-
GRF(general regulatory factor)蛋白质最先由MOORE等[1]在牛脑中发现,并根据淀粉凝胶电泳上的迁移特性命名。GRF蛋白质是一类高度保守的同源或异源的二聚体蛋白质,具有多种功能,广泛存在于真核生物中,如酵母Pichia guilliermondii、拟南芥Arabidopsis thaliana、水稻Oryza sativa、花生Arachis hypogaea等。已有研究[2]表明:GRF蛋白质家族通过与磷酸化的靶蛋白质相互作用参与植物信号传导、细胞定位、转录调控和应激反应等多种重要生命活动过程,在植物代谢调控和生物合成反应中发挥着重要作用,如拟南芥GRF蛋白质可以与感光系统中的蛋白质相互作用调节根系生长发育[3];葡萄Vitis vinifera GRF蛋白质参与冷热应激反应[4];木薯Manihot esculenta GRF蛋白质主要分布在细胞质中,作用于淀粉合成酶Ⅲ靶蛋白质,对淀粉的合成起到负调控作用[2];菊花Dendranthema morifolium GRF蛋白质参与开花和周期调控,盐、冷等胁迫响应过程[5];动物细胞中GRF蛋白质还可通过调节细胞周期,影响细胞凋亡,参与多种信号通路等方式来调控肿瘤进程[6]。GRF活化后可以使G2/M期阻滞从而起到负调控细胞周期,发挥抑制癌基因的作用[7]。在动物中GRF蛋白质的过表达可能转化为一种致癌因子,促进肿瘤的发生[8],还可能与肿瘤细胞耐药性有关[9]。毛竹Phyllostachys edulis用途广泛,笋和叶具有食用、药用价值;竹材多用于建筑制造、工艺品制作。毛竹林是一种重要的经济林,具有重要生态价值,其固碳作用机制在不同的生长阶段有所差异[10]。毛竹基因组草图已公布,且大量转录组数据也可以从公共数据库中获取[11]。目前根据毛竹全基因组数据进行基因家族分析已取得了一定的成果,如ZF-HD基因家族[12]、B3基因家族[13]、APX基因家族[14]等,也分析了毛竹快速生长期的基因表达[15-16]。但对于毛竹GRF基因家族的全基因组数据分析尚未有相关报道。本研究通过毛竹公开的相关测序结果,利用生物信息学的方法,从基因组及转录组数据入手,对毛竹GRF基因进行全基因组的鉴定与表达分析,拟为进一步明确GRF基因家族在毛竹重要生长发育过程中的功能解析提供依据。
HTML
-
毛竹基因组序列、编码序列(CDS)、蛋白质序列和基因组GFF注释文件均从以下站点ftp://parrot.genomics.cn/gigadb/pub/10.5524/100001_101000/100498/[12]下载。从Pfam数据库[17]中下载隐马可夫模型(HMM) PF00244.17的结构域数据,并以此结构域数据为种子模型,用HMMER[18]检索本地毛竹蛋白质数据库。在Excel 2018中,将E-value设置为≤1E−20,对检索结果排序整理,去除重复,获得候选基因。进一步从毛竹全基因组数据库中提取得到GRF家族成员的基因、CDS、蛋白质序列以及基因结构和位置信息;利用在线工具ProtParam(https://web.expasy.org/protparam/)、ProtScale(https://web.expasy.org/protscale/)[19]以及SignalP 4.1[20]在线分析GRF家族各成员理化性质等。
-
依据毛竹、拟南芥、水稻GRF家族成员蛋白质序列,分别通过ClustalW多重比对,用MEGA 7.0软件邻位连接(neighbor-Joining, NJ)法构建种内和种间系统进化树,自检值取1 000次抽样[21]。
-
根据毛竹全基因组的GFF注释文件基因位置信息,分析毛竹GRF家族的基因结构并绘制基因结构图;利用在线网站NCBI Conserve Domain(https://www.ncbi.nlm.nih.gov/cdd/)和MEME(https://www.ncbi.nlm.nih.gov/cdd/)对GRF家族成员的保守结构域(domain)和基序(motif)进行预测[22],并通过TBtools[23]将结果可视化。
-
提取毛竹GRF基因上游1 500 bp序列作为启动子序列信息,通过在线预测软件PlantCare[24]预测毛竹GRF基因的顺式作用元件,并整理预测结果,富集顺式作用元件,利用TBtools上的Simple Biosequence viewer功能进行可视化分析。
-
利用MCScanX[25]获取GRF家族种内、种间共线性关系,并用TBtools软件Amazing Super Circos[26]和Multipe Synteny Plot分别对种内和种间的结果可视化。
-
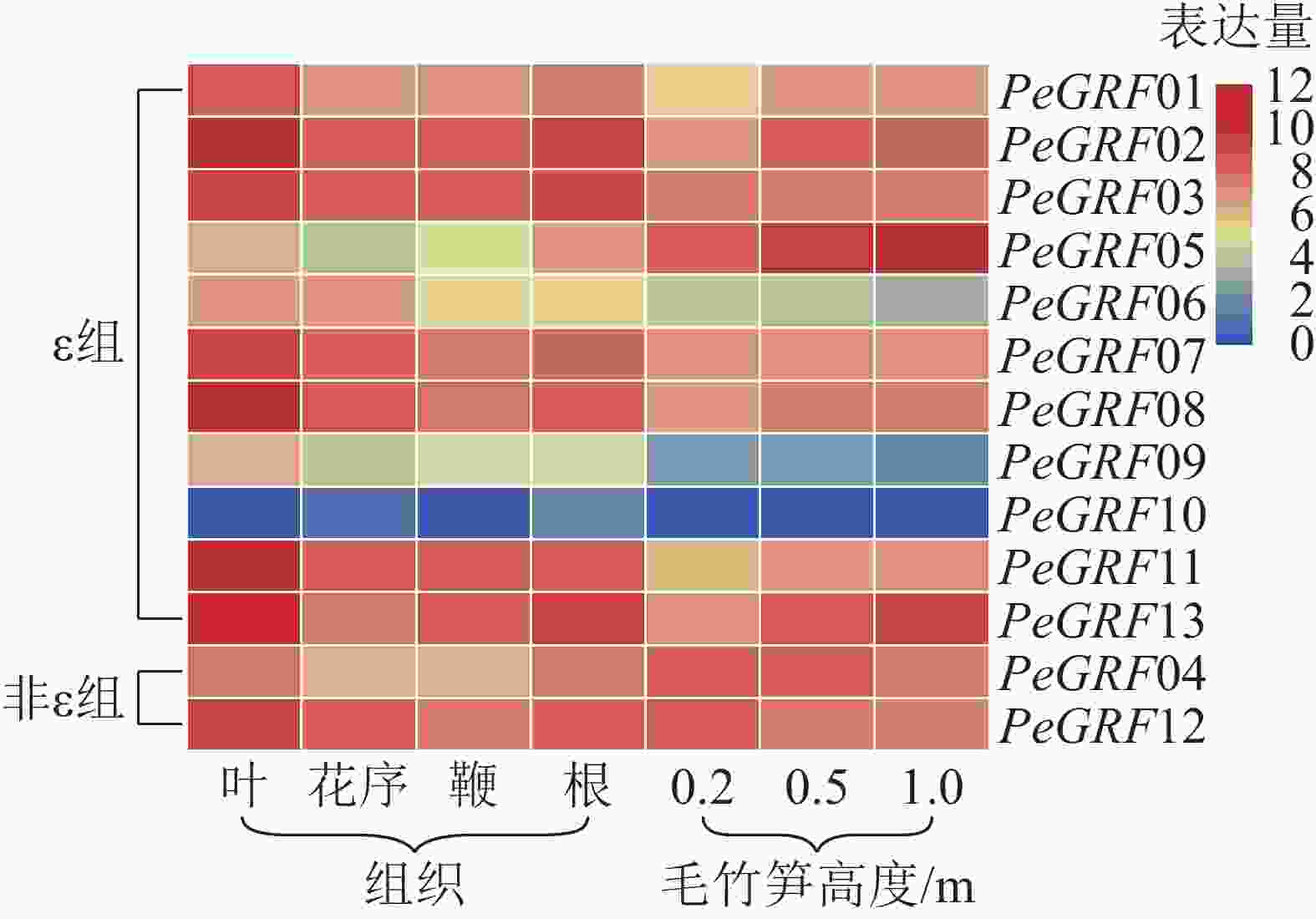
选取NCBI SRA数据库中毛竹不同组织器官:根(登录号为ERR105075、ERR105076),花序(登录号为ERR105069、ERR105070、ERR105071),叶(登录号为ERR105067、ERR105068、ERR105075),鞭(登录号为ERR105073、ERR105074)和笋不同生长高度:0.2 m(登录号为SRR6131114、SRR131113、SRR6131115),0.5 m(登录号为SRR131117、SRR6131118、SRR5710699)和1.0 m(登录号为SRR5710701、SRR5710702、SRR5710697)的转录组数据,分别计算毛竹GRF基因的TPM(transcripts per million reads)值表示基因的表达丰度。为方便统计,对每个表达数值取以2为底的对数(log2),使用TBtools Amazing Heatmap绘制基因表达热图,用对数转换预处理数据,再用正态标准化的方法处理数据。
-
利用SWISSMODEL(https://www.swissmodel.expasy.org/)在线软件[27]预测GRF蛋白质的3D结构。模建结果使用SAVES v5.0[19]进行评估。
1.1. 基因家族来源、鉴定及理化性质分析
1.2. 家族进化分析
1.3. 基因结构、基序和保守结构域预测
1.4. 启动子分析
1.5. 染色体定位及共线性分析
1.6. 基因表达分析
1.7. 蛋白质三级结构同源模建
-
根据植物GRF隐马可夫模型(PF00244.17)搜索毛竹相关基因组数据,获得相关GRF家族成员,然后通过E-value(≤1E−20)筛选、保守结构域、基序特征分析,去除相同转录本重复,最终筛选得到13个GRF家族成员(表1)。将获得13个GRF家族成员按照其在scaffold的分布先后顺序命名为PeGRF01~PeGRF13。进一步对PeGRF作蛋白质特性分析,13个GRF蛋白质中长度最短的为PeGRF10(256个氨基酸),最长的为PeGRF09(293个氨基酸),平均长度266.8个氨基酸;各GRF蛋白质等电点最小的为4.70(PeGRF02),最大的为5.29(PeGRF01),平均等电点为4.82;各GRF蛋白质分子量最小的为PeGRF04(28.65 kD),最大的为PeGRF09(32.41 kD),平均分子量为29.79 kD。
基因登录号 基因名称 等电点(pI) 平均分子量/kD 内含子数量/个 氨基酸数量/个 PH02Gene26029.t1 PeGRF01 5.29 32.20 5 286 PH02Gene21972.t1 PeGRF02 4.70 29.68 4 262 PH02Gene06378.t1 PeGRF03 4.79 29.14 4 263 PH02Gene19868.t1 PeGRF04 4.82 28.65 3 261 PH02Gene15394.t1 PeGRF05 4.76 31.08 4 274 PH02Gene31988.t1 PeGRF06 4.73 29.94 6 270 PH02Gene44376.t1 PeGRF07 4.79 29.15 4 263 PH02Gene09923.t1 PeGRF08 4.86 29.28 4 261 PH02Gene25395.t2 PeGRF09 4.72 32.41 5 293 PH02Gene13806.t1 PeGRF10 4.76 29.02 4 256 PH02Gene13908.t1 PeGRF11 4.82 29.15 4 260 PH02Gene26176.t1 PeGRF12 4.84 28.76 3 263 PH02Gene15240.t1 PeGRF13 4.75 28.77 4 256 Table 1. Characteristics of PeGRF family genes and their deduced proteins
-
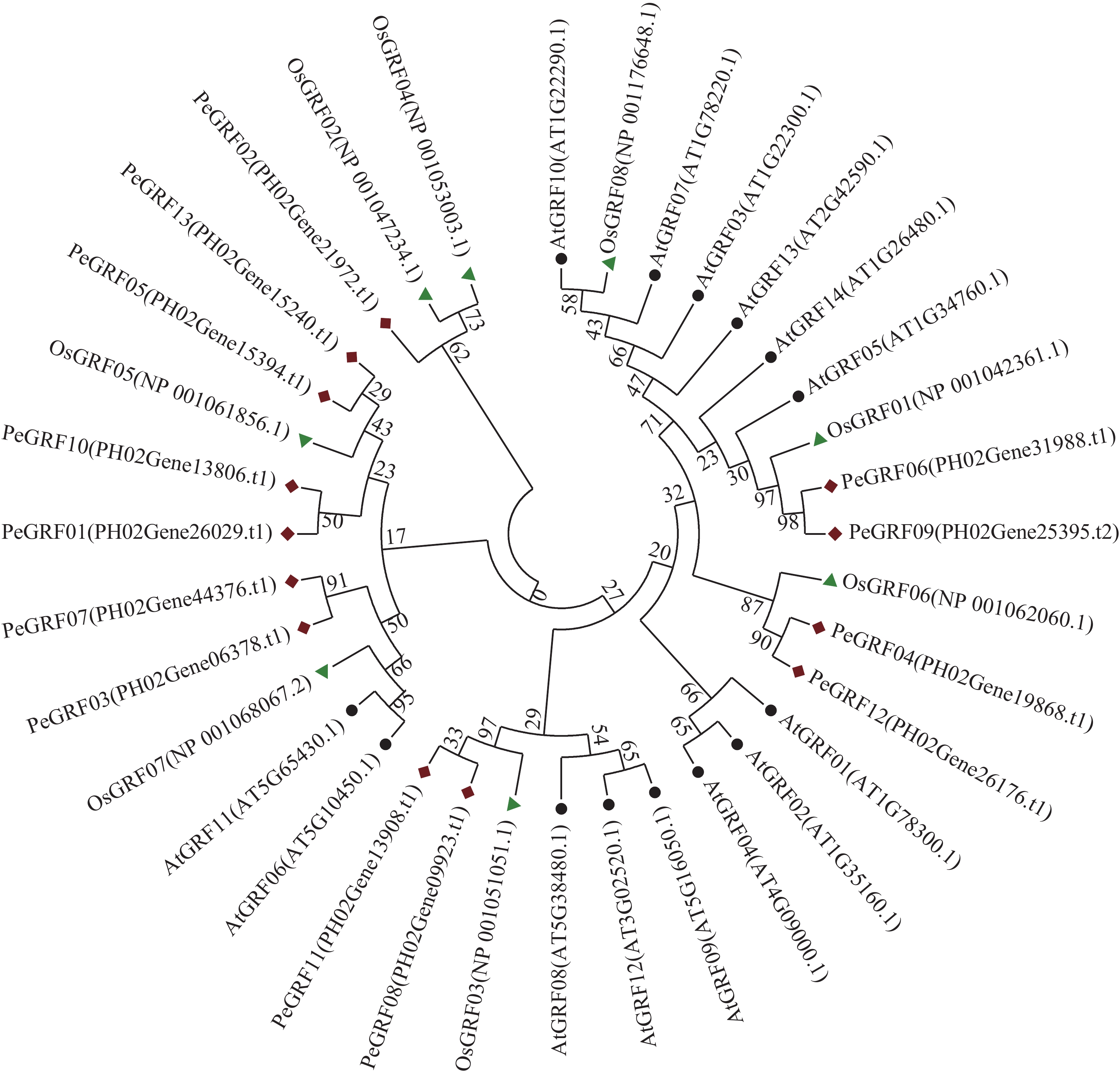
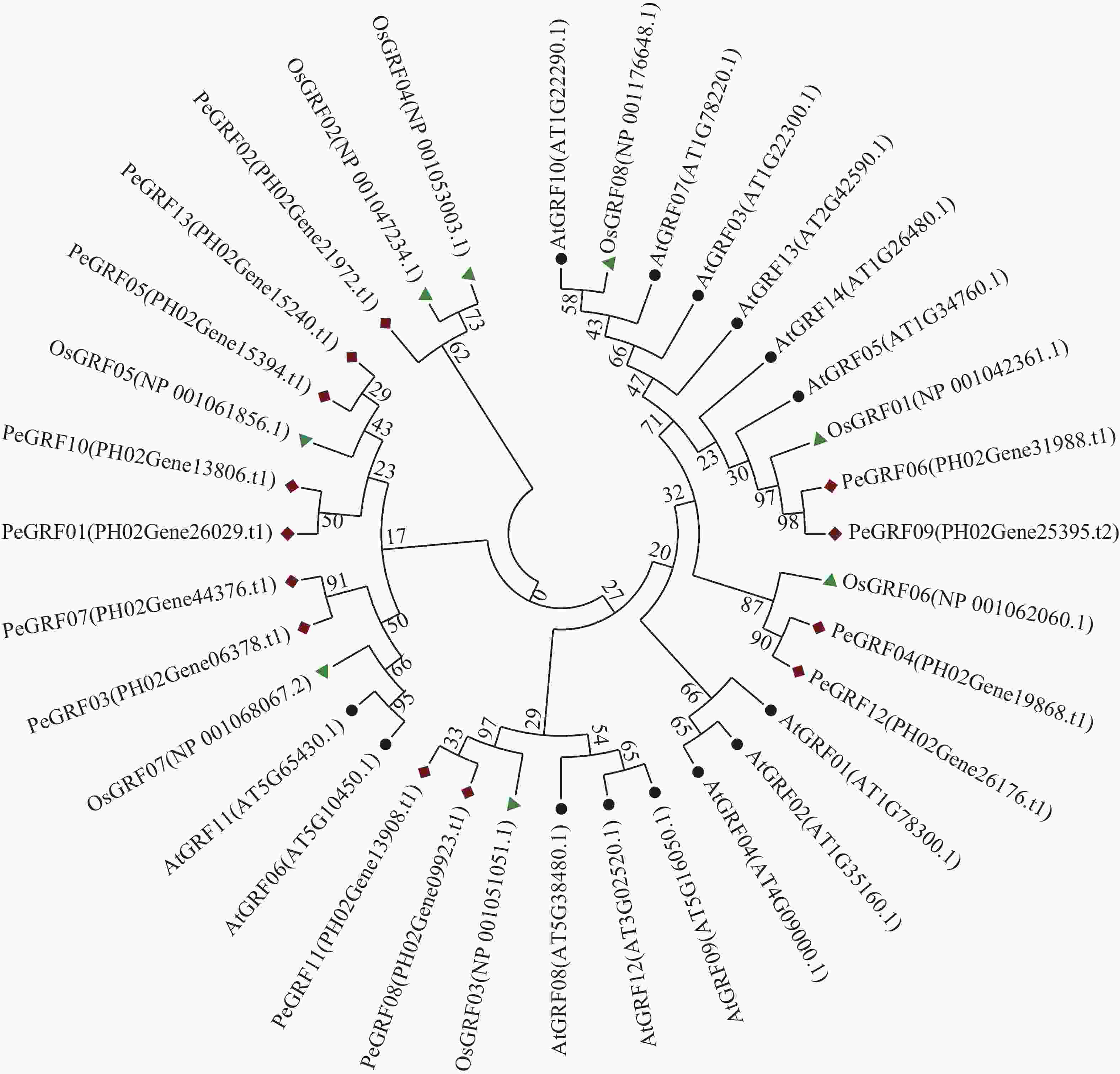
利用MEGA 7.0对13个毛竹GRF、14个拟南芥GRF和8个水稻GRF的氨基酸序列比对后,采用NJ法进行系统聚类分析(图1),绝大部分毛竹基因家族成员和水稻处于同一分支,表明毛竹与水稻的进化关系较近。
-
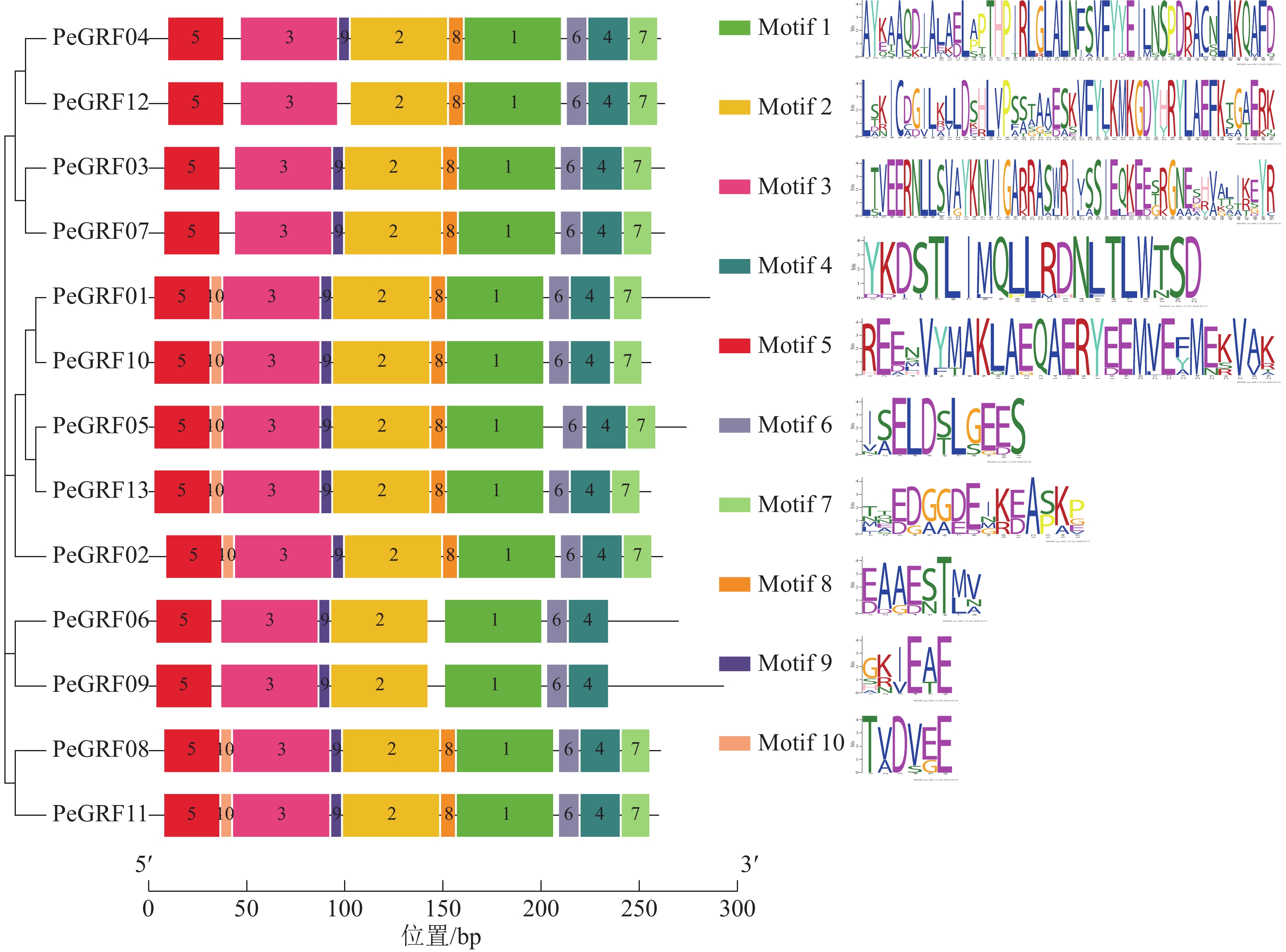
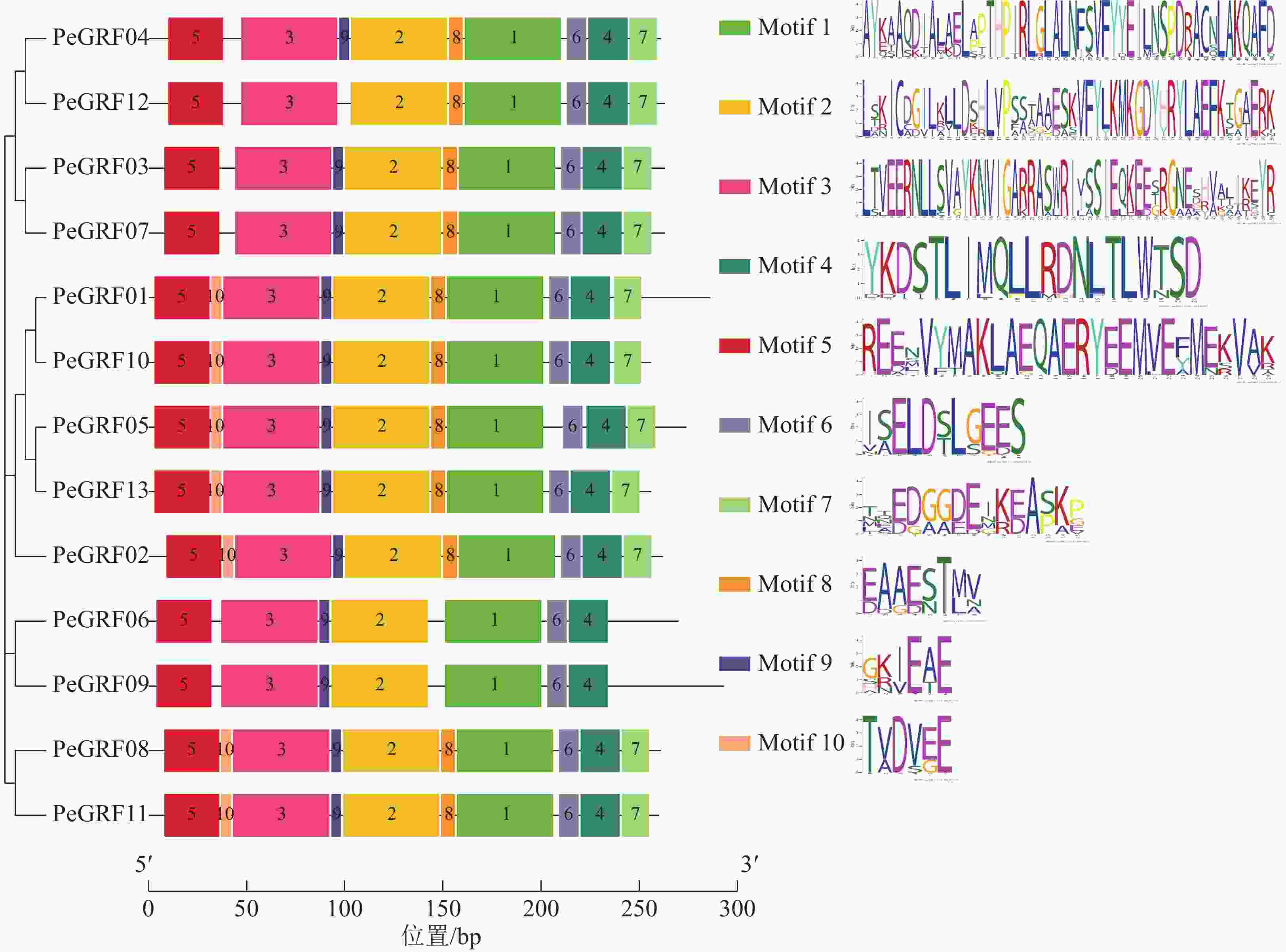
对毛竹GRF基因结构分析发现:内含子数量存在差异,非ε组成员都包含4个外显子和3个内含子,它们在位置上高度保守。ε组成员都具有不同于非ε组的内含子-外显子结构,具有2个额外的N-末端内含子[21]。利用NCBI-CDD对毛竹GRF基因进行保守结构域分析,PeGRF蛋白质均包含14/3/3结构域,毛竹GRF基因家族14/3/3结构域存在一定的保守性,但该结构域的分布位置有一定分化。利用MEME在线工具对该基因家族的保守基序预测,基数设置为10,结果显示(图2):Motif1~6在每个家族成员中均出现,属于高度保守结构,其余基序在家族成员中出现的频率及所在位置均存在一定的差异。
-
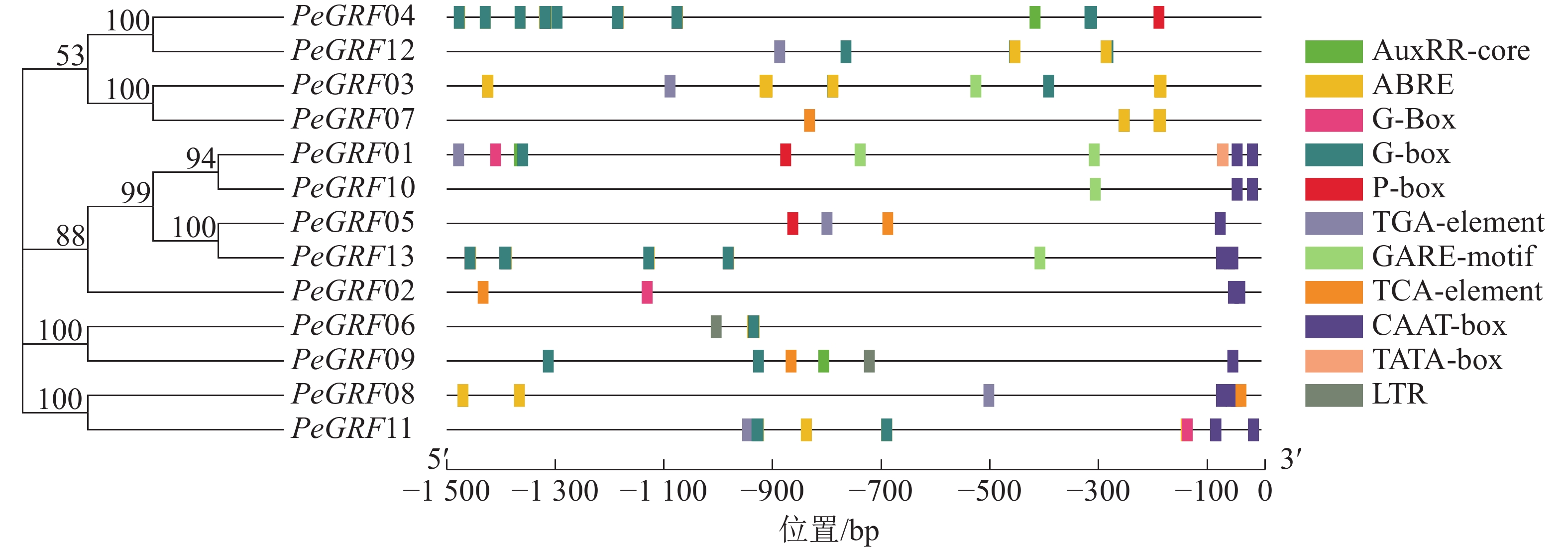
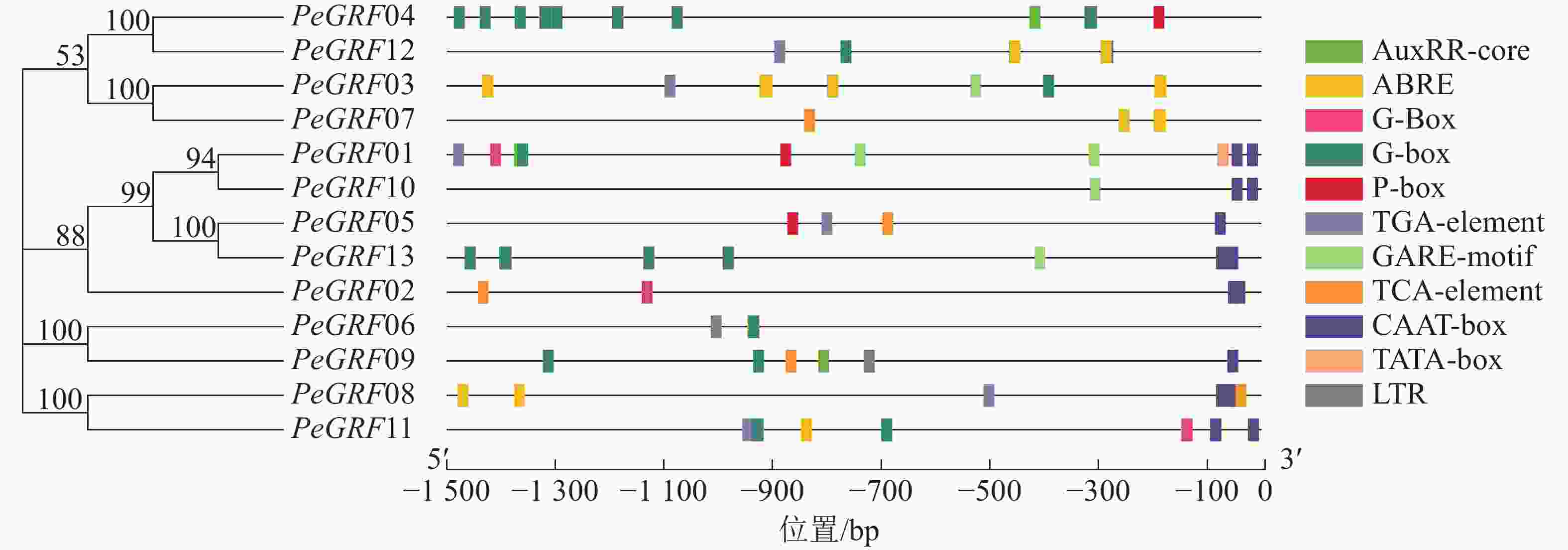
如图3所示:筛选出的部分典型的顺式调控元件,除核心启动子TATA-box(5个)和CAAT-box(16个)外,还有与激素相关的顺式调控元件,包括与赤霉素相关的GARE-motif(5个)、P-box(3个),与生长素有关的AuxRR-core(3个)、TGA-element(6个),与脱落酸有关的ABRE(42个),与水杨酸有关的TCA-element(5个);与外部条件有关的顺式调控元件,包括参与低温响应的LTR(2个)和光响应的G-box(48个)。推测毛竹GRF蛋白质家族可能参与激素和非生物胁迫响应,家族基因表达模式可能有所不同。
-
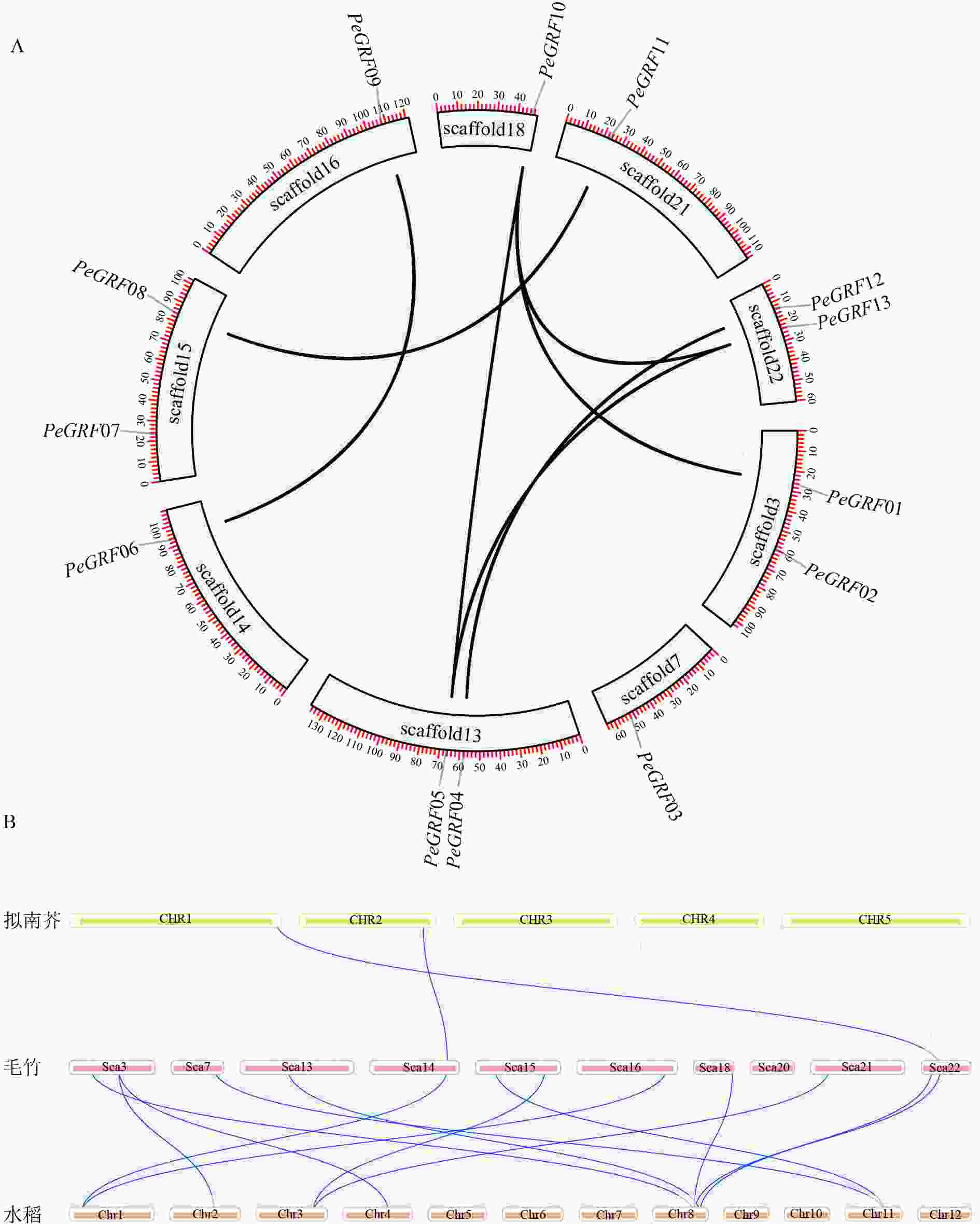
利用毛竹基因组GFF注释文件提取PeGRF在scaffold上的分布特征,结果显示:毛竹GRF基因在scaffold上分布不均匀,不同的scaffold基因分布密度不同,scaffold7、14、16、18和21仅包含1个PeGRF,scaffold3、13、15和22上分别包含2个。
利用TBtools工具,将毛竹GRF基因种内和种间的共线性关系进行了可视化分析。从图4A中可以看出:除PeGRF02、PeGRF03和PeGRF07不存在种内共线性关系外,其余家族基因成员间均有显著的共线性关系,说明GRF基因家族存在基因复制现象,推测在进化过程中GFR基因可能通过复制进行家族成员数量的扩张。但PeGRF不存在串联重复基因。物种间的共线性关系是反映不同物种来源于同一个祖先的现象。从图4B可以看出:毛竹与水稻的共线性关系要明显多于拟南芥,这可能与水稻和毛竹同属于禾本科Gramineae,进化关系较近有关。
-
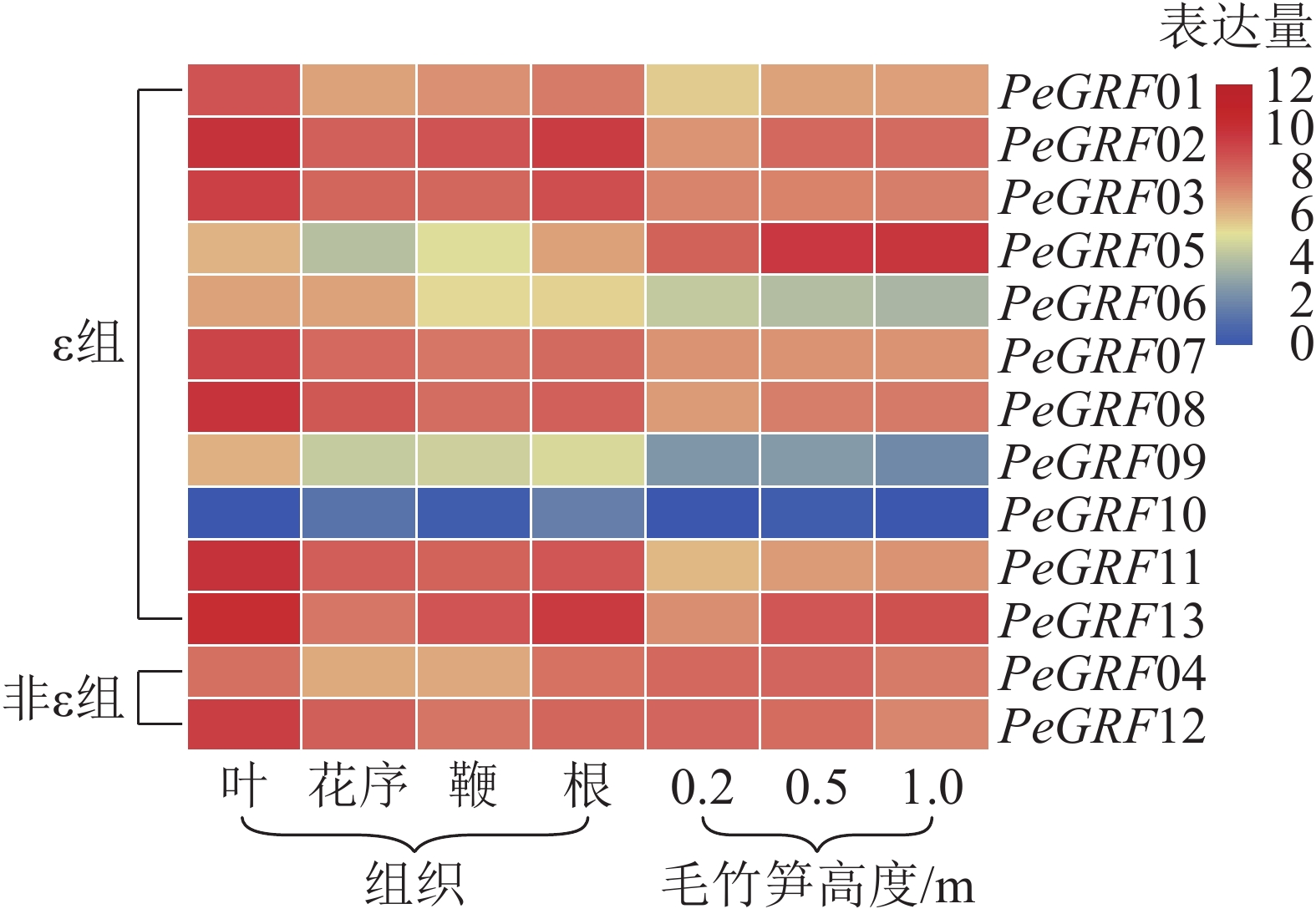
本研究基于毛竹RNA-Seq转录组数据,对毛竹不同组织(叶、花序、鞭及根)以及不同生长高度(0.2、0.5、1.0 m)的毛竹笋中的GRF表达量绘制热图。由图5可以看出:除PeGRF10,PeGRF09在不同组织和生长高度保持较低的表达量外,其他成员均有较高的表达量。在毛竹不同组织中,根和花序的表达量相对于叶和鞭要稍高;非ε组的GRF基因均有较高的表达。在竹笋的不同生长阶段,非ε组的GRF基因保持较高的表达水平;ε组不同的基因表达量有增有减,如PeGRF05在竹笋生长各个阶段均有较高的表达量,且随生长进程表达量不断增高;PeGRF06表达量随生长进程呈下降趋势。推测不同家族成员在参与组织器官发育的过程中发挥不同的作用,但其中的内在分子机制还值得进一步研究。
-
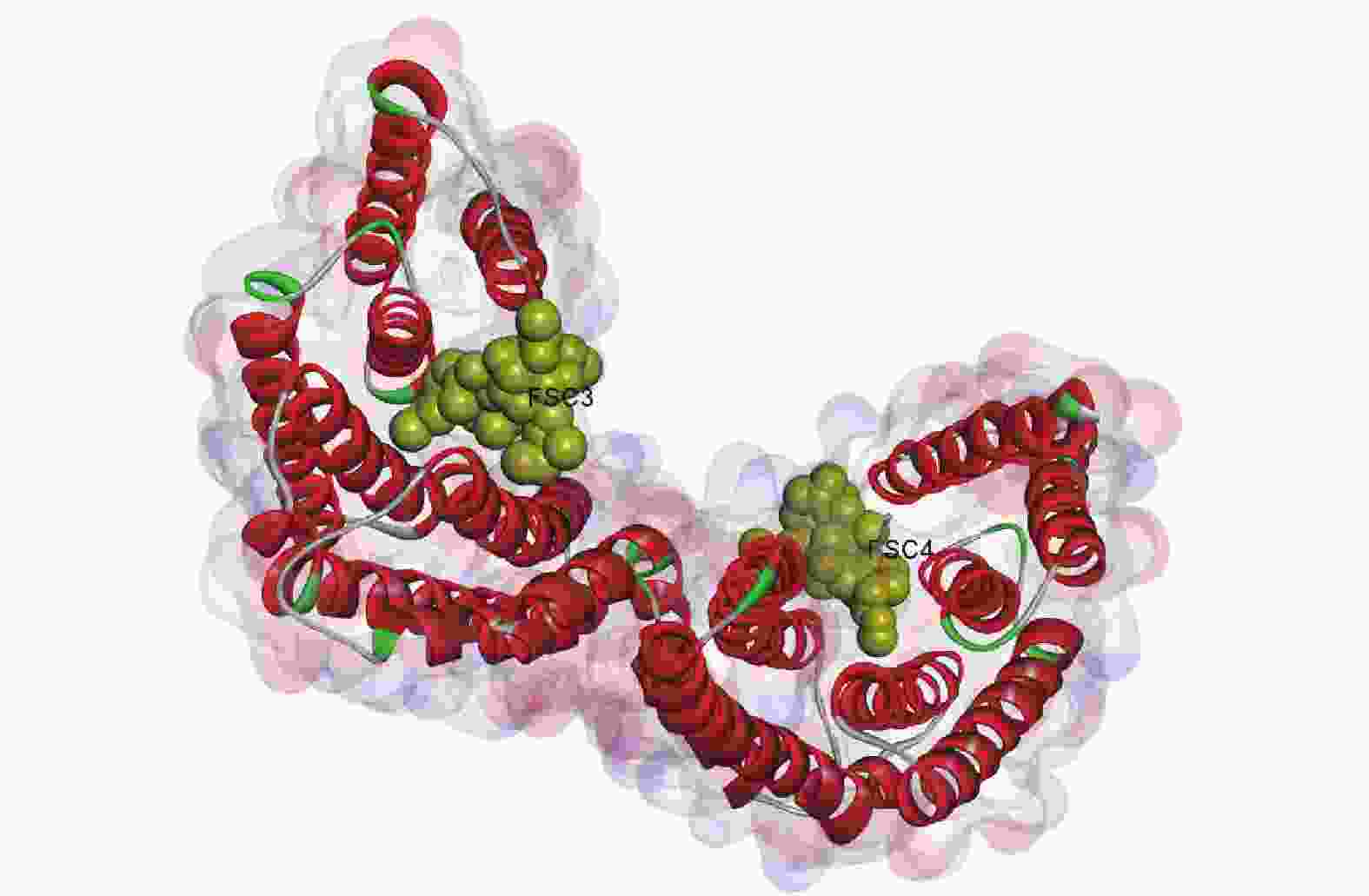
由图6所示:毛竹GRF蛋白质由2个单体连接而成,每个单体由反向平行的9个α螺旋组成,每个单体都存在与配体(FSC3、FEC4)相互作用的结合位点,2个FSC配体均与壳梭孢素有关,单体间构成同源或异源二聚体,总体呈“W”型[28-29]。
2.1. 基因家族成员鉴定及理化特性分析
2.2. GRF基因家族分类与进化树构建
2.3. GRF家族基因结构、基序及保守结构域
2.4. 启动子特征
2.5. 染色体分布及共线性分析
2.6. GRF家族基因表达模式
2.7. GRF家族蛋白质空间三级结构
-
物种基因组全序列的测定推动了生物信息学的迅速发展,在海量数据的基础上,利用生物信息学手段,对物种基因家族进行高效的统计分类和分析,预测基因家族的结构、功能及作用机制,将极大地推动相关功能基因的挖掘和农艺性状遗传的改良进程[30]。随着2018年第2版毛竹基因组数据的公布以及大量毛竹转录组数据的共享,毛竹GRF基因家族的生物信息学分析成为可能[11]。本研究通过全基因组数据分析发现:毛竹GRF家族成员共13个,数量多于水稻,可能的原因是毛竹染色体经过加倍,基因组数据远大于水稻;另外,共线性分析进一步证实:正是通过基因复制扩增,毛竹GRF在数量上有优势。毛竹GRF基因家族各成员间的理化性质存在一定的差异,但均含有14/3/3蛋白质结构域,其中有6种基序在每个成员中均出现。根据基因结构将PeGRF分为ε组和非ε组,其中ε组可能保留了祖先的蛋白质功能,这与PIOTROWSKI等[31]和WANG等[32]的研究结果相似。
大量研究表明GRF蛋白质参与激素信号的转导。如在拟南芥的研究中发现:GRF参与油菜素类激素(BR)调控细胞核发育的途径[33];在烟草Nicotiana tabacum中,GRF参与赤霉素(GA)生物合成调控[34];在水稻中,GRF表达同脱落酸(ABA)密切相关[35]。本研究发现:毛竹GRF顺式作用元件存在许多激素相关元件。由此可以推测毛竹GRF蛋白质可能介导激素信号的转导过程。但毛竹GRF同其他激素的相互关系还需进一步验证。
GRF蛋白质参与了植物的生长发育,特别是在花器官的发育中具有重要作用。PERTL等[36]证实随着百合Lilium brownii var. viridulum花粉管的生长,GRF蛋白质的表达量也明显增加。李兵娟[37]也证实雷竹Phyllostachys violascens GRF基因参与开花调控机制。本研究通过转录组数据分析发现:GRF蛋白质在花序组织中高表达,且表达量明显高于竹叶和竹鞭,这表明毛竹GRF基因可能参与花序的发育和调控。除此之外,在研究毛竹GRF顺式作用元件时还发现其启动子区域存在许多光响应元件,结合光周期对植物开花的作用机制以及在模式植物水稻上的研究[38],GRF基因可能是通过光响应元件接受外界环境信号从而触发其高表达,最终影响毛竹花的发育。由于受毛竹花发育相关材料的限制,该假设将在后续实验验证。
毛竹GRF蛋白质是以一个螺旋结构为主的同源二聚体,二聚体界面内包着多个疏水残基和多个极性残基,外周则由盐桥连接,三级结构呈“W”型,每个单体分别含有2个凹槽,可能用于结合配体靶蛋白质。毛竹GRF蛋白质序列在进化谱系中高度保守,并且与配体结合的氨基酸残基极端保守,这同SEHNKE等[28]发现的结果相似。另外,虽然毛竹GRF蛋白质的N端和C端同源性较低,但可能通过碱性簇维持空间构象的稳定[28]。PAUL等[39]在研究拟南芥GRF蛋白质时发现,GRF蛋白质还可以通过结合磷酸化的蛋白质,参与重力反应等生理过程。GRF蛋白质在进化上高度保守,毛竹PeGRF可能也具有相似的分子作用机制。但毛竹GRF蛋白质生物学功能与上述空间结构之间的关系还需进一步的探索。









 DownLoad:
DownLoad: