-
丹参Salvia miltiorrhiza是传统的中药材,其主要药用成分包括脂溶性的丹参酮类化合物和水溶性的酚酸类化合物。其中丹参酮是一类松香烷二萜化合物。20世纪30年代末,有学者首次发现丹参酮ⅡA和丹参酮ⅡB是天然的抗氧化剂。随后几十年,又连续发现了丹参酮Ⅰ、隐丹参酮、二氢丹参酮Ⅰ、异丹参酮、去甲基丹参酮等脂溶性有效成分。其药理作用主要表现在抗菌消炎、抗氧化、抗肿瘤、抗心肌缺氧、激素样活性等方面,并能够改善学习记忆能力,在临床上主要用于心脑血管疾病的治疗[1-2]。丹参有效成分产量较低,可利用资源有限,使其应用推广受到很大的限制。因此,筛选获得高丹参酮产量的优良品种或是通过其他方法解决药源问题是很有必要的。笔者从丹参酮生物合成途径及其分子调控机制等方面展开综述,同时总结了以往研究中存在的问题,并结合自身对研究前景进行了分析,为后续深入研究提供参考。
HTML
-
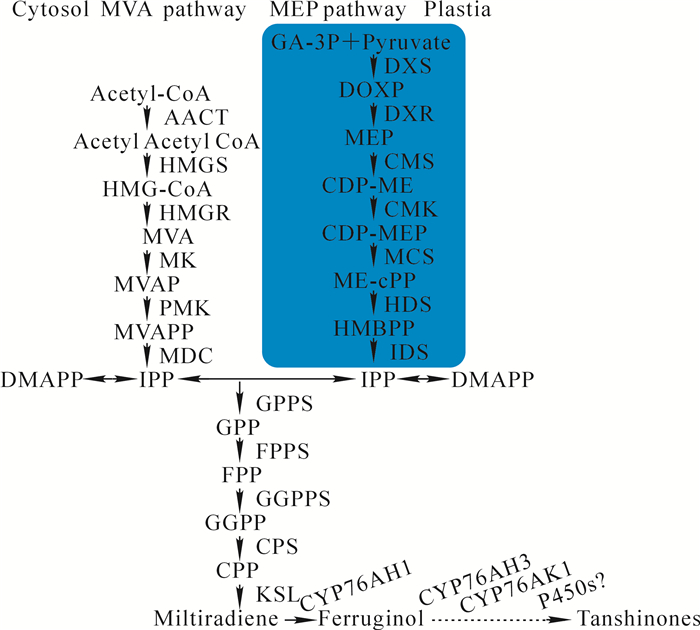
丹参酮是一类二萜类化合物,其代谢合成方式有2条途径(图 1),分别为位于细胞质中的甲羟戊酸途径(mevalonate, MVA)[3]和位于质体中的非甲羟戊酸途径(2-C-methyl-d-erythritol-4-phosphate, MEP)[4]。MVA合成途径的上游基因包括AACT,HMGS,HMGR,MK,PMK和MDC;MEP合成途径上游基因包括DXS,DXR,CMS,CMK,MCS,HDS和IDS;中游合成酶基因包括IPPI,GPPS,FPPS和GGPPS;下游合成酶基因包括CPS,KSL,CYP76AH1,CYP76AH3,CYP76AK1以及未知的P450s基因;在下游合成途径中柯巴基焦磷酸(copalyl diphosphate, CPP)经类贝壳杉烯合酶(KSL)催化形成丹参酮二烯(miltiradiene),丹参酮二烯经细胞色素P450家族蛋白CYP76AH1催化合成铁锈醇(ferruginol),铁锈醇经过CYP76AH3和CYP76AK1的多步催化生成11, 20-二羟基铁杉醇和11, 20-二羟基柳杉酚,最后在其他CYP家族或其他酶作用下产生次丹参酮(miltirone),进而合成丹参酮类成分[5]。目前,从铁杉醇合成丹参酮之间的多步催化反应步骤仍不清楚,需要进一步的研究。
-
诱导子能诱发植物次生代谢过程中酶的活性,从而增加次生代谢物的产量,有时可以诱导出新的代谢化合物。依据来源可划分为生物诱导子和非生物诱导子两大类。生物诱导子是指植物体在防御过程中为对抗微生物入侵而产生的防御性物质,如分生孢子、分解细胞壁的酶、细胞壁的碎片。非生物诱导子是指所有非植物细胞的固有成分,但能触发植物细胞形成抗毒素信号的物质[6]。
-
研究人员围绕生物诱导子对丹参酮合成的影响做了许多研究。晏琼等[6-7]利用酵母提取物(YE)、寡聚半乳糖醛酸和真菌诱导子等3种生物诱导子分别处理丹参毛状根,发现都能显著提高丹参酮的产量。施加100 g·L-1的真菌诱导子,总丹参酮的产量提升4.70倍;施加100 g·L-1的YE,总丹参酮的产量提升3.20倍;施加100 g·L-1的寡半乳糖醛酸,总丹参酮的产量提升4.20倍。GE等[8]考察了不同浓度的β-氨基丁酸(BABA)和YE单独或是协同对丹参酮代谢合成的影响,与对照相比最多可提高4.50倍。BABA+YE协同作用,在BABA处理3 d后再加入YE,对丹参酮的积累更加有效。YAN等[9]发现密旋链霉菌Streptomyces pactum处理丹参毛状根后,GGPPS,HMGR,DXS,DXR等4个关键酶基因的表达量显著提升,总丹参酮产量提高12.61倍[9]。研究发现,丹参毛状根中丹参酮ⅡA和隐丹参酮的积累和产量受YE诱导。丹参酮的产量经诱导处理6 d后测得的最大值为0.64 mg·g-1,其中HMGS,DXR,DXS2,CMK,IPPI,CPS等6个关键酶基因的表达量显著上调。YE和银离子(Ag+)共同诱导处理表明,丹参酮ⅡA和隐丹参酮的产量显著增加。丹参酮诱导处理9 d后测得的丹参酮的最大值为2.08 mg·g-1,其中AACT,HMGS,DXR,IPPI,CMK,FPPS,GGPPS,CPS等8个关键酶基因表达显著上调[10]。上述研究表明,生物诱导子对于丹参酮的合成具有促进作用。因此,新型高效生物诱导子的挖掘和开发利用将是后续重要研究方向之一。
-
非生物诱导子能引起或者促进植物细胞与防御机制有关的次生代谢产物的生物合成。目前,已证实的非生物诱导子约有100多种,主要包括茉莉酸甲酯(methyljasmonate,MeJA),茉莉酸(jasmonate,JA),水杨酸(alicylic acid,SA),重金属盐类和稀土元素。郭肖红等[11]研究发现,低浓度的铜离子(Cu2+),镁离子(Mg2+),锌离子(Zn2+),亚铁离子(Fe2+),锰离子(Mn2+)以及稀土元素均能促进丹参酮ⅡA的合成。KAI等[10]研究了0.10 mmol·L-1 MeJA诱导处理丹参毛状根后丹参酮ⅡA和隐丹参酮的积累变化。处理9 d后,HMGR,DXR,DXS2,GGPPS,CPS和KSL等基因的表达显著上调,丹参酮的产量达到最大值0.93 mg·g-1干质量(DW),相比于对照组0.16 mg·g-1(DW)增加了约5.78倍。HAO等[12]发现SA处理丹参毛状根36 h后,丹参酮产量上升1.63倍。银离子处理毛状根后,HMGR,DXS2,IPPI,GGPPS等基因的表达显著上调;丹参酮ⅡA和隐丹参酮的积累逐渐增加,处理9 d后测得的丹参酮的最大产量为0.35 mg·g-1,相比于对照组的0.16 mg·g-1(DW)增加约2.20倍。晏琼等[6-7]利用银离子(Ag+),钴离子(Co2+)和α-氨基异丁酸(AIB)等3种非生物诱导子分别处理丹参毛状根,通过检测丹参酮的产量发现,此3种非生物诱导子都能提高丹参酮的积累,但诱导能力比生物诱导子弱;其中50.00 mmol·L-1钴离子获得了最高的丹参酮产量(1.75 mg·g-1),是对照组产量的4.10倍;30.00 mmol·L-1银离子处理后总丹参酮的产量提高了2.10倍;50.00 mmol·L-1 AIB处理后总丹参酮产量提高了2.30倍。ZHOU等[13]利用金属元素0.01 mmol·L-1镧(La)对丹参毛状根进行诱导处理,结果表明丹参酮代谢途径上关键酶基因包括HMGR, HMGS, KSL, DXR, CMK的表达量均上调,丹参酮Ⅰ,丹参酮Ⅱ和隐丹参酮的产量均升高。由此可推测镧可能通过促进丹参酮代谢途径上游关键酶基因的表达而增加丹参酮的积累。房翠萍[14]研究了外源激素对丹参毛状根生长和丹参酮ⅡA生物合成的影响,结果显示在1/2MS培养基中单独施加2.00 mg·L-1 6-苄氨基腺嘌呤(6-BA)能使丹参酮ⅡA产量增加3.01倍;同时添加3.00 mg·L-1 6-BA与0.20 mg·L-1 NAA能使丹参酮ⅡA产量增加0.93倍;同时添加1.00 mg·L-1 6-BA与2 mg·L-1赤霉素(GA)能使丹参酮ⅡA浓度增加1.69倍;同时添加1.00 mg·L-1激动素(KT)与1.00 mg·L-1吲哚丁酸(IBA)能使丹参酮ⅡA产量增加0.21倍;此外还研究了pH值对丹参毛状根生长和丹参酮ⅡA生物合成的影响,结果发现培养基pH值为pH 9时,丹参酮产量最高。YANG等[15]利用聚乙二醇(PEG)和脱落酸(ABA)对丹参毛状根进行诱导处理,发现PEG和ABA通过增加毛状根中活性氧含量促进丹参酮的代谢合成,推测PEG和ABA可能通过ABA途径诱导丹参毛状根中内源茉莉酸(JA)的活性,从而刺激丹参酮的代谢积累[15-16](表 1)。
诱导子类型 诱导子名称 浓度或质量浓度 处理材料 受诱导的基因 丹参酮的变化 参考文献 非生物诱导子 Ag+ 30.00 mmol·L-1 毛状根 总丹参酮增加2.10倍 [7] Co2+ 50.00 mmol·L-1 毛状根 总丹参酮增加4.10倍 [7] YE 100.00 mg·L-1 毛状根 HMGR,DXR,DXS2, CMK,CPS,IPPI 6 d时达到最大值0.64 mg·g-1 [10] YE+Ag+ 100.00 mg·L-1+0.03 mmol·L-1 毛状根 AACT,HMGS,DXR,CMK,FPPS,CPS,GGPPS,IPPI 9 d时达到最大值2.08 mg·g-1 [10] Cu2+,Mg2+,Zn2+,Fe2+,Mn2+ 原MS培养基的0,0.5,1.0,1.5,2.0倍 丹参不定根 低产量的Cu2+,Mg2+,Zn2+以及较高产量Fe2+,Mn2+促进丹参酮ⅡA的合成 [11] MeJA 0.10 mmol·L-1 毛状根 HMGR,DXR,DXS2, GGPPS,CPS,KSL 总丹参酮增加5.78倍 [10] SA 10.00 mg·L-1 毛状根 GGPPS 总丹参酮增加1.60倍 [12] La 0.01 mmol·L-1 毛状根 HMGR,HMGS,DXR,CMK,KSL 总丹参酮增加1.50倍 [13] 6-BA 2.00 mg·L-1 毛状根 丹参酮ⅡA增加3.01倍 [14] 6-BA+NAA 3.00 mg·L-1+0.20 mg·L-1 毛状根 丹参酮ⅡA增加0.93倍 [14] 6-BA+GA 1.00 mg·L-1+2.00 mg·L-1 毛状根 丹参酮ⅡA增加1.69倍 [14] KT+IBA 1.00 mg·L-1+1.00 mg·L-1 毛状根 丹参酮ⅡA增加0.21倍 [14] PEG+ABA 20.00 g·L-1+0.20 mmol·L-1 毛状根 HMGR,DXS [15-16] 生物诱导子 真菌诱导子 100.00 g·L-1 毛状根 总丹参酮增加4.70倍 [7] 内生真菌 100.00 g·L-1 毛状根 总丹参酮增加3.20倍 [7] 寡聚半乳糖醛酸 100.00 g·L-1 毛状根 总丹参酮增加4.20倍 [7] YE 2.00 mmol·L-1 毛状根 总丹参酮增加4.50倍 [8] 1.00 mmol·L-1 BABA+YE 1.00 mmol·L-1+200mg·L-1 毛状根 总丹参酮增加9.40倍 [8] 密旋链霉菌 40.00g·L-1 毛状根 HMGR,DXR,DXS,GGPPS 总丹参酮增加12.60倍 [9] Table 1. Effect on the biosynthesis of tanshinone induced by elicitors in Salvia miltiorrhiza
2.1. 生物诱导子对丹参酮合成的影响
2.2. 非生物诱导子对丹参酮合成的影响
-
转录因子又称为反式作用元件,是能够特异结合真核基因启动子区域顺式作用元件的脱氧核糖核酸(DNA)结合蛋白,通过蛋白与基因之间、蛋白与蛋白之间的相互作用激活或抑制转录[17]。近年来,随着对植物转录因子研究和认识的不断深入,转录调控被认为是一种能有效调控植物次生代谢途径的新手段。超表达转录因子能够有效地克服多基因共转化存在的弊端,全面调节相关基因的表达,对整个代谢途径的调控效果要优于单纯提高单个或多个结构基因的表达。目前,研究人员对转录因子在丹参次生代谢调控方面进行了深入研究,研究较多的转录因子有基本螺旋-环-螺旋(basic helix-loop-helix,bHLH)、植物中髓细胞组织增生蛋白(myelocytomatosis,MYCs),WRKY和JAZ等几个类型(表 2)。
转录调节类型 转录因子名称 超表达载体类型 转基因子材料 丹参酮产量的变化 参考文献 正向调节 AtMYC2-like pCAMBIA2300+ 毛状根 提高5.40倍 [30] SmMYC2-like pCAMBIA2300+ 毛状根 提高3.50倍 [30] SmWRKY3 pCAMBIA2300+ 毛状根 提高1.83倍 [34] SmWRKY70 pCAMBIA2300+ 毛状根 提高6.31倍 [34] SmWRKY54 pCAMBIA2300+ 毛状根 提高2.23倍 [35] 反向调节 SmJAZ3 pCAMBIA2300+ 毛状根 下调94.0% [5] SmJAZ9 pCAMBIA2300+ 毛状根 下调80.6% [5] Table 2. Effect on the biosynthesis of tanshinone regulated by transcription factors in Salvia miltiorrhiza
-
植物bHLH转录因子家族因其成员含有高度保守的bHLH区域而被命名为bHLH转录因子。bHLH结构域由50~60个左右氨基酸组成,包含10~15个氨基酸组成的碱性氨基酸区和40个左右氨基酸构成的HLH区[18]。50%以上的植物碱性氨基酸区含有高度保守的His5-Glu9-Arg13序列,这使得碱性氨基酸区能够识别DNA上E-box(5′-CANNTG-3′)和G-box(5′-CACGTG-3′)位点,并与之结合[19-20]。由于HLH结构域中疏水氨基酸之间常常互作从而促进蛋白二聚体的形成,因此bHLH蛋白常以二聚体的形式发挥作用[21]。目前已报道的植物bHLH家族转录因子已有很多,其中拟南芥Arabidopsis thaliana中报道的bHLH家族成员有164个,水稻Oryza sativa中bHLH家族成员有180个[22],而烟草Nicotiana tabacum和葡萄Vitis vinifera中bHLH转录因子分别有190个和191个以上[23-24]。通过转录组测序,127个丹参bHLH类转录因子已被分离,其中7个成员被发现能响应甲基茉莉酸(MeJA)信号诱导。SmbHLH37,SmbHLH74和SmbHLH92被证实与丹参萜类物质的代谢合成密切相关[25]。由此推测,bHLH类转录调控因子在JA信号途径中调控丹参酮和丹酚酸的生物合成具有重要的作用。此外,丹参转录因子SmbHLH1和SmbHLH93的全长基因被相继克隆,分析表明此2个基因的表达与丹参酮的代谢合成密切相关[26-27],这些研究为从分子水平全面解析bHLH类转录因子调控丹参酮的代谢合成机制奠定了扎实的工作基础。
-
MYC类转录因子是植物茉莉酸类激素响应途径中的核心转录因子。MYC具有多种调节功能,广泛存在于动植物中。MYC类转录因子中MYC2研究最为深入。MYC2转录因子含有bHLH保守结构域,属于bHLH类转录因子家族成员。MYC2在N端含有1个JID结构域,与JAZ结合相关[28]。ZHOU等[29]研究发现,丹参SmMYC2a能和丹酚酸代谢途径的结构基因SmHCT6,SmCYP98A14的E-BOX互作;而SmMYC2b能和SmCYP98A14的E-BOX互作,直接调控丹酚酸而间接调控丹参酮的代谢合成。王晓荣[30]研究发现,SmMYC2-like能和SmJAZ3互作,在丹参毛状根中过表达AtMYC2后,丹参酮代谢合成关键酶基因SmGGPPS的表达上调,总丹参酮产量达16.00 mg·g-1(DW),高出对照5.40倍。而过表达SmMYC2-like后,总丹参酮产量增加3.50倍。
-
WRKY转录因子家族是高等植物十大转录因子家族成员之一。WRKY转录因子的保守结构域约由60个氨基酸残基组成,其中靠近氨基酸N端的7个保守氨基酸残基WRKYGQK为WRKY结构域的核心序列,它们的变异会导致DNA结合活性减弱或者丧失;羧基C末端的锌指类似结构在植物的进化中可能起到重要的作用[31-32]。
在植物进化过程中,WRKY转录因子家族成员不断扩大。LI等[33]克隆了61个WRKYs基因,发现其中42个WRKYs成员能够响应MeJA和银离子(Ag+)胁迫,与丹参酮的代谢合成密切相关。郝小龙[34]研究发现,在丹参毛状根中过表达SmWRKY3后,丹参酮合成关键酶基因SmCPS表达上调,总丹参酮产量达3.98 mg·g-1(DW),相比于对照提高了1.83倍;而抑制表达SmWRKY3后,SmCPS的表达下调,总丹参酮产量仅为1.98 mg·g-1(DW)。因此,WRKY转录因子能够促进丹参酮的合成,是丹参酮合成的正向调控因子。
-
含有ZIM结构域的基因称为JASMONATE ZIM-DOMAIN(JAZ)基因。ZIM结构域一般位于JAZ蛋白的中间,主要由28个氨基酸组成;在N端含有(TIF[F/Y]XG)结构域,在C末端有2个不变的丙氨酸。Jas结构域(SLX2FX2KRX2RX5PY)位于JAZ蛋白的C端,在JAZ蛋白中极其保守。研究发现,拟南芥的SCFCOI1复合体依赖JA-Ile,并通过Jas结构域与JAZ1,JAZ3,JAZ9,JAZ10蛋白结合;Jas结构域是JAZ蛋白核定位信号的关键结构。JAZ参与JA信号介导的次生代谢物的合成在拟南芥中研究得比较透彻。JAZ在拟南芥中有12个成员,其中至少8个JAZ蛋白成员都与MYC2相互作用。拟南芥MYC2的同源基因MYC2-LIKE成员JAM1,JAM2和JAM3也参与JA信号途径的次生代谢合成(表 2);其中JAM1和JAM2分别与6个不同的JAZ成员相互作用,而JAM3却不能与任何JAZ蛋白成员互作。近期丹参JAZ蛋白参与JA信号介导的次生代谢物的合成机制研究也取得一些进展。在丹参中超表达JAZ3和JAZ9基因能使毛状根中丹参酮的产量急剧降低;酵母双杂试验证实SmJAZ9蛋白能与bHLH类转录因子拟南芥的AtMYC2互作,而SmJAZ3不能与AtMYC2互作,推测SmJAZ9通过与bHLH类转录因子互作,SmJAZ3通过与其他bHLH类转录因子互作而调控丹参酮的生物合成[5]。研究证实将SmJAZ8遗传转化丹参,超表达SmJAZ8的转基因毛状根系表现出对JA信号不敏感的表型;SmJAZ8的超表达导致SmJAZ1,SmJAZ2和SmJAZ3的表达量急剧下调,从而提高丹参酮和花青素的代谢合成;究其原因发现,SmJAZ8蛋白跟拟南芥的AtJAZ8类似,缺少LPIARR的结构域,而此结构域是JA信号诱导JAZ蛋白降解释放正向调节因子的关键区域。可见拟南芥JAZ蛋白在JA信号介导的次生代谢合成作用机制的解析为在丹参中研究JAZ调控丹参酮和丹酚酸的合成提供了很好的参考。丹参JAZ家族其他成员的功能有待于进一步地解析。
3.1. bHLH类转录因子
3.2. MYC类转录因子
3.3. WRKY类转录因子
3.4. JAZ类转录因子
-
真核生物基因转录产生的mRNA前体大多数只按一种方式进行剪接,产生一种成熟的mRNA分子,只翻译成一种蛋白质。但有的基因mRNA前体通过不同的剪接方式(按不同的剪接位点)产生不同的mRNA剪接异构体,此过程称为可变剪接。RNA可变剪接不涉及遗传信息的永久性改变,是真核基因表达调控的重要手段,是调节基因表达和产生蛋白质多样性的重要机制,是导致人类基因和蛋白质数量变异的重要原因。
现如今可变剪接作为代谢和发育另一个重要的调节机制已在许多植物中被鉴定,如拟南芥[36-37],大豆Glycine max[38],蔓越橘Brachypodium distachyon[39]和水稻[40]。而在丹参中,利用短读高通量测序技术(NGS)和单分子实时(SMRT)长读测序技术,结合对根部各组织进行测序发现,KSL1,HDR,HMGR,AACT3,MK和PMK基因存在着选择性剪接调控的现象[41]。由此可见,丹参酮代谢合成途径中关键合成酶基因的可变剪接对丹参酮的代谢合成起到了重要的调节作用。某些转录调控因子是否也存在可变剪接的调控方式有待进一步研究。
-
丹参这种传统的中草药具有重要的药用价值。近年来野生丹参品质严重退化,濒临灭绝,导致丹参原药的供应极不稳定,解决药源问题已成为研究的热点。目前,对丹参酮次生代谢合成的研究重点主要包括以下几个方面:①丹参酮代谢合成途径的解析。分离丹参酮代谢合成酶或是修饰酶,通过底物与产物的鉴定,解析丹参酮次生代谢化合物的合成途径。②转录因子及其调控机制和调控网络的解析。转录因子可同时调控代谢途径上的1个或是多个基因的表达,在丹参酮次生代谢合成中起重要的调控作用。为阐明丹参酮代谢合成的调控机制,利用诱导子处理丹参,结合转录组学、蛋白质组学和代谢组学揭示候选转录因子的表达与产物代谢流的分配规律,以揭示转录因子间的表达与互作机制对丹参酮合成代谢的调控规律。③丹参酮基因工程的研究。在代谢途径和调控网络充分解析的基础上,实现代谢调控的可预测性。如采用时空特异性表达的启动子、多基因共转化技术,实现丹参有效成分在特异部位或是全株系大量积累和特定性状改良的目标。
随着研究的深入,进一步从转录调控水平全面解析丹参酮的代谢分子调控机制对于利用现代基因工程手段培育高品质丹参新品种/系具有重要的指导意义,也具有广阔的应用前景。因此,应从以下几个方面开展研究工作:①丹参酮代谢合成关键调控因子可变剪接的挖掘与分子机制解析。②非编码RNA在丹参酮代谢合成过程中的调控作用。③丹参酮合成生物学的研究。通过工程酵母生产丹参酮前体物,并利用有机半合成技术体外合成丹参酮,这将改变丹参药源生产方式,为丹参系列药物走向国际市场奠定工作基础,无疑将具有重要的应用前景。










 DownLoad:
DownLoad: