-
花青素具有极强的抗氧化活性,是近年来研究较多的抗氧化剂;查尔酮合成酶(chalcone synthase,CHS)是花青素生物合成途径上的第1个关键酶,催化1分子的香豆酰CoA与3分子的丙二酰CoA反应,生成4-羟基查尔酮。自1983年KREUZALER等[1]利用差异筛选和杂交筛选相结合的方法从欧芹Petroselinum hortense中获得了第1个CHS基因后,对查尔酮合成酶及其编码基因的研究工作就屡有报道[2-3]。研究发现,编码查尔酮合成酶(CHS)的基因组成了一个多基因家族,从葡萄Vitis vinifera中分离到的该家族的3个成员,分别被定名为CHS1,CHS2,CHS3[4];经氨基酸水平检测,发现CHS3与CHS1,CHS2的同源性均为89%,CHS1与CHS2同源性为96%,说明CHS家族成员之间具有较高的同源性。进一步研究发现,CHS1,CHS2主要在白葡萄、红葡萄的叶片及果皮的着色过程中表达,而CHS3主要在红葡萄果皮的着色过程中表达,表明CHS3主要参与花青素的合成,而CHS1,CHS2可能还参与合成其他代谢产物[5]。柑橘属Citrus植物中也有2个CHS的cDNA序列被克隆出[6-7],氨基酸序列上的同源性达到了86.6%,但具体功能不尽相同,说明CHS家族成员尽管在氨基酸序列上有很高的同源性,但功能上并不是完全重叠。从目前已经克隆到的上百个CHS基因序列[8-9]来看,CHS基因在植物体内的表达具有时空特异性,主要集中于花和果皮等部位,且在特定的时期才会表达;其表达量与花和果皮的颜色直接相关[10-12],许多果树如苹果Malus domestica[13],柑橘Citrus reticulata[14-15],沙梨Pyrus pyrifolia[16],葡萄[5]等,在受到紫外线照射[17]、降低温度[18]等物理刺激后,CHS基因的表达量增加,引起果实颜色加深。由此可见,CHS基因在花青素生物合成途径中扮演着十分重要的角色。蓝莓Vaccinium spp.为杜鹃花科Ericaceae越桔亚科Vaccinioideae越桔属Vaccinium植物,研究称其浆果花青素含量在水果中最为丰富。我们拟以红色突变兔眼蓝莓Vaccinium ashei为样品,克隆获得查尔酮合成酶基因(CHS),对获取的序列进行生物信息学分析,并与其他物种的相关基因进行对比,以期为今后高花青素含量品种的育种工作提供一定的生物信息学基础。
HTML
-
试材选用经组织培养8周的组培苗。液氮处理后,保存于-80 ℃备用。
-
试剂盒:RNAprep Pure植物总RNA提取试剂盒;PCR Mix;DNA胶回收试剂盒;pLB零背景快速克隆试剂盒。上述试剂盒均购自天根生化科技有限公司。cDNA反转录试剂盒(SMARTerTM RACE cDNA Amplification Kit),购自Clontech公司。
-
用RNAprep Pure植物总RNA提取试剂盒提取兔眼蓝莓总RNA。用核酸蛋白测定仪与琼脂糖凝胶电泳进行RNA的定量与质量检测,合格的RNA样品在-70 ℃中保存。按照SMARTerTM RACE cDNA Amplification Kit说明书进行操作, 以oligo(dT)寡苷酸为引物,获得cDNA模板。以反转录所得的cDNA为模板进行扩增,扩增引物如下:CHS(F1): 5′-CCACAAAGGCCATAAAGGAA-3′,CHS(R1): 5′-GGAGCTTGGTGAGCTGGTAG-3′,CHS(F2): 5′-CCACAAAGGCCATAAAGGAA-3′,CHS(R2): 5′-AGGAGCTTGGTGAGCTGGTA-3′。
-
用pLB零背景快速克隆试剂盒克隆目的片段。挑取1~3个能在含有氨苄青霉素的LB平板上生长的菌株并提取DNA;用CHS基因扩增引物进行聚合酶链式反应(PCR);把产物送出测序。
-
序列拼接由ContigExpress完成。依据http://www.ncbi.nlm.nih.gov/,,http://www.expasy.org/等网站提供的生物信息学软件对克隆的基因序列作在线分析。用NCBI-ORF Finder进行开放阅读框(ORF)的查找和翻译;基于美国生物技术信息中心(NCBI)的blast功能进行氨基酸序列同源性分析;用Clustal W作同源序列的比对,用MEGA 5.2在序列比对的基础上构建系统发育树。用ProtParamet在线工具分析基因组成及其理化性质:理化性质用Expasy,亚细胞定位用TargetP 1.1,信号肽预测用SignalP 4.1,跨膜结构域用TMHMM 2.0。分别利用SOPMA及Swiss-Mode完成蛋白二级结构和三级结构的预测。使用NCBI提供的Conserved Domain Search进行蛋白功能域的预测分析。
1.1. 实验材料
1.2. 主要试剂
1.3. 实验方法
1.3.1. 总RNA提取与检测
1.3.2. 目的片段的克隆
1.4. 数据分析
-
测序结束后,对序列进行拼接,得到长度为1 398 bp的CHS基因。含有1个完整的开放阅读框,长度为1 170 bp,并基于此基因序列推导其氨基酸序列(图 1)。
-
将兔眼蓝莓CHS基因核酸序列及其推导的氨基酸序列在NCBI中比对,选取与其同源性较高的16个物种(表 1),作进一步的进化判断及蛋白理化性质、结构功能分析。
序号 Genbank登录号 物种 核酸序列同源性/% 氨基酸序列同源性/% 1 KF157394.1 中华猕猴桃Actinidia chinensis 83 93 2 JN944575.1 红花油茶Camellia chekiangoleosa. 83 94 3 AB512766.1 山茶Camellia japonica 83 95 4 EU410483.1 舌甘橙Citrus sinensis 76 87 5 AY997297.1 草莓Fragaria ananassa 80 91 6 FJ554585.1 啤酒花Humulus lupulus 80 90 7 AY786996.1 苹果Malus domestica 82 93 8 KF438041.1 川桑(桑葚)Morus notabilis 82 93 9 GU990524.1 甜櫻桃Prunus avium 80 91 10 KF387513.1 白梨Pyrus bretschneideri 81 93 11 AJ413277.1 杜鹃Rhododendron simsii 90 97 12 AF400565.1 覆盆子Rubus idaeus 82 91 13 NM_001247104.1 番茄Lycopersicon esculentum 77 89 14 DQ286037.1 欧洲花楸Sorbus aucuparia 82 93 15 JN654702.1 高丛蓝莓Vaccinium corymbosum 83 87 16 JF808008.1 葡萄Vitis vinifera 81 93 Table 1. Nucleotide sequence and deduced amino acid sequence aligment for CHS
被比对的16个物种中,高丛蓝莓、苹果、草莓、覆盆子、欧洲花楸、甜樱桃、中华猕猴桃、甜橙、番茄和葡萄等物种花青素集中在果实,杜鹃、红花油茶、山茶等物种花青素集中在花瓣,白梨则发生了红茎突变。从表 1可知:就核酸序列同源性而言,与兔眼蓝莓同源性最高的是杜鹃,达90%;最低的是甜橙,仅为76%;核酸序列同源性平均为82%。与其氨基酸序列同源性最高的是杜鹃,达到97%;最低的是高丛蓝莓,为87%;氨基酸序列同源性平均为92%。
用Clustal W软件对兔眼蓝莓CHS基因编码的氨基酸序列与已知的CHS基因的氨基酸序列进行比对,再用MEGA 5.2软件采用邻接法(Neighbor-joining method, NJ法)做出系统进化树(图 2)。
系统树上物种聚类成支说明之间亲缘关系较近。由图 2可知:蔷薇科Rosaceae植物苹果、白梨、欧洲花楸、草莓、覆盆子和甜樱桃聚类为1个大分支,桑科Moraceae植物啤酒花和川桑聚类为1个小分支,山茶科Theaceae植物山茶和红花油茶聚为1个小分支,杜鹃花科植物杜鹃和兔眼蓝莓聚类为1个小分支,且遗传距离为0.009,相当接近。这与分类学上的结论是一致的。值得注意的是,高丛蓝莓与兔眼蓝莓在传统分类学上亲缘关系十分接近,是蓝莓下的2个栽培变种,但在系统树上却分别聚类在2个不同的大分支上。另一方面,甜橙是芸香科Rutaceae植物,番茄是茄科Solanaceae植物,但在系统树上,它们却聚类为1个小支。
-
Expasy在线软件对所得的查尔酮合成酶基因所编码蛋白质的理化性质分析结果表明:兔眼蓝莓中CHS基因预测编码氨基酸为389个,相对分子量42 555.2,等电点5.98,不稳定指数为36.42,是相对稳定的蛋白。
-
通过SignalP 4.1 Server对兔眼蓝莓CHS基因所编码蛋白质的信号肽进行预测,比对结果表明:查尔酮合成酶基因所编码蛋白质不具有分泌功能。
-
用TMHMM Server v.2.0对获得序列的CHS蛋白跨膜结构进行分析。结果表明:兔眼蓝莓CHS蛋白整条肽链都位于细胞膜外,说明其基因编码的多肽不存在跨膜结构;结合信号肽预测结果,可以推断兔眼蓝莓CHS蛋白在细胞质基质中合成后,直接作用于细胞质基质中的特定区域,行使其催化功能。对其他植物的跨膜结构分析也得到了一致的结果,CHS基因在大部分植物体内均不包含跨膜结构。
-
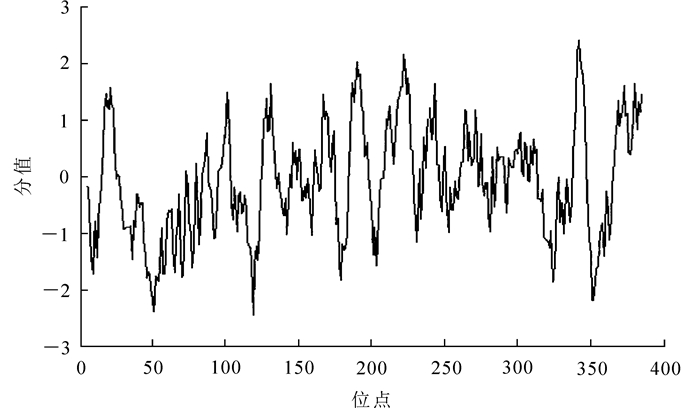
ProtScale预测的兔眼蓝莓CHS基因所编码蛋白质的亲疏水性,结果如图 3所示。兔眼蓝莓CHS蛋白多肽链第51位具有最低分值-2.478,说明亲水性最强;第342位具有最高分值2.422,说明疏水性最强。疏水性氨基酸均匀分布于整条肽链中,使整体表现为疏水性,但没有明显的疏水区域。ProtParam分析可知:兔眼蓝莓CHS蛋白的脂溶指数(Aliphatic Index)为91.49,总平均亲水性(GRAVY)为-0.071,预示其是一个脂溶性蛋白。与其他植物CHS蛋白的亲疏水性预测结果一致。
-
用TargetP 1.1对CHS蛋白进行的亚细胞定位结果显示:兔眼蓝莓CHS蛋白的叶绿体转运肽cTP,线粒体靶向肽mTP和分泌通路信号肽SP的数值都相当低(分别为0.029,0.235,0.069;阈值为0.000),可以认为兔眼蓝莓CHS蛋白并没有存在于叶绿体、线粒体中;而分泌通路信号肽数值跟信号肽预测一致,说明CHS蛋白并不是分泌蛋白。综合跨膜结构的预测结果,我们推测CHS蛋白应该位于细胞质基质中,在核糖体合成蛋白后,就在细胞质基质中发挥作用,参与催化对香豆酰辅酶A和丙二酰辅酶A合成查尔酮。对其他物种CHS蛋白的亚细胞定位也获得了类似的结果,不同物种中,CHS蛋白均位于细胞质基质中。
-
使用NCBI提供的Conserved Domain Search service(CD-Search)对兔眼蓝莓CHS蛋白功能域进行预测(图 4)。结果显示:兔眼蓝莓CHS蛋白属于CHS超家族,含有CHS超家族的结构域,即Pro16-Leu384,是一种植物特异的Ⅲ型聚酮合成酶(polyketide synthase,PKS)。此外,该蛋白还含有植物特异聚酮化合物合成酶(KAS-Ⅲ)功能域[8]和柚配基查尔酮合成酶(BcsA)功能域,后者参与类黄酮合成等代谢过程。
-
用SOPMA对CHS蛋白二级结构进行预测,发现兔眼蓝莓CHS蛋白二级结构中α-螺旋的比例为40.87%,β-折叠为18.25%,β-转角11.83%,随机卷曲29.05%;α-螺旋和随机卷曲是兔眼蓝莓CHS蛋白二级结构的主要结构原件,β-折叠和β-转角虽然占得比例小,但它们均匀分布在整个氨基酸序列上;α-螺旋为右手螺旋,没有3.010,2.270,4.416等左手螺旋结构,是较稳定的结构。这样的二级结构组成反映了兔眼蓝莓CHS蛋白是一种结构相对稳定的功能蛋白。
2.1. CHS基因序列及氨基酸序列推导
2.2. CHS氨基酸序列相似性比较与系统进化树分析
2.3. 查尔酮合成酶(CHS)的生物信息学分析
2.3.1. 查尔酮合成酶(CHS)化学性质分析
2.3.2. 查尔酮合成酶(CHS)信号肽分析
2.3.3. 查尔酮合成酶(CHS)跨膜结构分析
2.3.4. 查尔酮合成酶(CHS)亲疏水性分析
2.3.5. 查尔酮合成酶(CHS)亚细胞定位
2.3.6. 查尔酮合成酶(CHS)蛋白结构功能域分析
2.3.7. 查尔酮合成酶(CHS)蛋白二级结构
-
查尔酮合成酶(CHS)广泛存在于各种植物中,它是花色素苷生物合成途径中的第一个关键酶,在植物的花色素苷积累、细胞发育与分化、抗胁迫以及外源基因的表达等过程中都起着重要作用[19]。本研究采用同源克隆的方法,获得了兔眼蓝莓红色突变株的CHS基因。该基因全长为1 398 bp,包含1个长为1 170 bp的开放阅读框,编码389个氨基酸。序列比对分析显示,该基因与杜鹃、山茶、红花油茶等的CHS基因在核苷酸水平及氨基酸水平上有较高的同源性。生物信息学分析结果显示,兔眼蓝莓红色突变株CHS基因编码的蛋白质序列中没有信号肽,没有跨膜结构,侧面说明该基因编码的蛋白质没有参与跨膜转运。KATIYAR等[20]对拟南芥根组织中CHS酶和CHI酶的研究发现,此两者共定位于内质网和液泡膜上;本研究对CHS蛋白亚细胞定位的预测结果却表明,兔眼蓝莓红色突变株中CHS蛋白定位于细胞质,但尚有待试验的进一步确认。
FERRER等[9]和JEZ等[8]采用三维结构及定点突变分析研究查尔酮合成酶的空间结构,提出查尔酮合成酶催化中心由4个高度保守的残基(Cys164,His303,Asn336,Phe215)组成,是查尔酮合成酶的作用中心。生物信息学分析软件对兔眼蓝莓红色突变株CHS蛋白的功能域搜寻发现其含有CHS-like域,其CHS蛋白序列的第164位是半胱氨酸(Cys, C),215位是苯丙氨酸(Phe, F),303位是组氨酸(His, H),336位是天冬酰胺(Asn, N),与先前研究结果类似,这也说明CHS基因在高等植物中是高度保守的;兔眼蓝莓红色突变株CHS蛋白中的CHS-like域,也有可能是CHS酶的作用中心。
本研究获得的CHS基因及相关的生物学信息为今后的转基因及相关利用奠定了一定的基础。














 DownLoad:
DownLoad: