-
实时荧光定量聚合酶链式反应(quantitative real time fluorescence polymerase chain reaction,qRT-PCR)因具有灵敏度高、特异性强、精准性高、检测范围广等优点[1],已被广泛应用于植物基因表达的相关研究中。利用qRT-PCR分析基因相对定量表达时,为了消除不同组织细胞间初始模板量、核糖核酸(RNA)制备和反转录过程中的偏差,需要引入内参基因(reference gene)对表达结果进行校正[2-3]。实际研究中常选择的内参基因[4-8]只能在一定的试验范围内相对稳定表达,不能保证在所有试验条件下均能稳定表达[9-11]。如果盲目以一种看家基因作为任何试验条件下的内参基因,容易得到精确度低甚至错误的结果[12],因此,在利用qRT-PCR进行基因表达分析时,根据试验处理及不同实验材料的特点,优化和选择合适的内参基因就显得极其重要。桂花Osmanthus fragrans是中国十大名花之一,随着桂花分子生物学研究的不断深入和发展,基因表达分析已经广泛应用于揭示其观赏性状形成机制的研究中,因此,筛选稳定的内参基因在桂花重要基因表达分析中起着关键的作用。已有研究筛选出桂花不同品种、不同发育状态花序和不同温度处理条件中合适的内参基因[13],却未对不同组织中适合的内参基因进行筛选。本研究将以桂花‘堰虹桂’O. fragrans‘Yanhong Gui’不同组织(花蕾、盛开花序、嫩叶、成熟叶、1年生茎)为材料,利用qRT-PCR检测7个候选内参基因的表达水平,并利用软件geNorm[3],NormFinder[2]和BestKeeper[14]对各候选内参基因的表达稳定性进行评价,筛选出最稳定表达的内参基因,并研究桂花类胡萝卜素异构酶(OfCRTISO1)在不同组织中的表达模式,验证筛选得到的内参基因的可靠性,希冀为后续研究不同组织重要基因的定量表达提供依据。
HTML
-
浙江农林大学桂花资源圃10 a树龄地栽桂花‘堰虹桂’为试验材料,对其花蕾、盛开花序、嫩叶、成熟叶、1年生茎进行取样,液氮速冻后,于-80 ℃储藏备用。
-
采用RNAprep pure Plant Kit(天根,北京)按照说明书步骤提取各样品的总RNA。紫外分光光度计和10.0 g·L-1琼脂糖凝胶电泳检测总RNA浓度和质量。采用Reverse Transcriptase M-MLV(Takara,大连)按照说明书合成cDNA的第1链后,于-20 ℃储存备用。
-
本研究从前期构建的桂花转录组数据库中取肌动蛋白基因(OfACT),延伸因子1α蛋白基因(OfEF1α),NADP-异柠檬酸脱氢酶基因(OfIDH),GTP结合蛋白RAN1基因(OfRAN1),β微管蛋白基因(OfTUB),泛素结合酶E2基因(OfUBC2)和18S核糖体RNA基因(Of18S)等7种看家基因作为候选的内参基因。根据qRT-PCR引物设计原则,利用Primer Premier 5等软件设计各候选内参基因的qRT-PCR引物(引物序列见表 1),其中Of18S引物引自文献[5]。
基因名 上游引物序列(5'→3') 下游引物序列(5'→3') OfACT CCCAAGGCAAACAGAGAAAAAAT ACCCCATCACCAGAATCAAGAA OfEF1α CGTTTGCCACTTCAGGATGTCTA GTACCAGGTTTCAGGACTCCAGTTT OfIDH CTTGAAGCAGATGTGGAAGAGTC CTTTGTC CATC CTGGGAC CAGTC OfRAN1 AGAACCGACAGGTGAAGGCAA TGGCAAGGTACAGAAAGGGCT OfTUB AGAAGGGATGGATGGAATGGA GTCTTCTTCGTCCTCGGCAGT OfUBC2 TGTTGACAAAACCGATGGAAGGA GTGGAGTGTGGAGGATAAGGGTG Of18S AGCCTGAGAAACGGCTACCAC ATACGCTATTGGAGCTGGAA Table 1. Primer sequences of seven candidate reference genes in qRT-PCR analysis
-
利用两步法来进行qRT-PCR分析,在7300实时PCR仪(Applied Biosystems)运行。基于SYBR Green染料的qRT-PCR来检测内参基因的循环阈值Ct值,所用反应体系和程序参考Takara公司的SYB® Premix Ex TaqTM Ⅱ(Tli RNaseH Plus)试剂盒的说明书。PCR反应体系为20.0 μL,其中2×SYBR Premix Ex Taq Ⅱ(Tli RNaseH Plus)10.0 μL,上下游定量引物10.0 μmol·L-1各0.8 μL,模板cDNA 2.0 μL,50×ROX Reference Dye 0.4 μL,用灭菌双蒸水补齐至20.0 μL。qRT-PCR扩增采用两步法,即在95 ℃预变性30 s之后,先运行40个循环的95 ℃ 5 s,60 ℃ 31 s,再运行溶解曲线阶段的95 ℃ 15 s,60 ℃ 1 min,95 ℃ 30 s,60 ℃ 15 s。通过观察ABI 7300实时PCR仪在PCR反应后生成的溶解曲线为单一峰判定所用引物无非特异扩增;实验得到定量Ct值用于后期内参基因稳定性分析。
-
本研究采用geNorm[3],NormFinder[2]和BestKeeper[14]软件对7种候选内参基因在桂花不同组织中的表达稳定性进行统计学分析,从而筛选桂花组织基因表达中最合适的内参基因。
-
利用桂花不同组织中OfCRTISO1基因表达模式验证筛选的内参基因的稳定性,其表达上游引物为5′-GAAAAACAAGGGATTCTCGGA-3′,下游引物为5′-GCAGATAGTAGGCAAGGGTCAA-3′。
1.1. 材料
1.2. 方法
1.2.1. 不同组织材料总RNA提取与cDNA的第1链的合成
1.2.2. 内参基因的选择及特异性引物设计
1.2.3. 内参基因的qRT-PCR分析
1.2.4. 数据处理与分析
1.2.5. 桂花OfCRTISO1基因验证内参基因稳定性
-
以桂花嫩叶的cDNA为模板,利用7对引物均能扩增出与各候选内参基因预期大小一致的单一条带(结果未显示),说明这7对引物均能特异性地扩增相应片段,可以用于qRT-PCR分析。qRT-PCR溶解曲线分析表明:这7对引物的扩增产物单一,可用于下一步分析。将反转录的嫩叶cDNA样品以5倍浓度梯度稀释,以此为模板进行qRT-PCR扩增后制作7种内参基因的标准曲线,结果(表 2)显示:各内参基因的线性相关系数R2≥0.990 0,引物的扩增效率为97.7%~109.6%,符合qRT-PCR对扩增效率的要求。
基因名 扩增长度/bp PCR扩增效率/% 线性相关系数 OfACT 143 109.6 0.998 4 OfEF1α 89 97.7 0.997 4 OfIDH 118 101.8 0.995 2 OfRAN1 117 100.4 0.990 3 OfTUB 106 103.8 0.998 1 OfUBC2 75 97.7 0.994 8 Of18S 208 104.7 0.992 6 Table 2. Amplicon characteristics of seven candidate reference genes
-
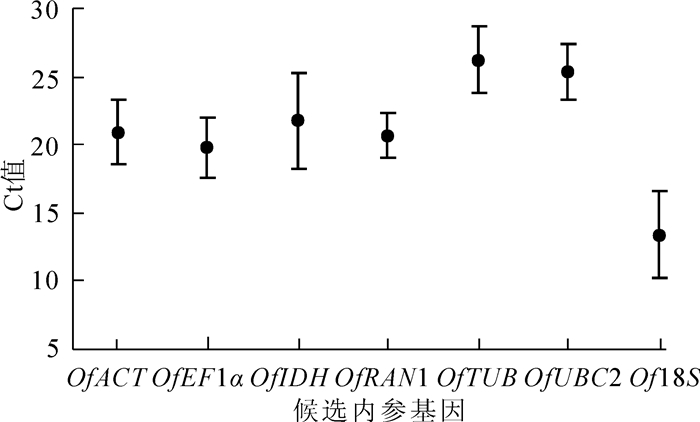
7个候选内参基因在不同组织中的Ct值平均为13.395~26.259(图 1),其中Of18S在所有样品中的转录水平最高,Ct值最小(13.395);而OfTUB基因的表达水平最低,Ct值最大(26.259)。从表达量变异系数看,OfRAN1最小,然后是OfUBC2,而OfIDH最大。
-
本研究首先采用geNorm软件对7个候选内参基因的表达稳定性进行分析(图 2),结果显示:在不同组织中,OfRAN1和OfUBC2基因的基因表达稳定值(M)最小(M=0.014 8),表示这2个基因表达最为稳定,其次是OfACT(M=0.038 4)和OfEF1α(M=0.045 3)。OfIDH,OfTUB和Of18S是稳定性排序靠后的3个候选内参基因,其中Of18S的M值最大(M=0.158 1),是最不稳定内参基因。

Figure 2. Expression stability values (M) of seven candidate reference genes as calculated by geNorm
利用geNorm程序对内参基因的配对差异值Vn/n+1进行分析(图 3),发现所检测的V2/3低于程序默认取舍值0.15,说明无需引入第3个基因进行校正,因此选择表达最为稳定的2个内参基因即OfRAN1和OfUBC2,以两者的几何平均值作为参照可更为准确地校正目的基因的表达。
此外,为保证结果分析的准确性,本研究还利用NormFinder和BestKeeper分析7个候选内参基因的表达稳定性(表 3),结果显示:NormFinder和BestKeeper的分析结果与geNorm分析的结果整体上一致。通过3个软件评价得到的稳定性排序中,OfRAN1和OfUBC2均是表达稳定性最高的2个基因,其次是OfACT和OfEF1α,表明这2个基因在不同组织中的表达较为稳定。而根据3个软件的分析,OfIDH,OfTUB和Of18S是表达稳定性最差的3个候选内参基因。综合3个软件对最佳内参基因和最差内参基因的评价(表 4),得到不同组织中最佳内参基因为OfRAN1和OfUBC2,而Of18S是最差内参基因。
基因名 NormFinder分析 BestKeeper分析 稳定值 排序 稳定值 排序 OfACT 0.010 2 1.678 4 OfEF1α 0.010 2 1.546 3 OfIDH 0.096 4 2.651 7 OfRAN1 0.005 1 1.168 1 OfTUB 0.084 3 2.080 5 OfUBC2 0.005 1 1.381 2 Of18S 0.227 5 2.272 6 Table 3. Gene expression stability of seven candidate reference genes by NormFinder and BestKeeper
分析软件 最佳内参基因 最差内参基因 geNorm OFRAN1和OFUBC2 Of18S NormFinder OFRAN1和OFUBC2 Of18S BestKeeper OFRAN1 OfIDH 综合结果 OFRAN1和OFUBC2 Of18S Table 4. Aggregated result of the optimal and worst
-
为了验证筛选的最佳内参基因稳定性是否准确,我们利用不同内参基因或组合校正桂花不同组织中OfCRTISO1基因的相对表达(图 4)。选择OfRAN1,OfUBC2,OfRAN1和OfUBC2基因组合进行校正的OfCRTISO1基因表达模式相同,均在盛开花序中表达量显著高于其他组织,在花蕾、嫩叶和成熟叶中的表达量相同,而在1年生茎中表达量最低。以稳定性较差的其他内参基因进行校正时,OfCRTISO1基因表达模式产生变化。上述结果表明:3个软件对内参基因表达稳定性的排序结果可靠,证实OfRAN1和OfUBC2为不同组织中最佳内参基因。
2.1. 候选内参基因引物评价
2.2. 候选内参基因的表达水平分析
2.3. 候选内参基因的表达稳定性分析
2.4. 桂花OfCRTISO1基因验证内参基因稳定性
-
理想的内参基因应在不同的基因型、不同发育阶段、不同组织器官、不同胁迫条件下均恒定表达,但事实上,并没有一个基因在所有试验条件下能稳定表达[15],因此,需要根据不同试验材料和不同试验处理条件下筛选合适的内参基因。本研究通过对7个候选内参基因在5个不同组织中的表达稳定性进行研究,筛选得到桂花不同组织中最佳内参基因为OfRAN1和OfUBC2,为后续桂花不同组织中目标基因表达规律的研究奠定了基础。
目前,3个统计分析软件geNorm,NormFinder和BestKeeper常用于确定不同植物材料或试验条件下最佳的内参基因[6, 13, 16-17]。然而,3个软件在许多物种中的分析结果存在差异,如芝麻Sesamum indicum[6],黄花蒿Artemisia annua[18]和牛奶子Elaeagnus umbellata[19]。在本研究中,3个软件的分析结果整体排序一致(图 2和表 3)。OfRAN1和OfUBC2基因在geNorm和NormFinder的稳定性排序中并列第一,而BestKeeper软件评价OfRAN1为表达最为稳定的内参基因,OfUBC2基因以微弱差距排列第二。综合3个软件分析结果确定OfRAN1和OfUBC2基因为桂花不同组织中最佳内参基因。有研究证实:选择2个或2个以上的内参基因作参照,可以减弱单一内参变化的影响,不同内参表达的相似性有助于增加结果的可靠度及精确度[3, 20-21],因此确定OfRAN1和OfUBC2基因组合为桂花不同组织中表达分析的内参基因可更为准确地校正桂花不同组织中目的基因的表达结果。
RAN(ras-related nuclear protein)是小G蛋白家族的一类,在真核生物进化过程中比较保守,在许多细胞生理进程过程中发挥着重要的作用[22-24]。RAN同源基因RAN3,是金鱼草Antirrhinum majus中qRT-PCR分析常用的内参基因[25-26]。通过对矮牵牛Petunia hybrida开花过程中相关候选内参基因的筛选,证实RAN1是矮牵牛中稳定表达的内参基因之一[27]。与ACT,EF1α和TUB等其他传统内参基因相比,将RAN基因作为内参基因进行研究和应用的报道较少。本研究证实RAN1在桂花不同组织中稳定表达,是最佳内参基因之一。此外,RAN1被证实是桂花不同品种中表达量最为稳定的内参基因[13]。与RAN基因相同,OfUBC2在桂花不同组织中也稳定表达,是另一个最佳内参基因。作为一个传统内参基因,UBC基因在拟南芥Arabidopsis thaliana[28-30],月季Rosa hybrida[31],侧柏Platycladus orientalis[32]等物种的内参基因筛选中均表现最佳。
本研究发现Of18S在桂花不同组织中表达最不稳定,不适合作为内参基因在桂花中应用。同样,在丹参Salvia miltiorrhiza[33],牡丹Paeonia suffruticosa[34],大白菜Brassica rapa ssp. pekinensis[8]和西瓜Citrullus lanatus[16]中,Of18S基因也被证实是表达最不稳定的内参基因。这可能与18S基因在样品中高丰度表达有关。












 DownLoad:
DownLoad:


