-
全球变暖导致植物在生长发育和环境适应性等方面面临更加多样化的挑战[1]。高温不仅影响植物表型变化,而且会引起植物体内细胞稳态失衡,生长和发育受到抑制,严重影响观赏植物的品质和产量[2-3]。如芍药Paeonia lactiflora在遭受高温胁迫后,植株出现萎蔫、畸形、变色等热害现象,甚至全株死亡[4]。在牡丹P. suffruticosa中也出现类似的表型现象[5]。另外,经过高温处理后,牡丹和月季Rosa chinensis植株内的脯氨酸、丙二醛、可溶性糖以及相对细胞膜透性显著升高,而净光合速率、超氧化物歧化酶和可溶性蛋白质含量等显著降低[6-8]。为了深入了解高温胁迫对观赏植物品质的影响,利用现代分子生物学以及转录组学等手段[9],挖掘关键基因并解析其调控耐热的分子机制,是培育和开发耐热优良品种的重要途径。对非洲菊Gerbera jamesonii切花高温处理后,转录组分析发现大部分单基因簇(unigene)可注释到细胞过程、代谢过程和结合功能等基因本体(GO)过程中[10];全基因组及代谢途径数据库(KEGG)中代谢功能途径中的单基因簇分布最多。在柳枝稷Panicum virgatum中发现[11]:长期高温胁迫条件下大量的基因发生差异表达,其中与ATP酶调节因子、伴侣结合和蛋白质折叠相关的基因显著上调,与蛋白质修饰、转录、磷和氮代谢途径相关的基因在热胁迫后显著下调。热激蛋白(heat shock protein, HSPs)是一类高温胁迫诱导的主要功能蛋白,其分子量为10~200 kDa,包括HSP60、HSP70、HSP90、HSP100和小分子量热激蛋白[12](small heat shock protein,sHSP)等5类。HSPs充当蛋白质质量控制的分子伴侣,作为分子伴侣和折叠催化剂细胞网络的中心成分,在热应激中被诱导并参与热应激过程中的信号转导[13];sHSP存在多数植物叶肉细胞中,在外界热诱导条件下的sHSP表达呈现上升的趋势[14-16]。在女萎Clematis apiifolia[17]中,高温处理后热激蛋白热胁迫后明显上调,而且sHSPs是对热胁迫反应最强烈的伴侣基因家族。HSPs不仅响应高温变化,还对水分、盐渗透和氧化胁迫等[12, 14]有响应。牡丹是芍药科Paeoniaceae芍药属Paeonia多年生落叶灌木,中国传统名花[18]。目前,关于牡丹基因的研究多集中于生长发育、花色基因挖掘以及抗氧化活性物质生物合成等方面[19-22],相关耐热研究报道较少。牡丹对高温较为敏感,夏季高温严重制约牡丹的生长和发育,致使花期缩短,观赏品质降低,因此,挖掘牡丹耐热基因对牡丹热胁迫分子机制探究,以及牡丹育种和推广具有重要的意义。
HTML
-
实验材料牡丹‘羽红’P. suffruticosa ‘Yuhong’保存在浙江农林大学智能实验大棚。取长势一致的1年生幼苗移栽至花盆缓苗,1株·盆−1,正常管理。后转移至人工气候箱中,在温度25 ℃,光12 h/暗12 h,光强4 000 lx,湿度80%条件下预培养2周。处理组和对照组分别置于40和25 ℃人工气候箱中处理24 h,选取相同位置的叶片,使用液氮速冻并保存于−80 ℃冰箱于后期备用,设置3个生物学重复。
-
使用Trizol试剂盒(天根,北京)提取牡丹叶片总RNA,按照说明书操作。用紫外分光光度计和质量分数1.5%琼脂糖凝胶电泳检查RNA质量,质量检测合格后,经百迈客生物科技有限公司(北京)使用Illumina HiSeq2500对6个独立样品进行双端文库构建以及RNA-Seq测序。
-
将测序获得的原始序列读长(raw data)进行过滤组装,得到高质量的有效序列读长(clean reads),然后使用Trinity进行序列组装[23-24]。所得单基因簇序列用基于BLAST算法的搜索和注释,对照非冗余蛋白质(Nr)数据库(www.ncbi.nlm.nih.gov/)、全基因组及代谢途径(KEGG)数据库(www.genome.jp/kegg/kaas/)、蛋白直系同源簇(COG)数据库(www.ncbi.nlm.nih.gov/COG/)以及瑞士蛋白质序列及注解(Swissprot)数据库(www.expasy.ch/SPORT)等数据库进行注释。使用Blast2Go软件(www.blast2go.com/)进行基因本体论(GO)功能分析。
-
使用Bowtie软件将测序所得的片段与非冗余基因序列库进行对比,结合RSEM(RNA-Seq by Expectation-Maximization)预估表达量水平[24-25]。在此基础上分析高温胁迫处理前后6组样本的差异表达基因,使用RPKM[每百万序列读长(Reads)中某基因每千长度序列读长值]统计基因的表达量;单基因簇表达丰度用FPKM值表示,以差异倍数|log2A|≥2。其中,A表示差异倍数或变化倍数(flod change),且错误发现率(false discovery rate)≤0.01为标准筛选差异表达基因。
-
使用Primer Premier 5.0软件设计荧光定量PCR引物(表1),Prime Script RT reagent Kit With gDNA Eraser试剂盒(TaKaRa,北京)进行实时荧光定量PCR反应。反应体系:上下游引物(10 μmol·L−1)各0.4 μL,cDNA 1.0 μL,ddH2O 3.2 μL,SYBR Premix Ex Taq 5.0 μL。总反应程序:95 ℃预变性30 s,95 ℃变性5 s,60 ℃复性30 s,共38个循环。以
$ {2^{ - \Delta \Delta {C_{\rm{t}}}}} $ 法[26-28]计算目的基因相对表达量,其中以牡丹泛素延伸蛋白基因(Ubiquitin)为内参基因[29],设3次生物学重复。基因编号 基因名称 引物序列(5′→3′) 退火温度/℃ C65824.graph_c0 PsHSP70 GCAGCCGTTCAGGCAGCAA 58.3 ACACCGCCAGCAGTTTCG C114411.graph_c0 Ps4CL1 AGGCTATGCTACTCACGC 53.5 TCATCCTCGGCAATCTG C106006.graph_c0 PsCAT5 GAGTGTTGCGTTAATAGGCG 56.5 GCACTTGGACAGGGAAGGTAT C111201.graph_c0 PsCHL1 GAGTGCTGAGATTTTAAGTGGC 56.7 CAACCCTTTCTACCTGACGA C120725.graph_c0 PsAMP1 ATTGTACGGCAGAACAGGC 55.3 GTGTAGCTCAGAACCCATTAGAC C112526.graph_c0 PsSAMDC GTCCAGGCGAATGTTCTAATG 56.7 GTCGAAGGTCTTCAATGCTG C119988.graph_c0 PsRPS4 GGCTTTCATCCGATTTGTTG 55.4 ATGCCACCTGTCCTTACCTG C97637.graph_c0 PsCCD4 GTGAGGCATTTGCTTTCCG 55.3 TGCTGGGTTCATTCCTATCTG C111758.graph_c0 PsPPIL1 TCCCCTCCAACTATTCCTCC 52.2 AACTCAACCCCTTCCGATT C120729.graph_c1 Hypothetical protein CACATGAAGAGGACGACCAA 56.5 CCACATACATCAACCCAGGAC Ubiquitin Ubiquitin GACCTATACCAAGCCGAAG 55.1 CGTTCCAGCACCACAATC Table 1. Real-time fluorescence quantitative gene primer information
1.1. 植物材料
1.2. 叶片总RNA提取及cDNA文库构建
1.3. 转录组测序及数据分析
1.4. 基因表达量及差异基因分析
1.5. 实时荧光定量PCR
-
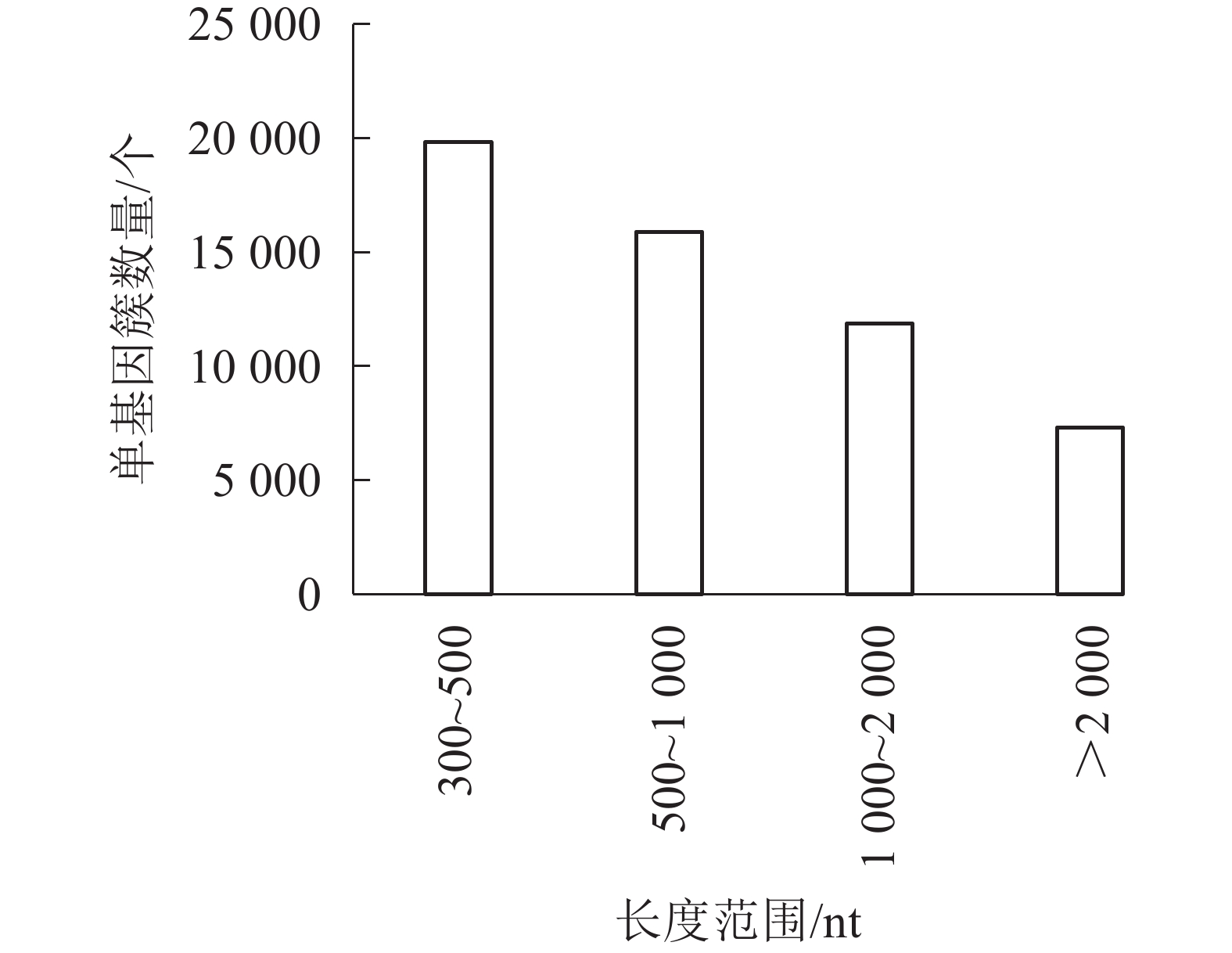
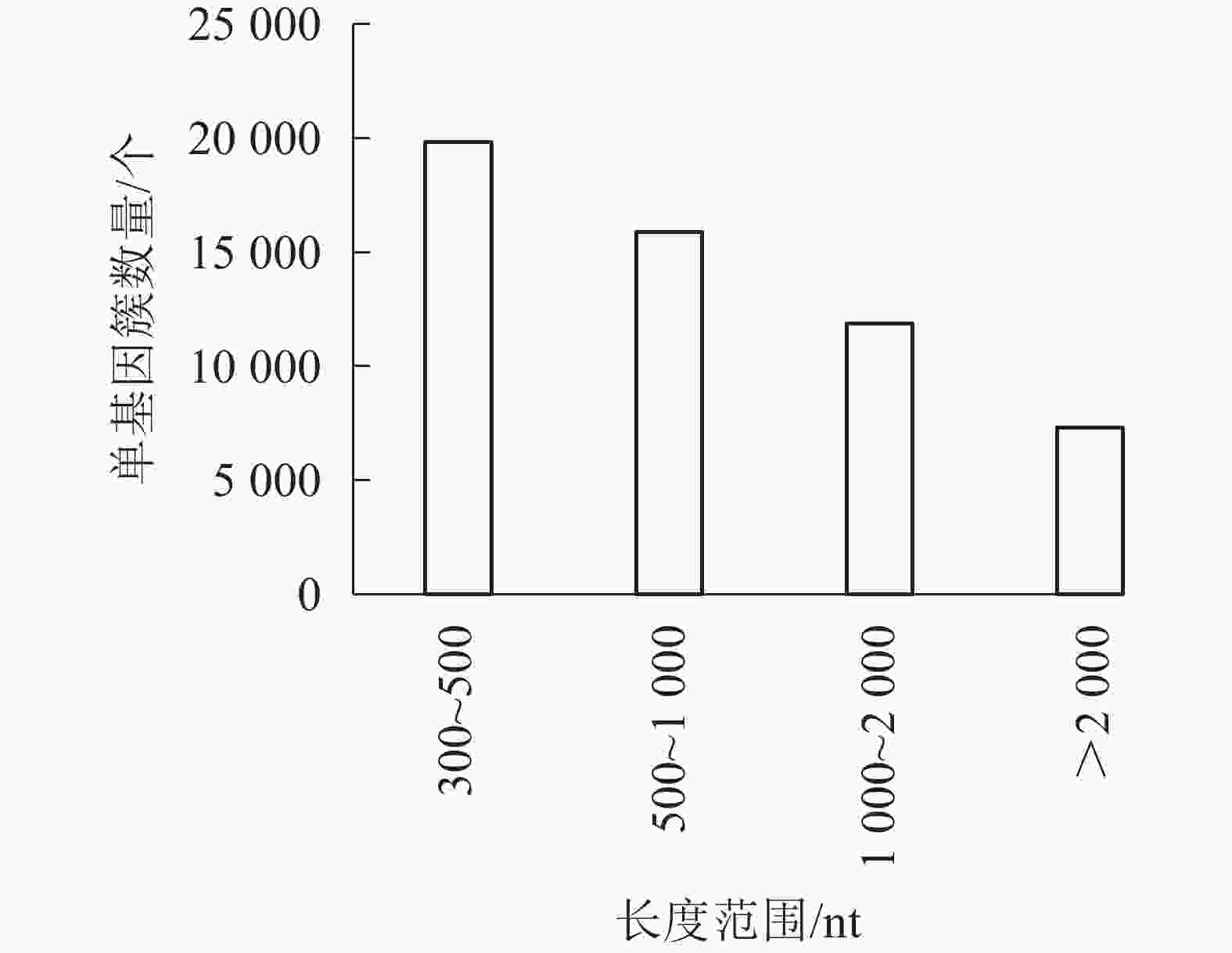
对照组(ck1、ck2和ck3)与高温(40 ℃)处理组(TE1、TE2和TE3)样本经过测序后,获得45.97 Gb的有效序列读长,且6个样品Q30碱基百分比值均大于等于96.71%(表2)。转录本序列数量(transcript number)总计151 939条,非冗余基因序列数量(unigene number)54 935条,有较高的组装完整性。经拼接后片段重叠群N50长度为1 561 nt,平均长度为1 042 nt,长度为300~500 nt的单基因簇数量占组装总量的36.1%,有19 834条。长度500~1000 nt的数量次之,占总量的28.94%。大于2 000 nt的占总量的13.33%(图1)。
样品名称 读取碱基数/个 总碱基数/个 GC碱基数
比例/%≥Q30碱基
百分比/%ck1 24 156 058 7 200 895 958 44.96 96.78 ck2 24 256 024 7 236 093 266 44.76 96.89 ck3 25 256 661 7 530 655 892 44.53 96.74 TE1 30 430 627 9 077 213 032 43.68 96.79 TE2 22 967 108 6 856 944 378 43.95 96.73 TE3 27 110 006 8 071 678 010 43.68 96.71 合计 154 176 484 45 973 480 536 Table 2. Statistical table of sample sequencing data evaluation
-
将单基因簇基因序列分别在各数据库注释,其中以非冗余蛋白质数据库(Nr)获得注释的比例最高,达到98.76%(表3)。
数据库 注释 300≤长度<
1 000 nt长度≥
1 000 nt数量 百分比/% COG 6 915 28.44 1 698 5 217 GO 14 568 59.93 5 011 9 557 KEGG 8 416 34.62 2 813 5 603 KOG 13 669 56.23 4 761 8 908 Pfam 15 108 62.15 4 096 11 012 Swissprot 15 426 63.46 4 825 10 601 Eggnog 21 798 89.67 7 891 13 907 Nr 24 009 98.76 9 509 14 500 总数 24 309 100.00 9 731 14 578 Table 3. Unigene annotation statistics
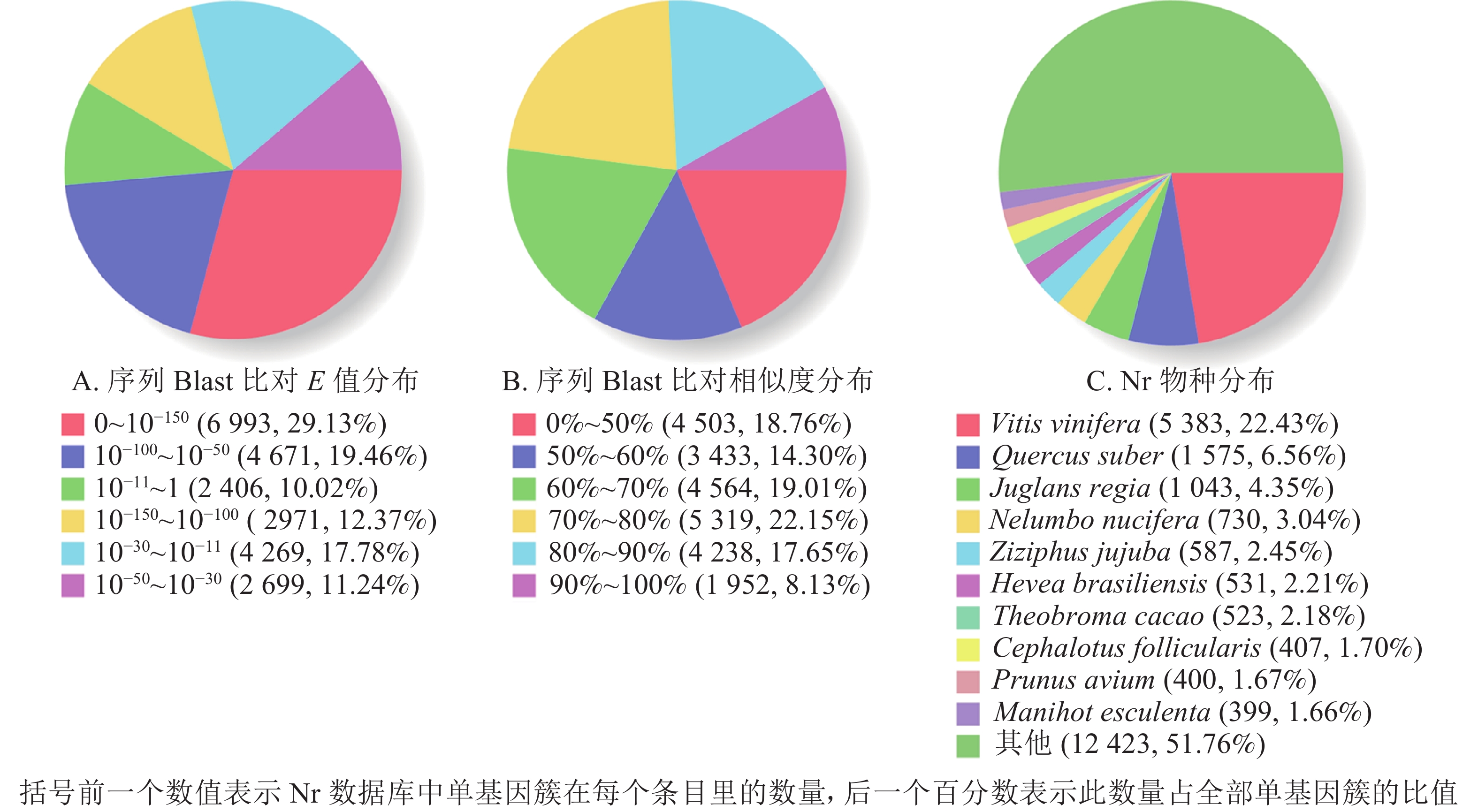
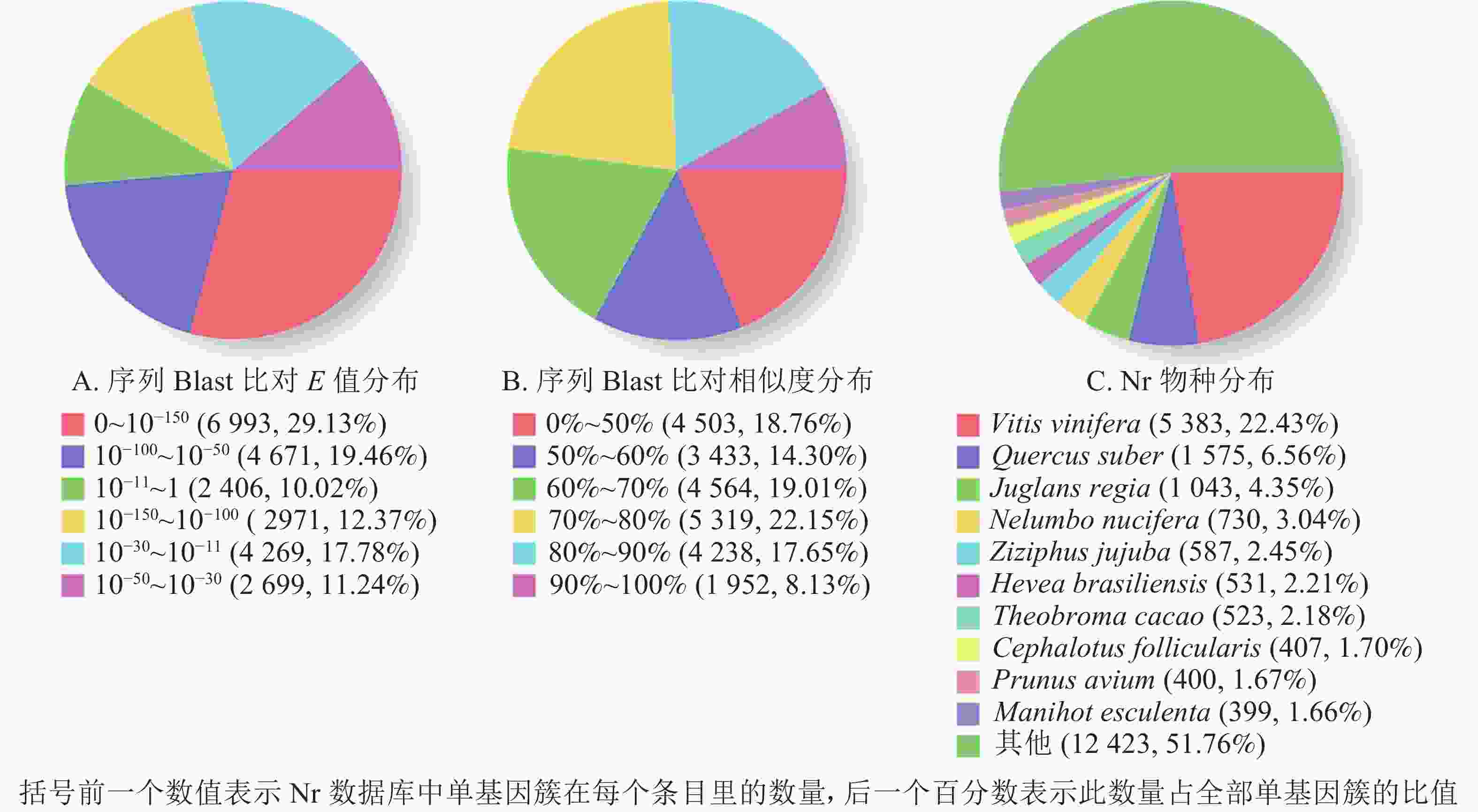
根据非冗余蛋白质数据库中的物种注释对比,做出对应注释图和E值分布图,大部分序列(29.13%)E值为0~10−150 (图2A),25.78%的序列相似度大于80.00%(图2B);另外,22.43%的单基因簇基因可注释到葡萄Vitis vinifera,6.56%的单基因簇序列可以注释到欧洲栓皮栎Quercus suber(图2C)。
-
以差异倍数≥2和错误发现率≤0.01为标准,共筛选获得7 673个差异基因,其中4 220个基因上调表达,3 453个基因下调表达。利用Web基因本体论注释软件进行基因本体富集分析(表4),有6 443个差异基因富集于生物过程,主要集中于代谢过程(1 702个,占26.42%,GO:0008152)、细胞进程(1 489个,占23.11%,GO:0009987)和单一生物进程(1 148个,占17.82%,GO:0044699);有6 491个差异基因富集于细胞组分,主要集中于细胞(1 316个,占20.27%,GO:0005623)、细胞组分(1 302个,占20.06%,GO:0044464)和细胞膜(1 232个,占18.98%,GO:0016020);有3 812个差异基因富集于分子功能,主要分布在催化活性(1 719个,占39.95%,GO:0003824)、结合活性(1 523个,占35.09%,GO:0005488)和转运活性(291个,占7.63%,GO:0005215)等过程。
注释本体
类别编号 功能类型 基因注释数量 比例/% 注释本体
类别编号 功能类型 基因注释数量 比例/% 生物过程 1 代谢过程 1 702 26.42 细胞组分 26 复杂大分子复合体 242 3.73 2 细胞进程 1 489 23.11 27 细胞外区域 63 0.97 3 单一生物进程 1 148 17.82 28 细胞连接 54 0.83 4 生物调节 457 7.09 29 共质体 53 0.82 5 定位 436 6.77 30 腔上包膜 48 0.74 6 应激反应 366 5.68 31 超分子复合物 17 0.26 7 细胞组分及生物合成 213 3.31 32 细胞外区域部分 8 0.12 8 信号转导 128 1.99 33 病毒体 4 0.06 9 发育过程 123 1.91 34 病毒体部分 4 0.06 10 多细胞机体进程 122 1.89 35 拟核 2 0.03 11 繁殖 74 1.15 分子功能 36 催化活性 1 719 39.95 12 繁殖进程 74 1.15 37 结合 1 523 35.09 13 有机体进程 49 0.16 38 转运活性 291 7.63 14 生化解毒 35 0.54 39 结构分子活性 58 1.52 15 生长 15 0.23 40 核酸结合转录因子活性 46 1.21 16 免疫系统进程 9 0.14 41 分子功能调节器 46 1.21 17 细胞活动 2 0.03 42 信号传感器活动 35 0.92 18 节律进程 1 0.02 43 抗氧化活性 27 0.71 19 生物黏附 0 0 44 分子传感器活性 26 0.68 细胞组分 20 细胞 1 316 20.27 45 电子载体活动 21 0.55 21 细胞组分 1 302 20.06 46 转录因子活性 14 0.37 22 细胞膜 1 232 18.98 47 养分储藏活性 4 0.10 23 细胞膜组成 935 14.40 48 蛋白质标签 1 0.03 24 细胞器 866 13.34 49 翻译调节器活动 1 0.03 25 细胞器成分 345 5.32 50 金属伴侣蛋白活性 0 0 说明:生物过程编号为1~19;细胞组分编号为20~35;分子功能编号为36~50 Table 4. Difference expression gene GO annotation statistics
将高温组(TE1、TE2、TE3)与对照组(ck1、ck2、ck3)相比,进行全基因组及代谢途径调控网络富集分析,共有949个差异基因(different expression gene set,DEGs)可注释到全基因组及代谢途径通路中(表5)。集中在碳代谢途径(carbon metabolism)和内质网蛋白质加工(protein processing in endoplasmic reticulum)的差异基因数量较多(99个和59个),其次是苯丙烷类生物合成(phenylpropanoid biosynthesis)和糖酵解/糖异生途径(glycolysis/gluconeogenesis)。表明在高温刺激后,牡丹中的多种代谢途径、生物合成途径和内质网蛋白加工途径等均发生变化,积极参与应激响应。
全基因组及代谢途径通路术语 基因数目/个 全基因组及代谢途径编号 校正后P值 苯丙烷生物合成 47 ko00940 1.00E−07 内质网蛋白质加工 59 ko04141 5.50E−02 脂肪酸延伸率 14 ko00062 3.41E−05 萜类骨架的生物合成 26 ko00900 6.79E−05 光合生物中的碳固定 38 ko00710 7.39E−05 氮代谢 15 ko00910 7.84E−05 碳代谢 99 ko01200 1.23E−04 光合作用 25 ko00195 3.69E−04 类黄酮生物合成 12 ko00941 5.23E−04 脂肪酸降解 22 ko00071 9.32E−04 亚油酸代谢 6 ko00591 7.97E−03 α-亚麻酸代谢 18 ko00592 9.71E−03 鞘脂代谢 12 ko00600 9.81E−03 丙酮酸代谢 37 ko00620 1.11E−02 氰氨基酸代谢 17 ko00460 1.12E−02 缬氨酸、亮氨酸和异亮氨酸的降解 19 ko00280 1.30E−02 类固醇生物合成 12 ko00100 1.33E−02 脂肪酸代谢 25 ko01212 1.61E−02 谷胱甘肽代谢 27 ko00480 1.65E−02 其他聚糖降解 10 ko00511 1.70E−02 光合作用-天线蛋白质 8 ko00196 9.17E−03 糖酵解/糖异生 41 ko00010 2.24E−02 油菜素类固醇生物合成 6 ko00905 2.34E−02 甘氨酸、丝氨酸和苏氨酸的代谢 21 ko00260 5.25E−02 Table 5. Analysis of metabolic pathways for gene expression of differences under high temperature treatment
-
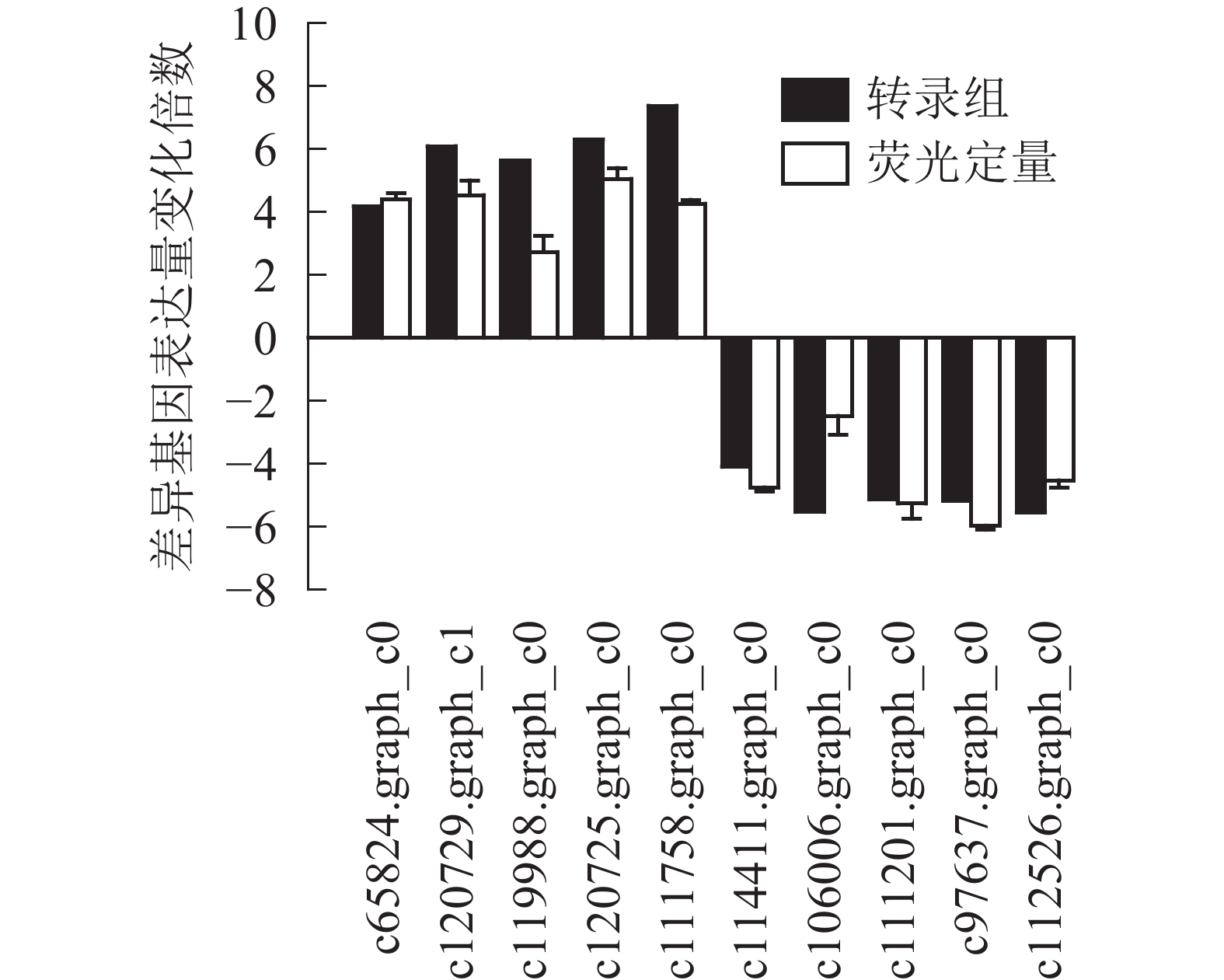
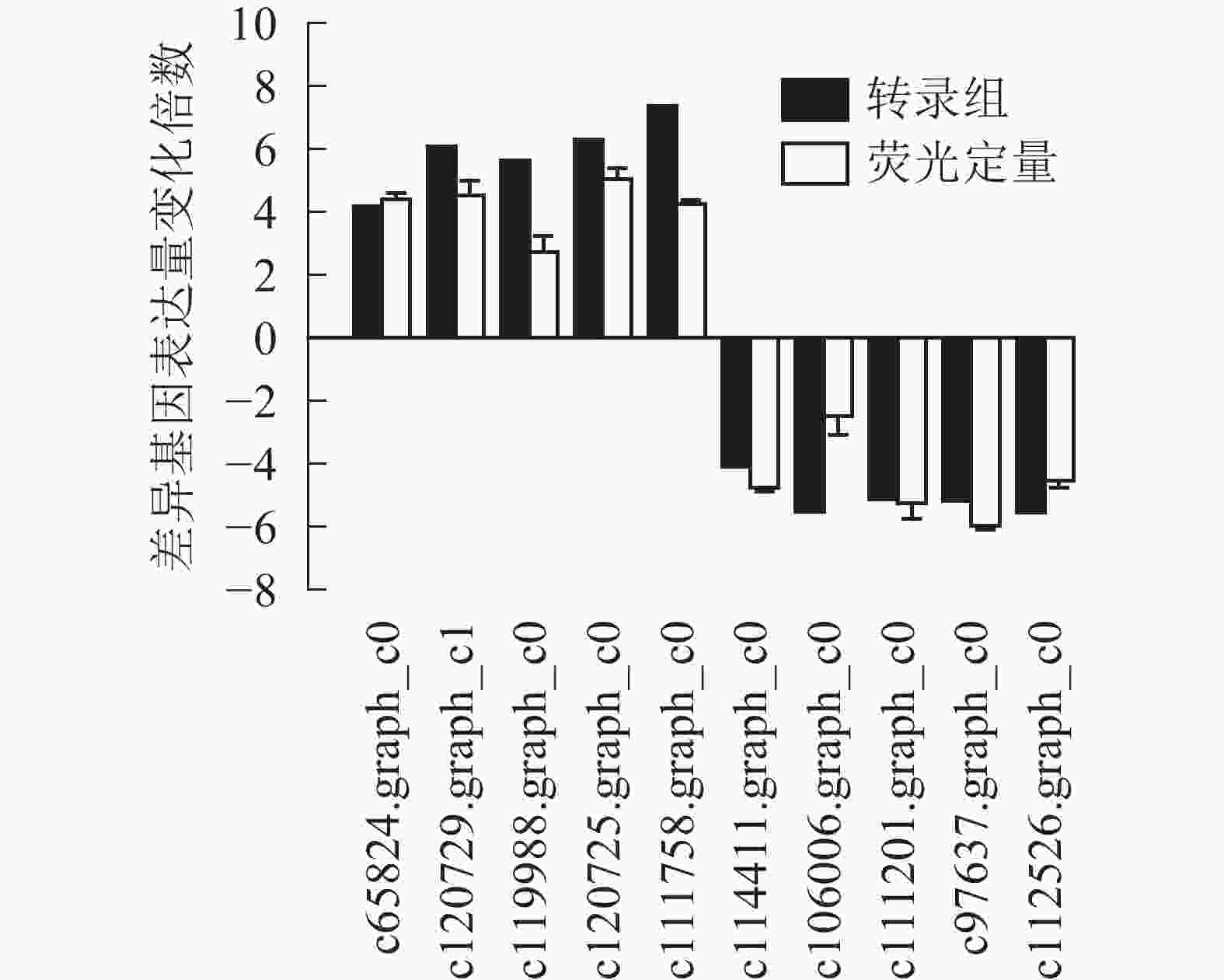
为了进一步验证转录组数据,随机选取10个上调和下调的差异基因,分别是热激蛋白70(PsHSP70,c65824.graph_c0)、核糖体蛋白S4(PsRPS4,c119988.graph_c0)、酸性黏多糖(PsAMP1,c120725.graph_c0)、肽基脯氨酰顺反异构酶(PsPPIL1,c111758.graph_c0)、4-香豆酸辅酶A连接酶(Ps4CL1,c114411.graph_c0)、阳离子氨基酸转运蛋白5(PsCAT5,c106006.graph_c0)、叶绿素酶(PsCHL1,c111201.graph_c0)、类胡萝卜素裂解双加氧酶(PsCCD4,c97637.graph_c0)、腺苷甲硫氨酸脱羧酶样原酶(PsSAMDC,C112526.graph_c0)以及假定蛋白(c120729.graph_c1),使用qRT-PCR的方法进行验证分析(图3),两者在变化趋势和差异倍数总体相符,表明测序结果相对可信。
-
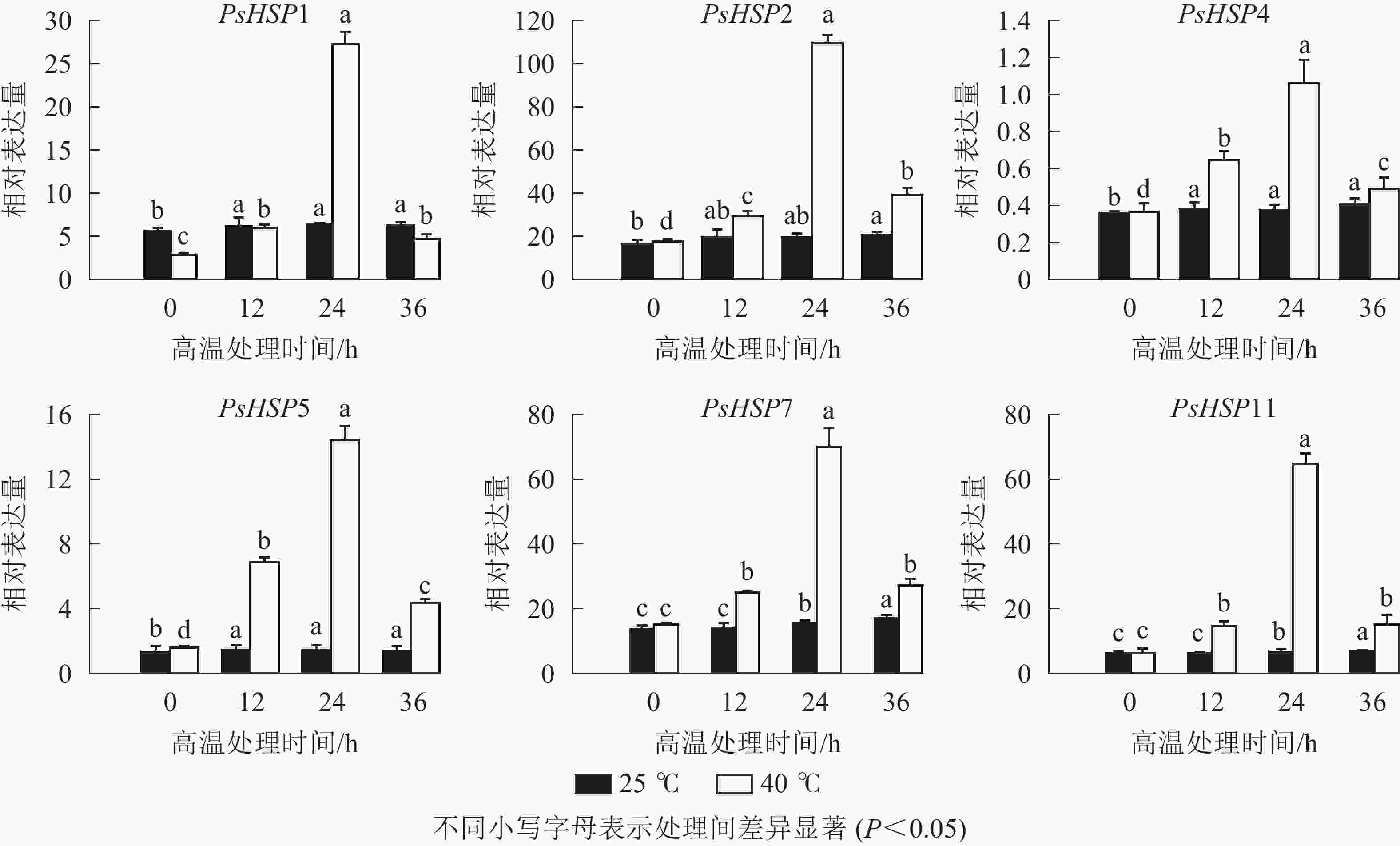
分析测序发现:17个热激蛋白基因在高温胁迫后显著上调表达(表6),这些热激蛋白多定位在细胞核、线粒体、叶绿体和细胞质等部位。选取6个PsHSP (PsHSP1、PsHSP2、PsHSP4、PsHSP5、PsHSP7、PsHSP11)分别在25和40 ℃处理下进行时空表达分析(图4)。在40 ℃高温处理下,6个PsHSP表达呈上升趋势,且全部于24 h达到最大值,之后便呈现下降趋势;25 ℃条件下,6个PsHSP表达变化相对稳定。
基因编号 基因命名 基因表达量 log2A 亚细胞位置预测 对照组25 ℃ 高温组40 ℃ c112621.graph_c0 PsHSP1 2.61 295.98 6.83 内质网 c107551.graph_c0 PsHSP2 59.46 2 254.78 5.24 细胞核 c114303.graph_c1 PsHSP3 161.17 440.53 1.45 线粒体 c112152.graph_c0 PsHSP4 11.64 162.01 3.80 线粒体 c117526.graph_c2 PsHSP5 7.06 172.15 4.61 细胞核 c103807.graph_c0 PsHSP6 10.79 26.40 1.29 线粒体 c116391.graph_c0 PsHSP7 150.15 2 659.72 4.15 叶绿体/细胞质 c100675.graph_c0 PsHSP8 21.49 215.13 3.32 叶绿体/细胞质 c117374.graph_c1 PsHSP9 100.54 591.55 2.88 叶绿体 c114184.graph_c0 PsHSP10 11.60 136.37 3.56 叶绿体/细胞质 c119657.graph_c2 PsHSP11 63.38 3 980.17 5.97 细胞质 c100778.graph_c0 PsHSP12 102.14 614.30 2.59 叶绿体/线粒体 c120033.graph_c1 PsHSP13 127.21 608.07 2.26 细胞核 c114516.graph_c1 PsHSP14 5.79 28.50 2.30 细胞膜 c118563.graph_c0 PsHSP15 9.93 58.46 2.56 叶绿体 c111652.graph_c0 PsHSP16 15.34 34.67 1.18 细胞核 c65824.graph_c0 PsHSP17 33.61 873.89 4.19 线粒体 Table 6. Peony leaves respond to high temperature heat shock DEGs
2.1. 测序数据组装结果
2.2. 转录组单基因簇的数据库功能注释
2.3. 响应高温胁迫的差异基因功能富集分析
2.4. 实时荧光定量分析
2.5. 高温胁迫下牡丹‘羽红’热激蛋白HSP基因挖掘及表达分析
-
全球变暖使得世界上许多地区农业受到不同程度的热害影响。近年来,植物耐热性研究逐渐受到关注。短期或持续高温胁迫引起植物形态和生理生化方面的改变,影响植物生长发育的同时也导致农林经济产量下降[1, 30-31]。高温是限制牡丹生长的重要因素且严重影响其观赏性。通过RNA-Seq测序发现:牡丹‘羽红’高温胁迫后共有7 673个基因差异表达,这些差异基因在基因本体注释主要集中在代谢过程、细胞过程和单一生物过程;全基因组及代谢途径数据库富集分析发现:大部分DEGs富集在碳代谢、内质网蛋白质的加工途径;同时,苯丙烷生物合成、光合生物中的碳固定、糖酵解/糖异生和丙酮酸代谢等途径均受到高温胁迫的影响。
极端温度可以造成植物体内蛋白质功能障碍导致生长发育受限,热激蛋白HSPs参与植物细胞膜稳定及相关代谢过程[12, 30],在响应和调节高温胁迫的过程中发挥重要作用[32-34]。芍药PlHSP70基因过表达拟南芥Arabidopsis thaliana发现,PlHSP70对拟南芥高温的耐受性有正向调节作用,高温处理后转基因株系的叶绿素荧光值明显高于野生型植株,此外还具有更加完整的细胞膜、叶绿体和淀粉颗粒。百合Lilium brownie LlHSP70基因异源转化拟南芥,LlHSP70可以增强转基因植株的高温耐性;烟草Nicotiana tabacum Class Ⅰ 类热激蛋白TLHS1基因在烟草幼苗中的表达水平与耐热性有较强的相关性,TLHS1转至烟草幼苗经过高温胁迫1~4 h后,子叶开张率比转反义转基因烟草幼苗高出1倍以上。本研究发现:高温胁迫后,共有17个PsHSP基因在牡丹中上调表达,且PsHSP1、PsHSP2、PsHSP4、PsHSP5、PsHSP7和PsHSP11在高温处理后表达上调3倍以上;同时PsHSPs时空表达也发现,PsHSP随热胁迫时间增加而表达量上升,24 h后达到最大值后便逐渐下降,说明PsHSP基因可能在短期内迅速响应高温胁迫并参与牡丹的耐热调控。综上,通过转录组测序发现,高温显著影响牡丹的代谢和生物合成等过程,从而抑制牡丹的生长和发育。PsHSP基因可在短期内迅速响应高温胁迫并参与牡丹的耐热调控。本研究可为进一步探究牡丹耐高温胁迫的分子机制提供依据。










 DownLoad:
DownLoad: