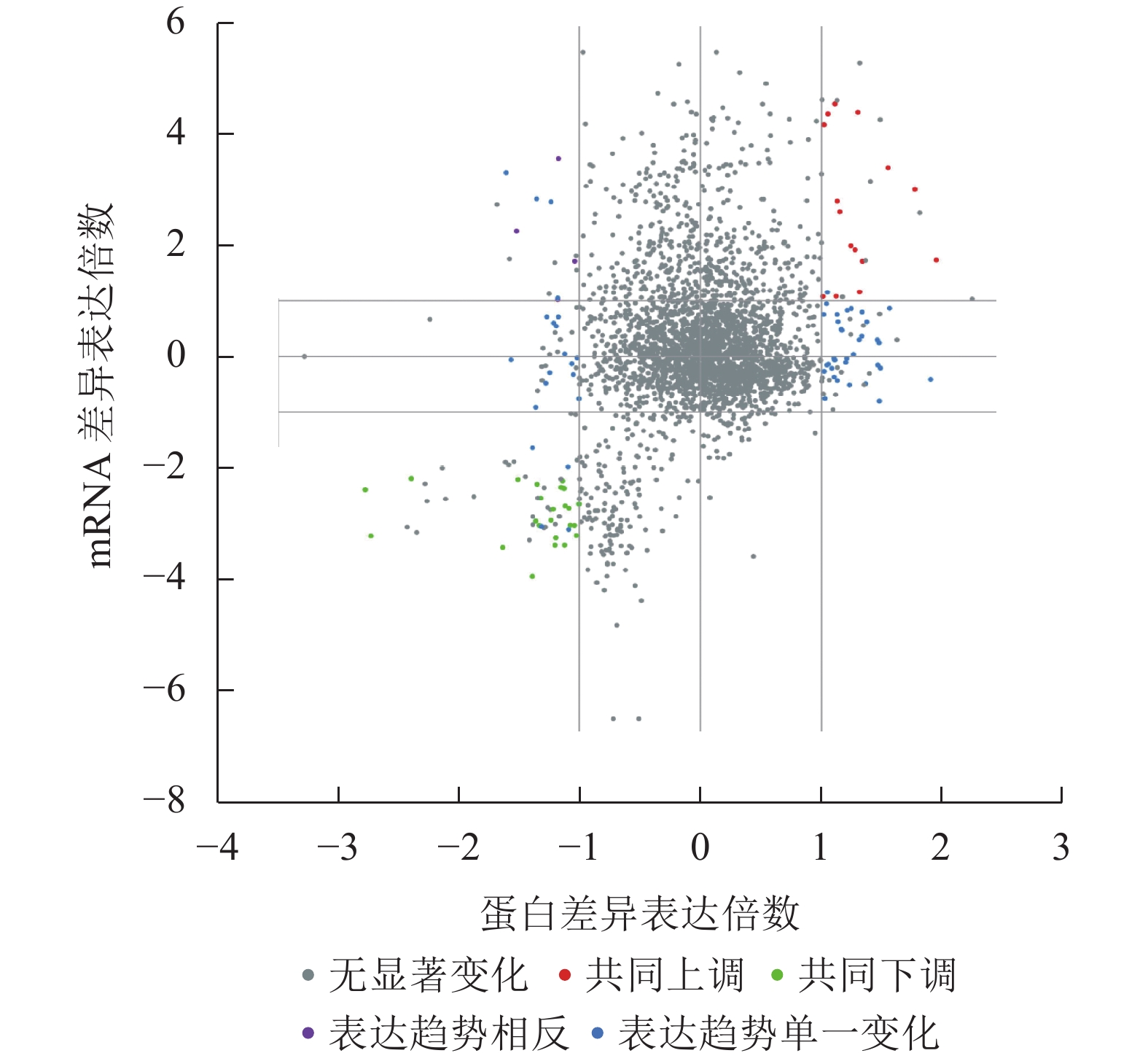
结果(图1和图2)发现:胚根突出前后种子的差异表达mRNA与蛋白可划分为16个象限,包括5种类型。其中,转录和翻译两者同时无差异的蛋白(基因)最多;其次为仅在蛋白或者mRNA一种水平下差异表达的蛋白(基因);再次为同时下调的差异蛋白(基因),有24个;再次为同时上调的差异蛋白(基因),有15个;而表达趋势相反的差异蛋白(基因)最少,仅为3个。上述结果说明:黑暗条件在转录和翻译水平同时上调或者下调的蛋白(基因)数量相对较少,而更多的基因是在转录或者翻译单一水平表达参与调控胚根突出。
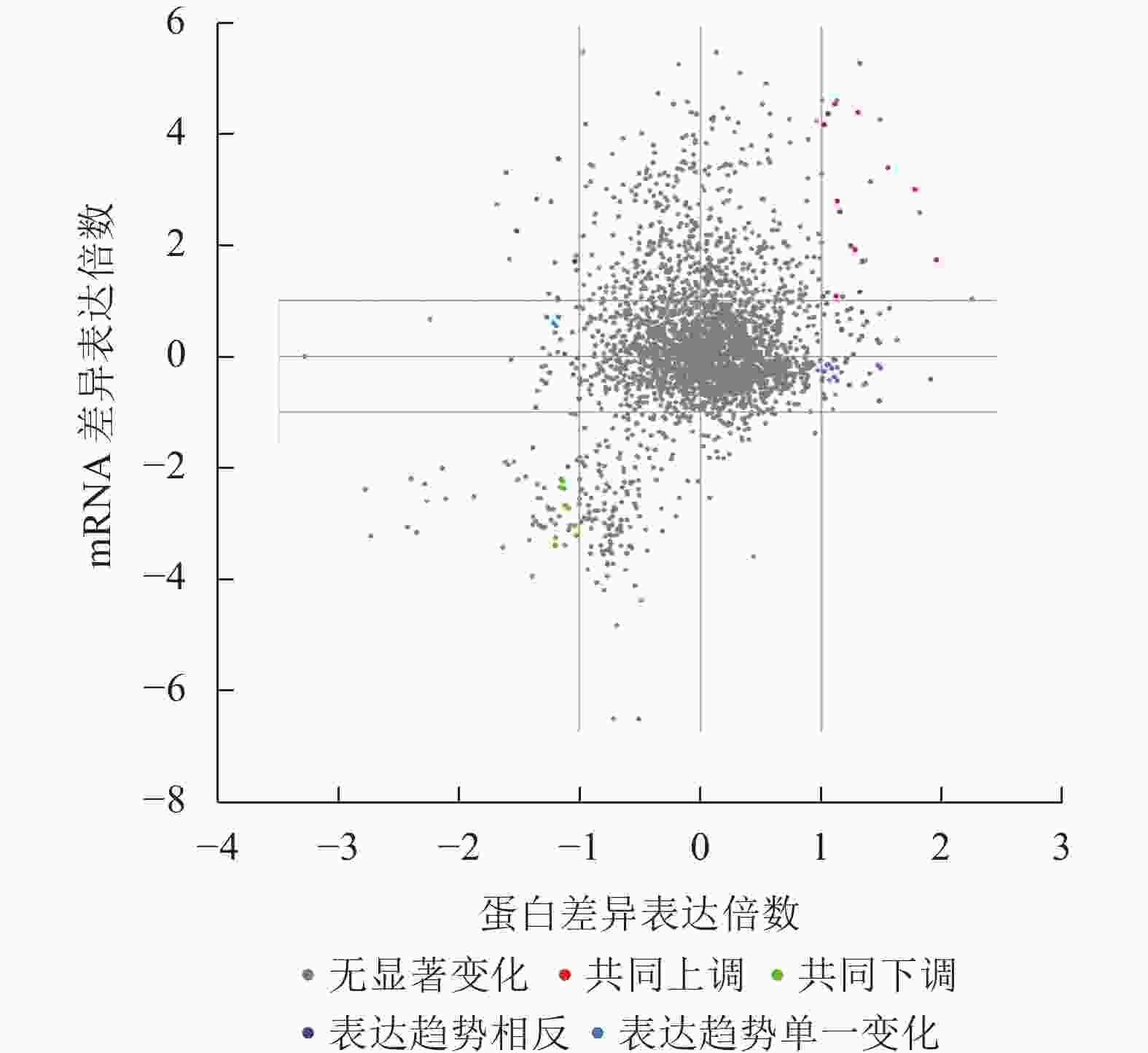
图 1 差异表达基因和差异表达蛋白象限图
Figure 1. Quadrant map of differentially expressed genes and differentially expressed proteins

图 2 共上调(下调)差异表达基因(蛋白)维恩分析示意图
Figure 2. Schematic diagram of Wien analysis of co-upregulated (co-downregulated) differentially expressed genes (proteins)
本研究重点关注了胚根突出后转录和翻译共同上调或者下调的蛋白(基因),共同上调表达的蛋白(基因)有β-葡糖苷酶(BoGH3B)、内甘露聚糖-1,4-β-甘露糖苷酶1 (MAN1)、甲基转移酶 PMT21 (ERD3)、MLP-like protein 31 (MLP31)、脂肪酸结合蛋白1 (FAP1)、非特异性脂质转移蛋白C、60伴侣蛋白2β亚基、叶绿体(CPN60B2)、线粒体解偶联蛋白1 (PUMP1)、豇豆球蛋白(CYSEP)、过氧化物酶24 (PER24)、B8网状内皮素蛋白(RTNLB8)等(图2和表1)。而共同下调表达的蛋白(基因)有坏死稳定素1 (NEC1)、母体植株开花时间调控蛋白(MFT)、胚胎晚期丰度蛋白63 (ECP63)、Peroxygenase (SOP1)、跨膜蛋白205 (TMEM205)、胚胎晚期丰度蛋白25 (LE25)、种子维生素内含蛋白(SBP65)、应激蛋白(At3g01520)、莨菪碱6-双加氧酶(H6H)、聚二磷酸腺苷核糖聚合酶3 (PARP3)等(图2和表1)。
表 1 差异表达基因和差异表达蛋白联合分析
Table 1. Combined analysis of differentially expressed genes and differentially expressed proteins
登录号 转录差异 翻译差异 基因名称 上下调基因/蛋白 倍数 P 倍数 P Nitab4.5_0000010g0130.1 3.325 5 0.001 7 3.853 8 0.000 4 BoGH3B 上调 Nitab4.5_0000023g0130.1 20.474 7 0.000 0 2.071 3 0.000 9 MAN1 上调 Nitab4.5_0000108g0440.1 2.120 7 0.001 4 2.171 9 0.014 5 ERD3 上调 Nitab4.5_0001238g0070.1 6.056 6 0.009 8 2.218 9 0.005 6 MLP31 上调 Nitab4.5_0001550g0100.1 3.766 1 0.000 0 2.421 4 0.000 4 FAP1 上调 Nitab4.5_0006538g0070.1 2.239 3 0.031 9 2.485 4 0.010 9 CPN60B2 上调 Nitab4.5_0008198g0010.1 2.110 4 0.001 6 2.014 6 0.034 4 PUMP1 上调 Nitab4.5_0010931g0010.1 20.901 6 0.000 0 2.461 1 0.001 5 CYSEP 上调 Nitab4.5_0014169g0010.1 6.893 3 0.016 7 2.187 9 0.003 1 PER24 上调 Nitab4.5_0014487g0010.1 3.953 8 0.000 0 2.362 0 0.011 5 RTNLB8 上调 Nitab4.5_0000541g0010.1 0.064 3 0.000 0 0.381 9 0.006 4 NEC1 下调 Nitab4.5_0000649g0080.1 0.095 6 0.046 2 0.459 1 0.018 2 MFT 下调 Nitab4.5_0000680g0110.1 0.196 4 0.000 0 0.450 5 0.031 1 ECP63 下调 Nitab4.5_0001051g0080.1 0.159 4 0.001 4 0.498 9 0.009 4 SOP1 下调 Nitab4.5_0001378g0010.1 0.155 3 0.000 0 0.460 8 0.026 6 TMEM205 下调 Nitab4.5_0001538g0010.1 0.107 0 0.009 4 0.151 8 0.014 4 LE25 下调 Nitab4.5_0002674g0020.1 0.149 6 0.000 1 0.430 3 0.007 4 SBP65 下调 Nitab4.5_0003715g0060.1 0.151 4 0.033 7 0.470 9 0.000 1 At3g01520 下调 Nitab4.5_0004232g0030.1 0.095 5 0.000 0 0.435 2 0.001 6 H6H 下调 Nitab4.5_0005388g0040.1 0.104 5 0.001 0 0.437 4 0.034 1 At3g06035 下调 Nitab4.5_0017202g0010.1 0.122 7 0.000 0 0.475 3 0.001 9 PARP3 下调 -
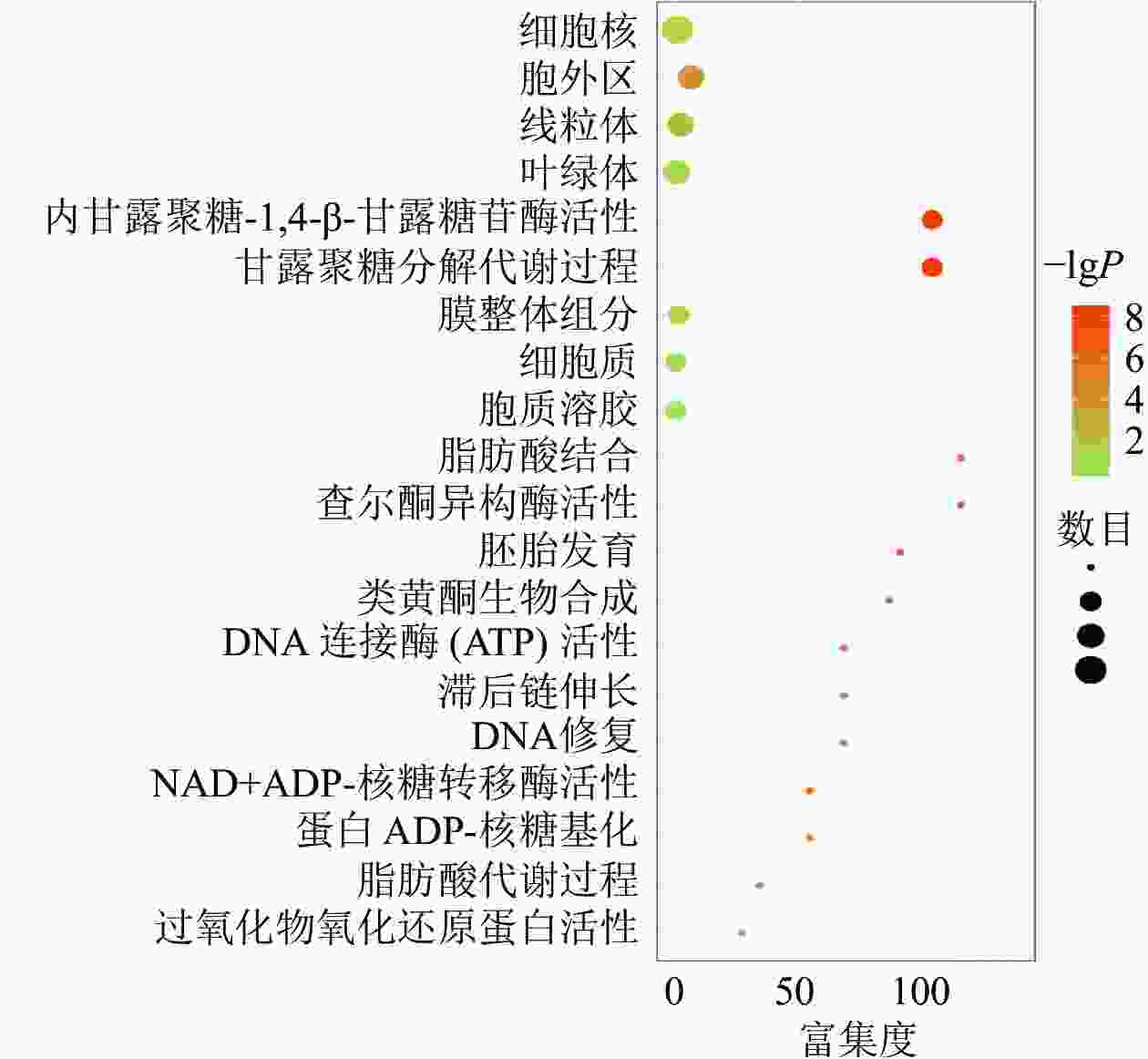
对同时上调或下调的蛋白(基因)进行GO和KEGG富集,富集到的信号通路主要包括果糖和甘露糖代谢、甘露聚糖分解、内甘露聚糖-1,4-β-甘露糖苷酶活性等,发生功能的细胞成分包括细胞外区域、线粒体、膜的组成部分和叶绿体等(图3)。说明在黑暗下种子萌发可能依赖于甘露聚糖分解代谢,而线粒体和叶绿体2个细胞器在种子暗萌发中可能均具有一定的作用。
-
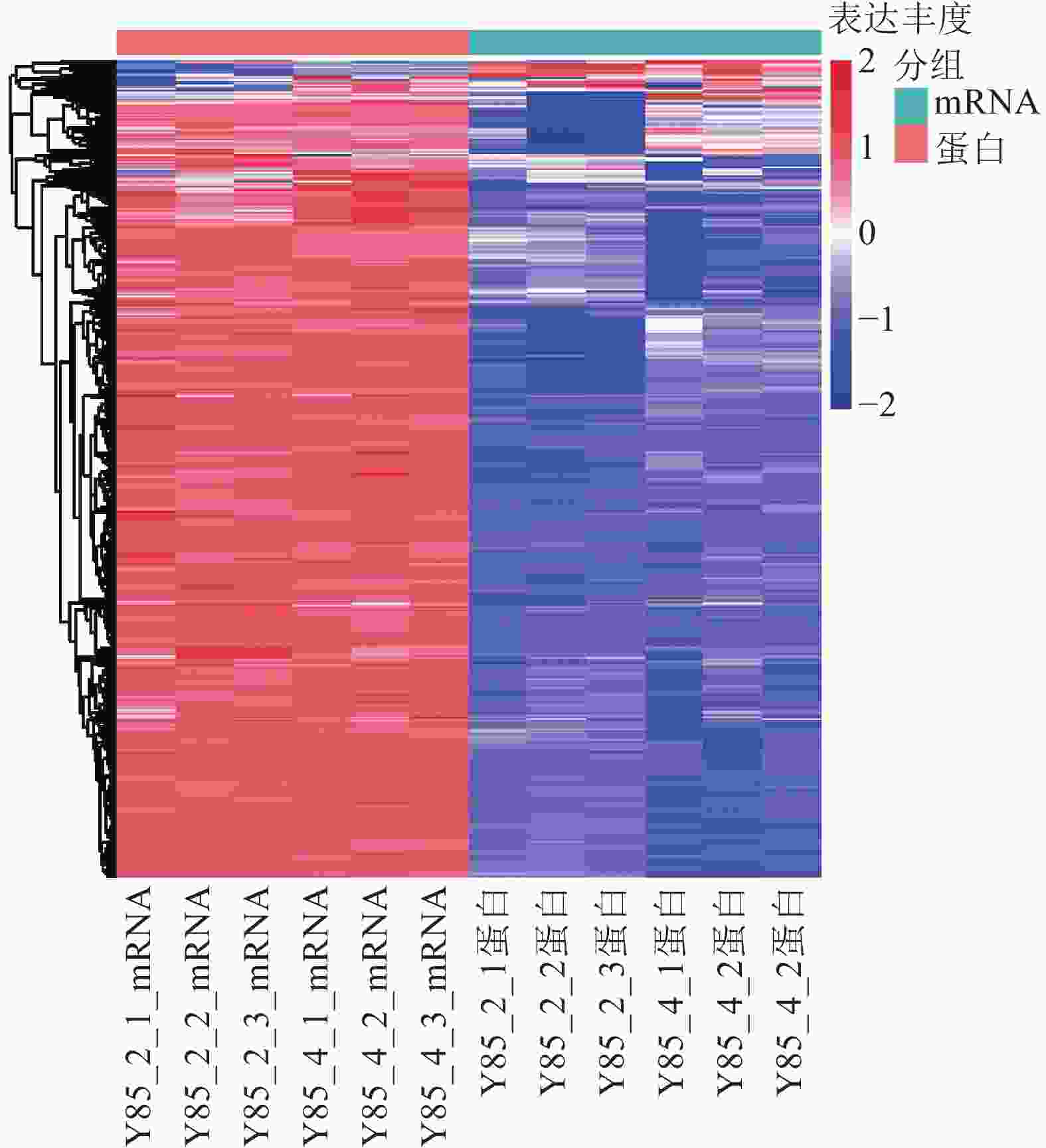
对差异蛋白和差异mRNA/基因进行非监督层次聚类(图4)表明:转录组和蛋白组测序样品的重复性较好。同一处理样品可以通过聚类出现在同一个簇中,聚在同一个簇中的差异蛋白或基因可能具有相似的生物学功能。通过 RT-PCR进一步验证了转录组测序的准确性。由图4可知:第2天转录组测序结果与RT-PCR结果的皮尔逊相关系数为0.44,而第4天转录组测序结果与RT-PCR测序的皮尔逊相关系数为0.96,但随机挑选的10个基因RT-PCR验证结果与转录组测序结果表达趋势一致(图5),因此认为测序结果是可靠的。
-
光照是调控种子萌发的重要环境信号,对于嗜光性种子,未完成后熟其种子萌发依赖于光照,如拟南芥、生菜Lactuca sativa和烟草种子等;对于厌光性种子,其萌发被光照所抑制,如番茄和岩芥菜Brassica juncea种子等;对于光中性种子,光照与否种子均可萌发,如玉米Zea mays、水稻Oryza sativa、小麦Triticum aestivum等主要粮食作物种子[16]。DONG等[8]研究发现烟草种子萌发普遍对光比较敏感,新采收种子暗发芽率普遍<50%。宋碧清等[17]也表明:烟草种子萌发存在光敏感类型,但不敏感品种居多,这可能是由于栽培环境差异导致。DONG等[8]研究发现:烟草种子萌发光敏感型不仅存在基因型差异,而且存在强烈的母体环境效应,在遮阴环境下成熟的浅光休眠烟草种子萌发率显著提高。与光中性种子类似,浅光休眠烟草种子在光和暗环境下均可萌发,但光可以提高萌发比率和加快萌发速度[18]。本研究也发现:浅光休眠烟草种子在暗下可萌发,发芽率达50%以上,这与上述研究结果一致。
光通过影响ABA和GA信号和水平来控制种子萌发。在拟南芥中,黑暗条件下PIF1强烈抑制种子萌发。光照下,PHYA和(或)PHYB被光激活,进而促进PIF1的降解,导致GA/ABA升高,GA信号被激发,促进种子萌发[19−20]。本研究未发现光敏色素协同植物激素ABA和GA信号通路基因(蛋白)参与调控烟草种子暗萌发。2个阶段差异表达基因达到数千个,而差异蛋白仅数百个,且前人在拟南芥种子萌发时研究表明:光主要在转录层面调控ABA和GA代谢和信号基因表达[19−20]。因此,本研究对转录组数据进行单独分析,但也未富集到光介导的ABA和GA信号通路。说明浅光休眠烟草种子暗萌发可能存在一种新的调控机制。
在种子暗萌发前后多个基因和蛋白表达水平发生了共表达。LEA 广泛参与种子成熟脱水的调控,在种子萌发期间迅速消失[21]。本研究发现:ECP63、LE25和SBP65等3类胚胎晚期丰度蛋白(基因)在烟草种子萌发后表达量显著下调。MFT (MOTHER-OF-FT-AND-TFL 1)抑制拟南芥种子萌发[22]。而本研究也发现:MFT抑制烟草种子萌发,这与拟南芥种子研究结果一致。β-1,3-葡聚糖酶是烟草种子萌发时胚乳破裂的关键酶,它的表达介于种皮破裂后和胚乳破裂前[23]。番茄种子萌发过程中胚根突破种皮需要水解酶软化胚乳帽,与此过程有关的蛋白是 expansion和内β-甘露聚糖酶[24]。本研究发现:水解酶MAN1无论是转录还是翻译水平在胚乳破裂前显著上调表达,这与上述研究结果类似,说明水解酶亦参与调控黑暗条件下的烟草种子萌发。此外,本研究还筛选到烟草种子暗萌发的正调控因子BoGH3B、ERD3、MLP31、FAP1、CPN60B2、PUMP1、CYSEP、PER24、RTNLB8等和负调控因子NEC1、SOP1、TMEM205、At3g01520、H6H、PARP3等,关于这些基因(蛋白)调控种子萌发的功能还未见报道,可通过遗传学实验进一步证实。
-
本研究通过整合转录组和蛋白组数据,发现烟草萌发前后蛋白和mRNA表达差异存在5种类型,其中共同上调表达的蛋白(基因)有BoGH3B、MAN1、ERD3、 MLP31、FAP1等,共同下调表达的蛋白(基因)有NEC1、MFT、ECP63、SOP1、LE25、SBP65等。基因富集分析和调控网络预测表明:在黑暗下,种子萌发依赖于甘露聚糖分解代谢,线粒体和叶绿体2个细胞器在种子暗萌发中具有关键作用。
Analysis of molecular networks for dark germination of shallow photodormant Nicotiana tabacum seeds based on transcriptomic and proteomic data
-
摘要:
目的 旨在挖掘浅光休眠烟草Nicotiana tabacum种子在黑暗下的调控基因和分子网络。 方法 以浅光休眠烟草Y85种子为实验材料,通过整合转录组和蛋白组数据,挖掘其暗萌发的分子网络。 结果 胚根突出前后蛋白和(或) mRNA表达差异存在5种差异类型,其中共同上调表达的蛋白(基因)有BoGH3B、MAN1、ERD3、MLP31、FAP1等,共同下调表达的蛋白(基因)有NEC1、MFT、ECP63、SOP1、LE25、SBP65等。上述差异蛋白(基因)富集的信号通路包括果糖和甘露糖代谢、甘露聚糖分解、内甘露聚糖-1,4-β-甘露糖苷酶活性;而发生功能的细胞成分包括线粒体、膜组成部分和叶绿体。 结论 通过整合转录组和蛋白组数据初步构建了浅光休眠烟草Y85种子的暗萌发调控网络。图5表1参24 Abstract:Objective This study aims to explore the regulatory genes and molecular network for shallow photodormant Nicotiana tabacum in the dark. Method Shallow photodormant N. tabacum Y85 seeds were used as experimental materials, and the molecular network of dark germination was studied by integrating transcriptome and proteome data. Result There were five types of differences in the expression of protein or mRNA expression before and after radicle protrusion, among which the jointly up-regulated proteins (genes) were BoGH3B, MAN1, ERD3, MLP31, FAP1, etc., and the jointly down-regulated proteins (genes) were NEC1, MFT, ECP63, SOP1, LE25, SBP65, and so on. The signal pathways enriched by the above proteins (genes) included fructose and mannose metabolism, mannan decomposition, endomannan-1,4-β-mannosidase activity. Functional cellular components included mitochondria, membrane components and chloroplasts. Conclusion By integrating transcriptomic and proteomic data, the dark germination regulatory network for shallow photodormant Y85 seeds was preliminarily constructed. [Ch, 5 fig. 1 tab. 24 ref.] -
Key words:
- Nicotiana tabacum /
- photodormant seed /
- dark germination /
- transcriptome /
- proteome /
- joint analysis
-
表 1 差异表达基因和差异表达蛋白联合分析
Table 1. Combined analysis of differentially expressed genes and differentially expressed proteins
登录号 转录差异 翻译差异 基因名称 上下调基因/蛋白 倍数 P 倍数 P Nitab4.5_0000010g0130.1 3.325 5 0.001 7 3.853 8 0.000 4 BoGH3B 上调 Nitab4.5_0000023g0130.1 20.474 7 0.000 0 2.071 3 0.000 9 MAN1 上调 Nitab4.5_0000108g0440.1 2.120 7 0.001 4 2.171 9 0.014 5 ERD3 上调 Nitab4.5_0001238g0070.1 6.056 6 0.009 8 2.218 9 0.005 6 MLP31 上调 Nitab4.5_0001550g0100.1 3.766 1 0.000 0 2.421 4 0.000 4 FAP1 上调 Nitab4.5_0006538g0070.1 2.239 3 0.031 9 2.485 4 0.010 9 CPN60B2 上调 Nitab4.5_0008198g0010.1 2.110 4 0.001 6 2.014 6 0.034 4 PUMP1 上调 Nitab4.5_0010931g0010.1 20.901 6 0.000 0 2.461 1 0.001 5 CYSEP 上调 Nitab4.5_0014169g0010.1 6.893 3 0.016 7 2.187 9 0.003 1 PER24 上调 Nitab4.5_0014487g0010.1 3.953 8 0.000 0 2.362 0 0.011 5 RTNLB8 上调 Nitab4.5_0000541g0010.1 0.064 3 0.000 0 0.381 9 0.006 4 NEC1 下调 Nitab4.5_0000649g0080.1 0.095 6 0.046 2 0.459 1 0.018 2 MFT 下调 Nitab4.5_0000680g0110.1 0.196 4 0.000 0 0.450 5 0.031 1 ECP63 下调 Nitab4.5_0001051g0080.1 0.159 4 0.001 4 0.498 9 0.009 4 SOP1 下调 Nitab4.5_0001378g0010.1 0.155 3 0.000 0 0.460 8 0.026 6 TMEM205 下调 Nitab4.5_0001538g0010.1 0.107 0 0.009 4 0.151 8 0.014 4 LE25 下调 Nitab4.5_0002674g0020.1 0.149 6 0.000 1 0.430 3 0.007 4 SBP65 下调 Nitab4.5_0003715g0060.1 0.151 4 0.033 7 0.470 9 0.000 1 At3g01520 下调 Nitab4.5_0004232g0030.1 0.095 5 0.000 0 0.435 2 0.001 6 H6H 下调 Nitab4.5_0005388g0040.1 0.104 5 0.001 0 0.437 4 0.034 1 At3g06035 下调 Nitab4.5_0017202g0010.1 0.122 7 0.000 0 0.475 3 0.001 9 PARP3 下调 -
[1] MAMZ B, MÜLLER K, KUCERA B, et al. Water uptake and distribution in germinating tobacco seeds investigated in vivo by nuclear magnetic resonance imaging [J]. Plant Physiology, 2005, 138: 1538 − 1551. [2] OH E, KANG H, YAMAGUCHI S, et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis [J]. The Plant Cell, 2009, 21: 403 − 419. [3] OH E, YAMAGUCHI S, HU Jianhong, et al. PIL5, a phytochrome-interacting Bhlh protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds [J]. The Plant Cell, 2007, 19: 1192 − 1208. [4] PARK J, LEE N, KIM W, et al. ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds [J]. The Plant Cell, 2011, 23: 1404 − 1415. [5] SHINOMURA T, NAGATANI A, CHORY J, et al. The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A [J]. Plant Physiology, 1994, 104: 363 − 371. [6] SHINOMURA T, NAGATANI A, HANZAWA H, et al. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana [J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93: 8129 − 8133. [7] APPENROTH K J, LENK G, GOLDAU L, et al. Tomato seed germination: regulation of different response modes by phytochrome B2 and phytochrome A [J]. Plant Cell and Environment, 2006, 29: 701 − 709. [8] DONG Shuai, LIU Yiling, ZHANG Min, et al. Maternal light environment interacts with genotype in regulating seed photodormancy in tobacco[J/OL]. Environment and Experimental Botany, 2022, 194: 104745[2022-09-07]. doi: 10.1016/j.envexpbot.2021.104745. [9] BOLGER A M, LOHSE M, USADEL B. Trimmomatic: a flexible trimmer for illumina sequence data [J]. Bioinformatics, 2014, 30(15): 2114 − 2120. [10] KIM D, LANGMEAD B, SALZBERG S L. HISAT: a fast spliced aligner with low memory requirements [J/OL]. Nature Methods, 2015, 12(4)[2022-09-07]. doi: 10.1038/NMETH.3317. [11] TRAPNELL C, WILLIAMS B A, PERTEA G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation [J]. Nature Biotechnology, 2010, 28(5): 511 − 515. [12] ROBERTS A, TRAPNELL C, DONAGHEY J, et al. Improving RNA-Seq expression estimates by correcting for fragment bias [J/OL]. Genome Biology, 2011, 12(3): R22[2022-09-07]. http://genomebiology.com/2011/12/3/R22. [13] LOVE M I, SONESON C, PATRO R. Swimming downstream: statistical analysis of differential transcript usage following salmon quantification [J/OL]. F1000Research, 2018, 7: 952[2022-09-07]. https://f1000research.com/articles/7-952. [14] WIESE S, REIDEGELD K A, MEYER H E, et al. Protein labeling by ITRAQ: a new tool for quantitative mass spectrometry in proteome research [J]. Proteomics, 2007, 7(3): 340 − 350. [15] LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method [J]. Methods, 2001, 25(4): 402 − 408. [16] 李振华, 徐如宏, 任明见, 等. 光敏色素感知光温信号调控种子休眠与萌发研究进展[J]. 植物生理学报, 2019, 55(5): 539 − 546. LI Zhenhua, XU Ruhong, REN Mingjian, et al. Advances in phytochrome regulating seed dormancy and germination by sensing light and temperature signals [J]. Plant Physiology Journal, 2019, 55(5): 539 − 546. [17] 宋碧清, 郑昀晔, 马文广, 等. 贮藏温度对烟草种子后熟的影响研究[J]. 种子, 2017, 36(4): 77 − 79. SONG Biqing, ZHENG Yunye, MA Wenguang, et al. Effects of storage temperature on after-ripening of tobacco seed [J]. Seeds, 2017, 36(4): 77 − 79. [18] LIU Qiyuan, LI Zhenhua, ZHANG Min, et al. Systematic analysis of photo/sko-regulated germination and post-germination development of shallow photodormant seeds in Nicotiana tabacum L. [J/OL]. Frontiers in Plant Science, 2022, 13: 1042981[2022-09-07]. doi: 10.3389/fpls.2022.1042981. [19] de WIT M, GALVÃO V C, FANKHAUSER C. Light-mediated hormonal regulation of plant growth and development [J]. Annual Review of Plant Biology, 2016, 67(1): 513 − 537. [20] 杨立文, 刘双荣, 林荣呈. 光信号与激素调控种子休眠和萌发研究进展[J]. 植物学报, 2019, 54(5): 569 − 581. YANG Liwen, LIU Shuangrong, LIN Rongcheng. Advances in light and hormones in regulating seed dormancy and germination [J]. Chinese Bulletin of Botany, 2019, 54(5): 569 − 581. [21] ALBAN C, JOB D, DOUCE R. Biotin metabolism in plants [J]. Annual Review of Plant Biology, 2000, 51: 17 − 47. [22] VAISTIJ F E, BARROS-GALVÃO T, COLE A F, et al. MOTHER-OF-FT-AND-TFL1 represses seed germination under far-red light by modulating phytohormone responses in Arabidopsis thaliana [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115: 8442 − 8447. [23] LEUBNER-METZGER G. Functions and regulation of β-1, 3-glucanase during seed germination, dormancy release and after-ripenning [J]. Seed Science Research, 2003, 13(1): 17 − 34. [24] NONOGAKI H, GEE O H, BRADFORD K J. A germination specific endomannanase gene is expressed in the micropylar endosperm cap of tomato seeds [J]. Plant Physiology, 2000, 123(4): 1235 − 1246. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.20220515






 下载:
下载: