-
梅花Prunus mume为蔷薇科Rosaceae李属Prunus落叶乔木,其独特的观赏特性与深厚的文化内涵使它成为重要的景观树种[1]。在观赏树木评价体系中,树体构型、枝姿形态特征与花器官具有同等重要的观赏价值[2]。树形结构的形成与分枝特性密切相关[3]。然而,目前关于梅花分枝性状的遗传调控机制仍不清晰,这限制了梅花品种改良与应用进程。
植物分枝作为决定株型建成的关键因素,其腋芽发育过程表现出显著的表型可塑性[4]。腋芽的激活受多层级网络调控[5],涉及内源激素信号通路、遗传调控模块与环境因子的协同互作[6]。现有研究表明:生长素[7]、细胞分裂素[8]和独脚金内酯[9]构成调控腋芽激活的核心激素调控系统,同时受光照强度[10]、营养状况[11]、水分条件[12]、温度波动[13]及生物胁迫[14]等环境信号的显著调节。植物分枝形成的分子调控机制十分复杂,近年来已经确定了几个调节腋芽分枝的转录因子,其中BRC1[15]、BRC2[16]、MYB2[17]、TB1[18]、SPL[19]在腋芽生长和分枝中起抑制作用,NAC[20]、WOX[21]、WRKY[22]刺激腋芽形成并促进分枝。
AP2/ERF是植物中广泛存在的一类转录因子超家族,该家族蛋白含有AP2/ERF结构域,第1个AP2基因是在模式植物拟南芥Arabidopsis thaliana中被发现的[23]。根据AP2/ERF类转录因子结构域的数量及结合序列,将其分为5个亚家族:AP2、ERF、DREB、RAV和Soloist[24],各亚家族呈现显著的功能分化。AP2/ERF家族主要在植物抵御寒冷[25]、干旱[26]、盐碱[27]、高温[28]等非生物胁迫以及生物胁迫[29]等方面发挥重要作用,同时在植物生长发育调控方面的作用也正在被挖掘。例如水稻Oryza sativa DSP基因调控分蘖和穗型发育[30],OsEATB影响植株高度和穗长[31]。杨树Populus trichocarpa PtAIL1促进不定根发生[32]。此外,AP2/ERF成员通过整合激素与代谢信号调控芽器官发育。卷丹Lilium lancifolium LlERF12通过感知乙烯信号调控珠芽形成[33]。烟草Nicotiana tabacum NtESR2则通过维持生长素运输稳态保障叶芽形态建成[34]。拟南芥EBE基因通过调控细胞增殖影响腋芽发育和芽分枝[35]。黑杨P. nigra PnANTL1和PnANTL2刺激细胞扩增使叶长增加[36];ERF114在欧洲油菜Brassica napus中通过重塑顶端生长素梯度增加分枝数[37];ERF115则通过茉莉酸-细胞分裂素信号互作抑制拟南芥不定根发生[38],ERF109在拟南芥中由损伤诱导调节根再生[39],表明该家族在分生组织调控中的功能多样性。上述研究阐释了AP2/ERF转录因子通过时空特异性调控激素网络、代谢通路及分生组织活性从而协调植物发育可塑性的分子机制。
梅花作为重要的早春木本观赏植物,以直枝类为主,垂枝类和龙游类品种极少,因此株型是梅花育种的重要方向。AP2/ERF转录因子家族在植物分枝调控中具有重要作用,前期研究在梅花中鉴定出了116个AP2/ERF家族成员,细分为3个家族(AP2、ERF和RAV)及1个单独成员Soloist,PmERF011属于ERF家族[40],在其他物种中的同源基因被发现参与植物生长发育,但梅花PmERF011的生物学功能仍不清楚。为此,本研究克隆了梅花PmERF011基因并进行基因时空表达模式、转基因功能验证分析,以明确它在梅花分枝调控中的作用。
-
梅花‘早绿萼’‘Zao Lve’1年生枝条取自北京林业大学温室;用于RNA提取的茎段、叶片等材料经液氮速冻后保存于−80 ℃冰箱中。野生型拟南芥(Col-0)和pCAMBIA2300载体由北京林业大学园林学院花卉种质创新与分子育种北京市重点实验室保存。
-
通过龙游梅基因组获取PmERF011的核酸和蛋白序列(https://www.rosaceae.org/Analysis/13114608);通过ExPASy(https://web.expasy.org/protparam/)网站分析其分子量、氨基酸长度、理论等电点、不稳定指数和亲水指数等,并利用Cell-PLoc 2.0(http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/)在线分析工具对PmERF011转录因子进行亚细胞定位预测。通过Sopma (https://npsa.lyon.inserm.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html)预测蛋白质二级结构,观察α-螺旋、β-折叠和无规卷曲等的组成比例;利用Swiss model (https://swissmodel.expasy.org/)预测蛋白质三级结构。提取PmERF011上游2 000 bp作为启动子序列,利用在线软件PlantCARE(https://www.plantcare.co.uk/)对PmERF011启动子序列进行顺式作用元件的预测分析,进而推测目的基因主要功能。通过美国生物技术信息中心(NCBI)网站对PmERF011蛋白序列进行Blast操作,找到梅花近缘物种ERF家族中与PmERF011蛋白相近的序列,使用ESPript 3.0(https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi)软件将PmERF011蛋白序列与通过NCBI-blast操作得到的蛋白序列进行多重比对,确定相似区域和变异位点。同时使用MEGA 7.0 (https://www.megasoftware.net/)软件构建系统发育树,生成算法采用邻接法。
-
为检测梅花PmERF011基因在不同组织的表达水平,选取长势健壮且一致的‘早绿萼’3株,收集幼叶、成熟叶、老叶、根、树皮、木质部、形成层、韧皮部、嫩茎、半木质茎、完全木质茎11种组织材料,其中形成层组织分离方法:枝条剥去外层树皮后,在荧光体视显微镜(Leica M165FC)下,使用无菌解剖刀沿去皮木质部交界处轻轻刮取形成层薄壁细胞(厚度为0.2~0.5 mm),尽量避免木质部组织污染。样品分离后立即置于液氮中速冻,确保RNA完整性。液氮速冻后分为2个部分:一部分送广州基迪奥生物科技有限公司利用Illumina HiSeq 2500进行转录组测序,另一部分保存于−80 ℃冰箱中用于实时荧光定量PCR (RT-qPCR)检测。利用RNAprep Pure多糖多酚植物总RNA提取试剂盒(DP441,天根生物技术公司)提取梅花上述11种组织总RNA,并利用NanoDrop-1000测定样品RNA浓度和质量,进一步使用质量分数为1%的琼脂糖凝胶电泳检测样品RNA的完整性。参照PrimeScript RT reagent Mix with gDNA Eraser试剂盒操作流程将梅花总RNA反转录为cDNA。
-
根据转录组测序数据结果,使用TBtools绘制组织特异性表达热图。同时利用IDT在线软件(https://sg.idtdna.com/scitools/Applications/RealTimePCR/)设计梅花PmERF011基因特异性引物(表1)。TB Green嵌合荧光法用于RT-qPCR。使用PikoReal系统(Thermo Fisher Scientific)进行RT-qPCR分析。反应体系为:TB Green Premix Ex Taq II(2×)10.0 μL,上游引物0.8 μL,下游引物0.8 μL,cDNA 2.0 μL和ddH2O 6.4 μL;反应程序为:95 ℃预变性30 s;95 ℃反应 5 s,60 ℃反应15 s,40个循环;溶解曲线60~95 ℃。选用梅花PmPP2A基因作为内参基因。采用3个生物学重复和3个技术重复,目的基因表达量采用2−△△Ct法计算。
类别 序列(5′→3′) 类别 序列(5′→3′) 克隆正向引物 TGCTCTCAGGAATTGCAAGTG RT-qPCR反向引物 TCTAATCTCAGCCACCCACTTC 克隆反向引物 TGAACTCCTTGTCATCCTTGATGA 卡那霉素基因正向引物 AAGATGGATTGCACGCAGGT RT-qPCR正向引物 AAACCAGCAGCAACAGCAAC 卡那霉素基因反向引物 TCACGGGTAGCCAACGCT Table 1. Primer sequences
-
根据梅花参考基因组[41],获得PmERF011基因序列。利用在线软件Primer Premier 5 (https://www.premierbiosoft.com/prim erdesign/)设计特异性克隆引物,送至北京擎科生物工程股份有限公司合成,引物序列见表1。克隆模板为‘早绿萼’cDNA,使用TaKaRa Prime Star Max高保真酶进行PCR反应,琼脂糖凝胶电泳检验条带正确,使用购自天根生化科技有限公司(北京)的普通DNA产物纯化试剂盒(DP 204)对目的片段纯化回收。与pTOPO克隆载体连接,转化至大肠埃希菌Escherichia coli DH5α感受态细胞。将感受态细胞涂布于含有50 mg·L−1卡那霉素的LB固体培养基上,于37 ℃恒温培养箱中倒置过夜培养。挑取阳性单菌落进行鉴定,经菌落检测后送至北京擎科生物工程股份有限公司测序,测序正确的菌株保存为pTOPO-PmERF011载体。
-
利用本课题组保存的pCAMBIA 2300载体构建PmERF011过表达载体。用TaKaRa限制性内切酶Xba Ⅰ Quickcut、Sac Ⅰ Quickcut进行双酶切,使其线性化。利用带有同源臂的克隆引物,以pTOPO-PmERF011质粒作为模板,进行PCR反应,对产物进行分离纯化,将双酶切线性化后的载体与DNA片段进行重组,转化大肠埃希菌,挑取单克隆进行PCR鉴定。将阳性PCR菌液进行送测,最后获得正确的重组质粒。采用液氮冻融法将测序成功的质粒转化至农杆菌Agrobacterium tumefaciens GV3101感受态中,进行菌液PCR检测,将成功转化的农杆菌于−80 ℃保存。
-
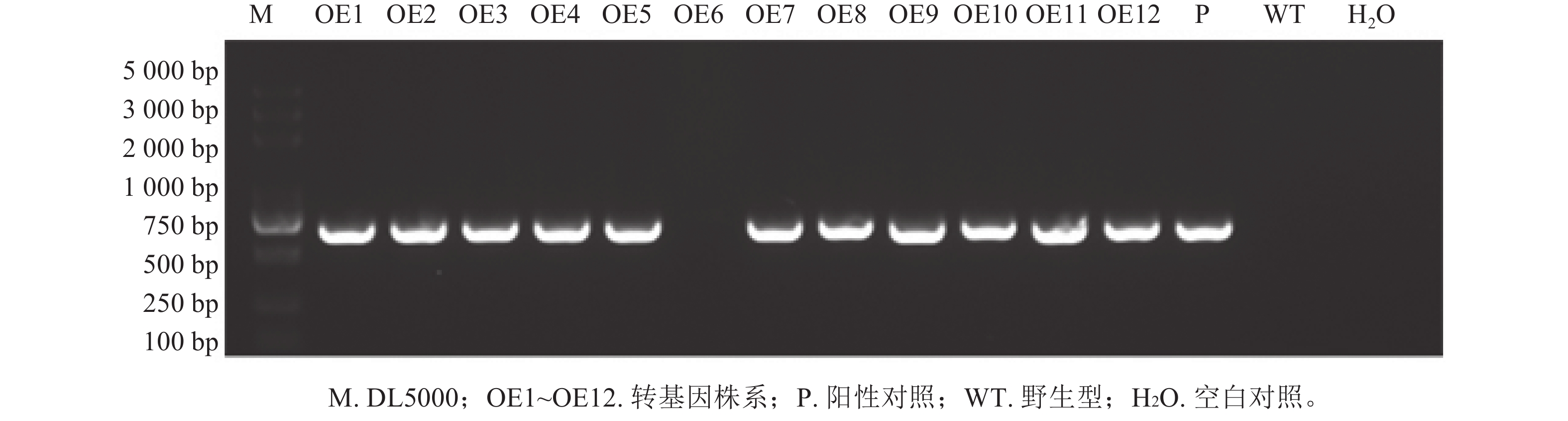
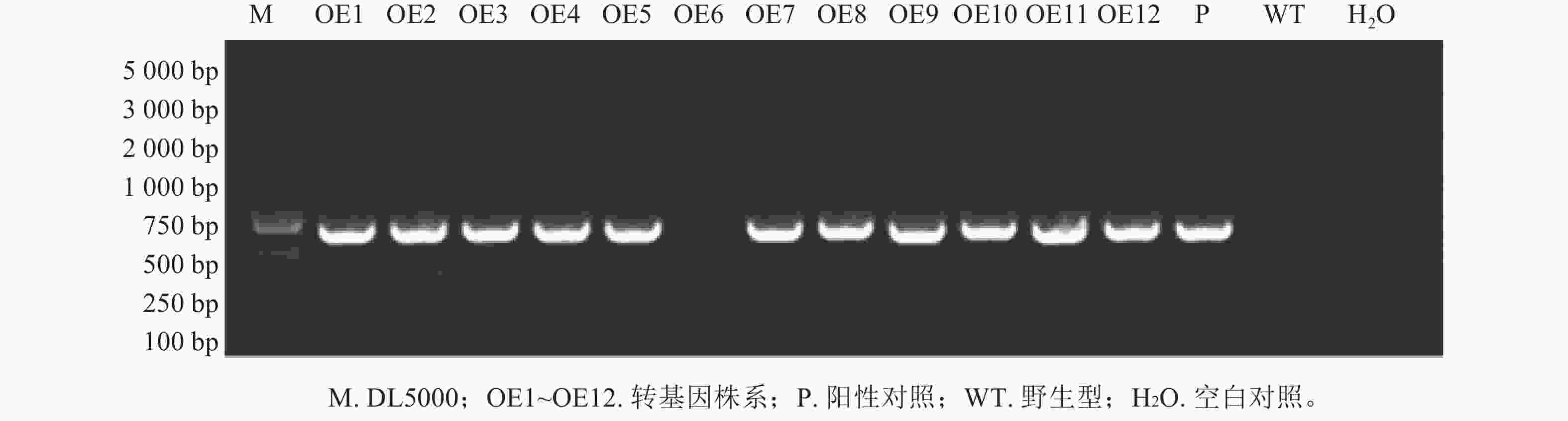
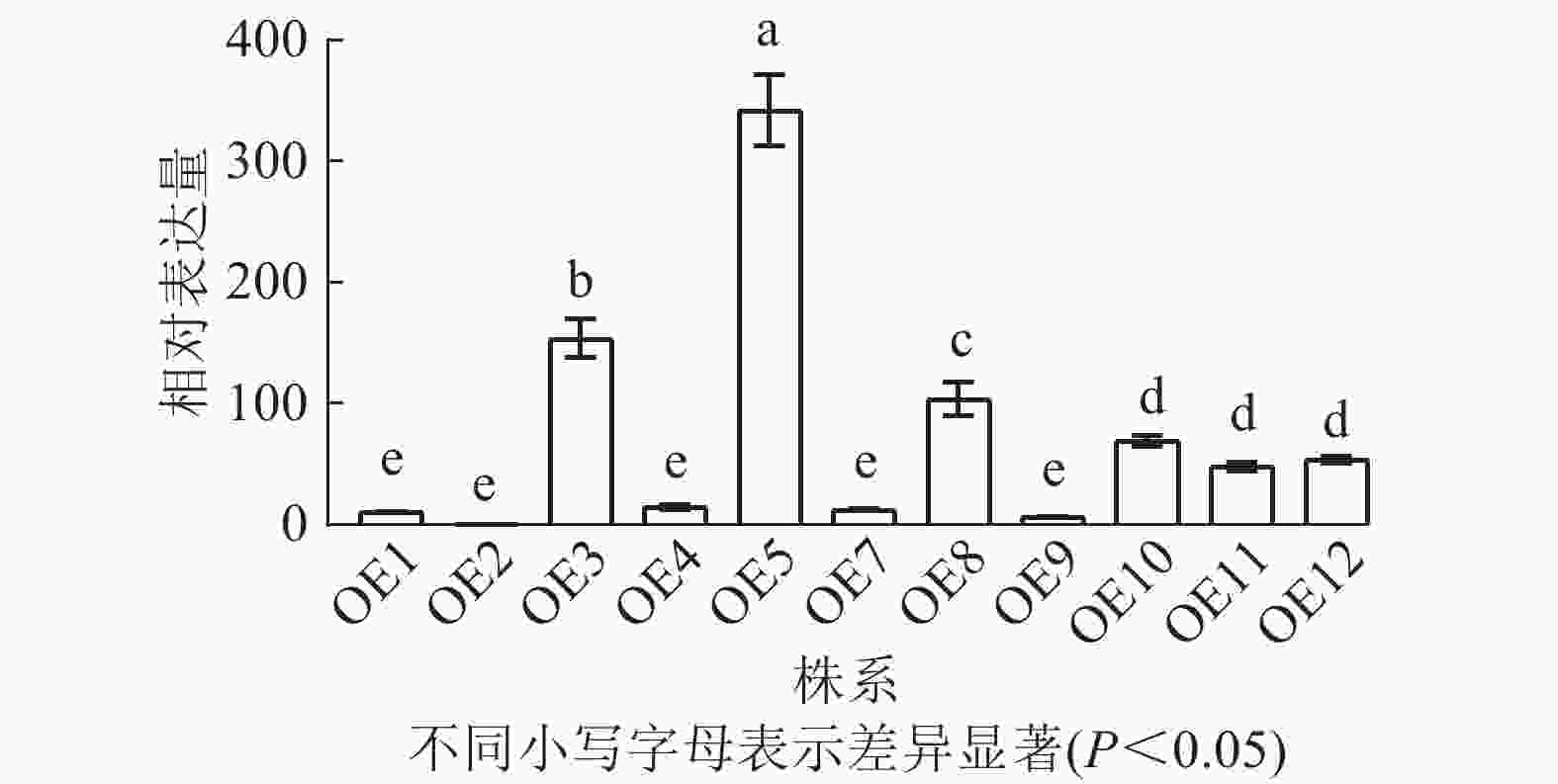
将构建好的植物表达载体pCAMBIA2300-PmERF011转入农杆菌GV3101感受态,选取初花期拟南芥植株,利用花序浸染法转化拟南芥。收集T0代种子后,播种于含50 μg·mL−1卡那霉素的1/2 MS培养基,获得了12株抗性苗,命名为OE1~OE12,10 d后移栽至基质。取健壮植株叶片,通过液氮研磨提取DNA,以PmERF011克隆引物和卡那霉素基因引物分别进行PCR鉴定阳性植株,发现OE6为假阳性(图1)。后续结合RT-qPCR分析PmERF011基因表达量,筛选出高表达转基因株系用于后续表型观察。比较T2代各株系与野生型(WT)的叶片数量和分支数量,每株系至少测量10株,计算平均值±标准差,与WT进行t检验(P<0.05)。
-
通过ExPASy在线分析平台对PmERF011蛋白进行生物信息学预测,结果表明:该蛋白由175个氨基酸组成,其理论分子质量为19.844 kDa,理论等电点为9.39。氨基酸组成分析显示:丝氨酸(Ser,11.4%)与谷氨酰胺(Gln,8.6%)为该蛋白的主要组成氨基酸。根据Guruprasad不稳定系数评估标准(阈值40),PmERF011的不稳定指数为58.88,表明其属于不稳定蛋白。电荷特性分析发现:该蛋白含20个带负电荷残基[天冬氨酸(Asp)和谷氨酸(Glu)]与26个带正电荷残基[精氨酸(Arg)和赖氨酸(Lys)],净电荷为+6,符合碱性蛋白特征。此外,其总平均亲水性(GRAVY指数)为−1.107,这证实PmERF011为典型亲水性蛋白。Cell-PLoc 2.0在线分析工具进行亚细胞定位分析结果表明:PmERF011位于细胞核和细胞质中。
-
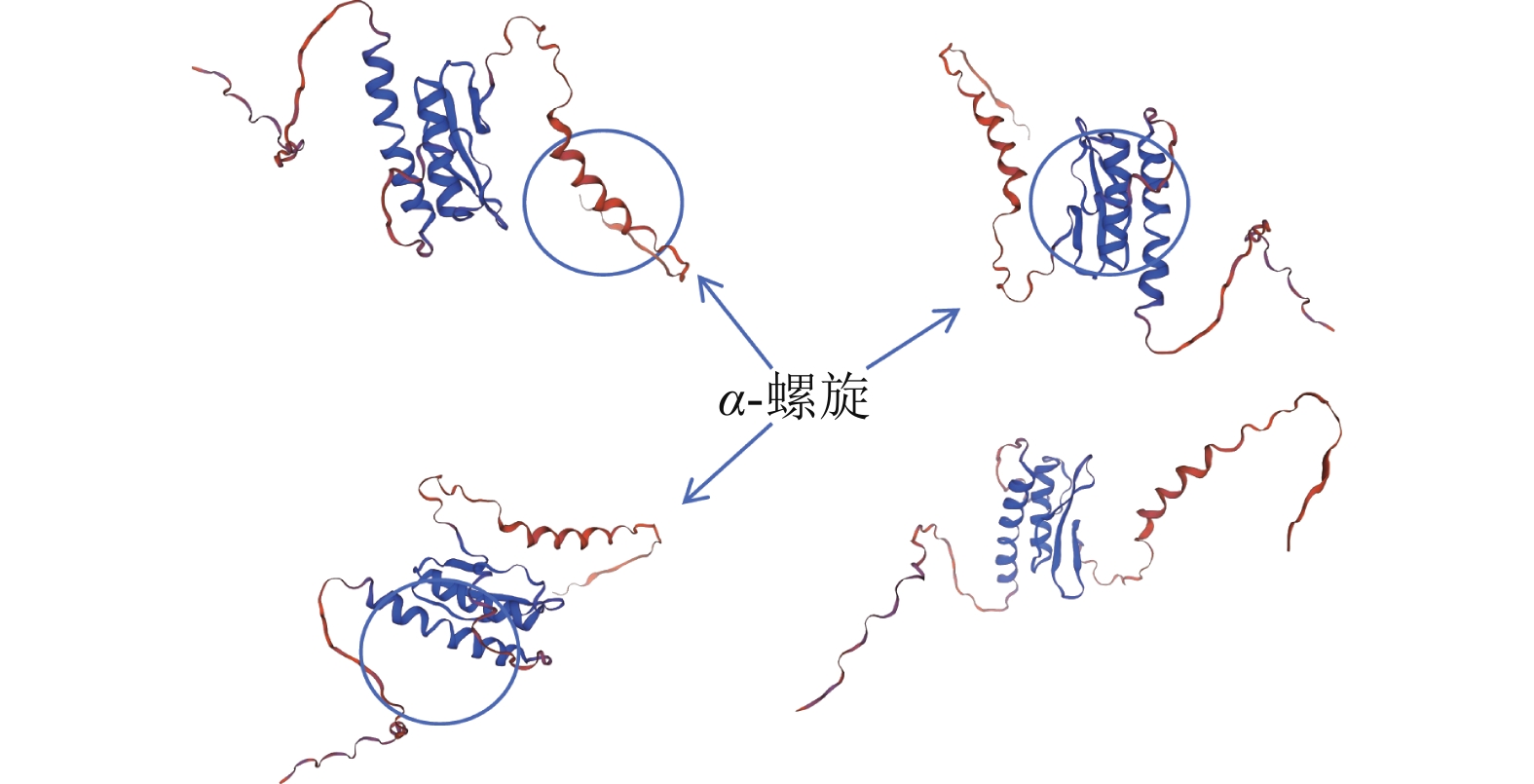
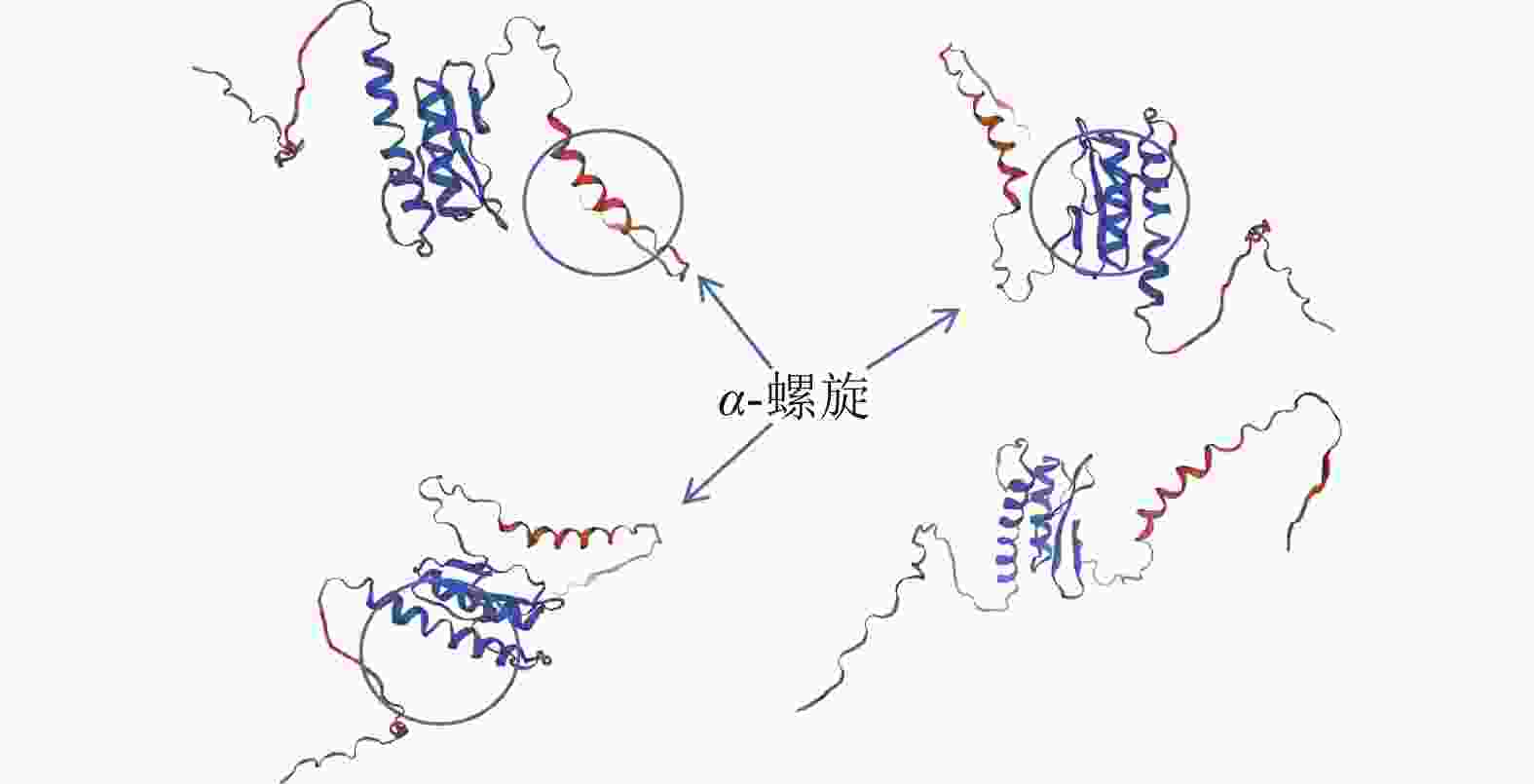
通过NCBI网站对PmERF011蛋白结构域分析,发现48~111位氨基酸有1个AP2/ERF保守结构域。进一步利用Sopma在线网站预测其二级结构发现:α-螺旋与延伸链集中于58~134位氨基酸,占比为24%;无规则卷曲结构分布其中及两侧,占比最高约76%。未预测到β-转角及β-折叠。基于Swiss model网站预测的蛋白质三级结构(图2)可知:PmERF011具有3个显著的α-螺旋,与二级结构预测结果一致,进一步支持它为典型AP2/ERF家族成员。
-
利用在线软件PlantCARE对PmERF011基因启动子的顺式作用元件进行预测。除常见的启动子和增强子区域中核心的顺式作用元件TATA-box、CAAT-box外,启动子序列包含多个参与光反应、生长发育及厌氧诱导的元件;植物激素响应元件包括参与脱落酸反应及茉莉酸甲酯响应的元件(表2)。
元件名称 核心序列 数量 生物学功能 元件名称 核心序列 数量 生物学功能 ABRE ACGTG 1 参与脱落酸反应 GATA-motif AAGGATAAGG 2 光响应元件的一部分 ARE AAACCA 3 参与厌氧诱导 GC-motif CCCCCG 1 参与缺氧特异性诱导 Box 4 ATTAAT 2 参与光反应 MRE AACCTAA 1 MYB结合位点(参与光响应) Box II ACACGTAGA 1 光响应元件的一部分 O2-site GATGACATGG 1 参与玉米蛋白代谢调控 CAT-box GCCACT 1 参与分生组织表达 P-box CCTTTTG 1 赤霉素响应元件 CGTCA-motif CGTCA 1 参与茉莉酸甲酯响应 Sp1 GGGCGG 1 光响应元件的一部分 G-box CACGAC 2 参与光反应 TGA-element AACGAC 1 生长素响应元件 G-box TAACACGTAG 1 参与光反应 TGACG-motif TGACG 1 参与茉莉酸甲酯响应 G-box TACGTG 1 参与光反应 Table 2. Analysis of promoter cis-acting elements
-
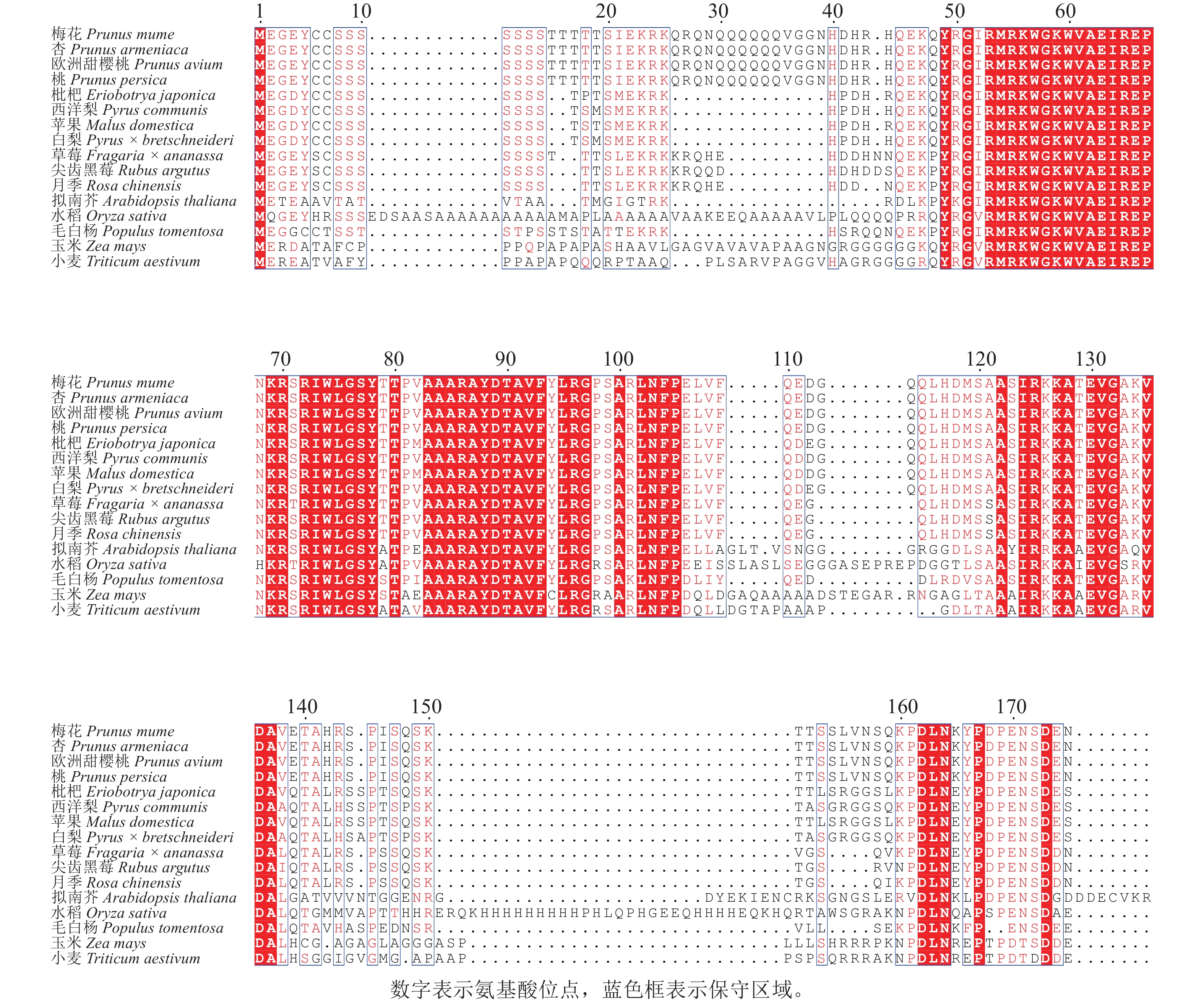
为探究PmERF011的进化关系及保守性,通过NCBI-BLAST对其与近缘物种的ERF家族蛋白进行同源性分析,涵盖杏Prunus armeniaca、桃Prunus persica、草莓Fragaria × ananassa、毛白杨Populus trichocarpa、欧洲甜樱桃Prunus avium、枇杷Eriobotrya japonica、西洋梨Pyrus communis、苹果Malus domestica、白梨Pyrus × bretschneideri、尖齿黑莓Rubus argutus、月季Rosa chinensis、水稻、拟南芥、玉米Zea mays、小麦Triticum aestivum共15个物种。同源序列比对结果显示:梅花PmERF011与桃、杏、欧洲甜樱桃的ERF011蛋白序列完全相同,表明蔷薇科李属内该基因高度保守,与拟南芥、水稻、草莓、毛白杨、水稻、枇杷、西洋梨、苹果、白梨、尖齿黑莓、月季、玉米、小麦蛋白序列的AP2/ERF结构域同源性较高(图3)。
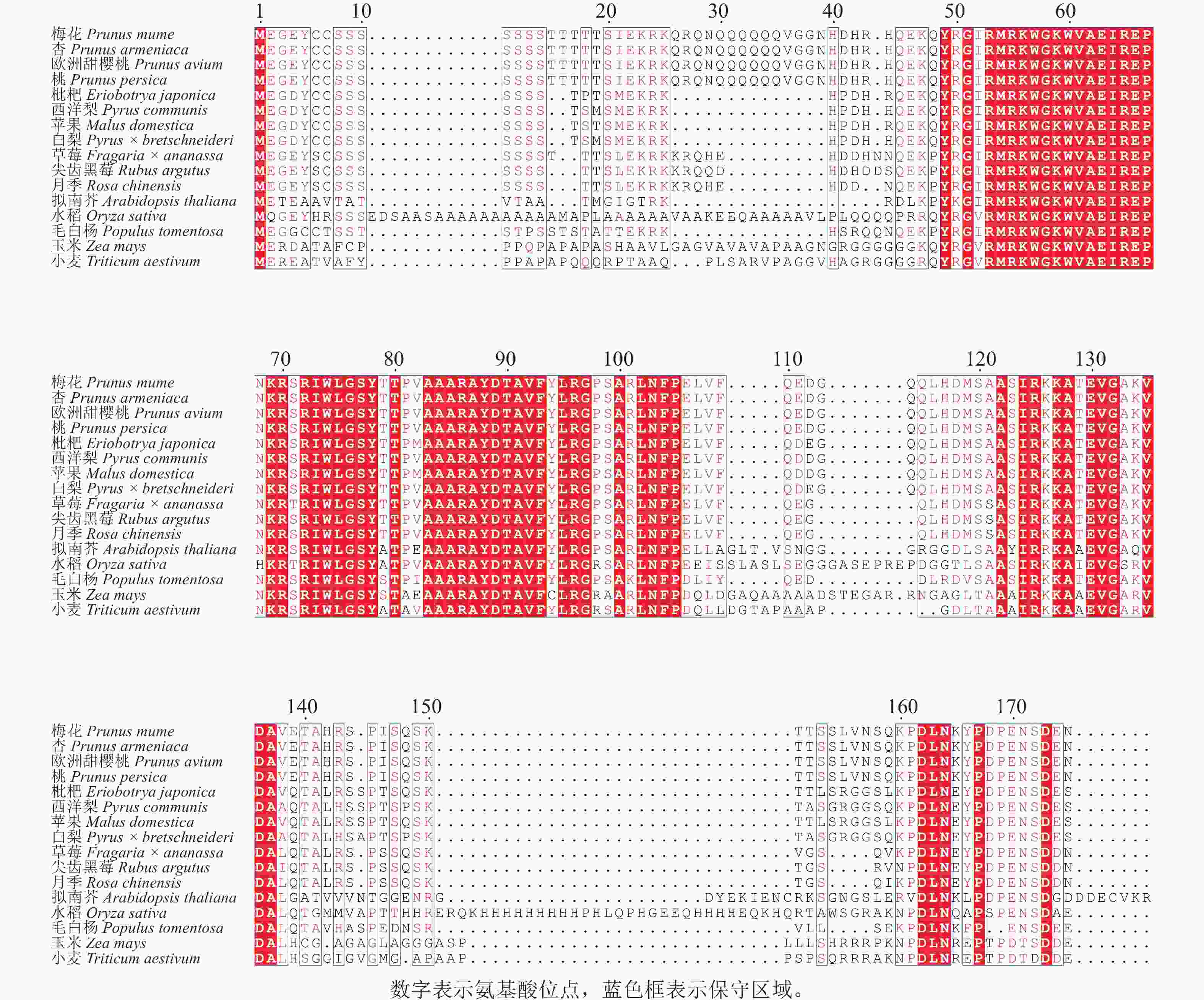
Figure 3. Comparison analysis of the protein homologous sequences of PmERF011 and its close relatives
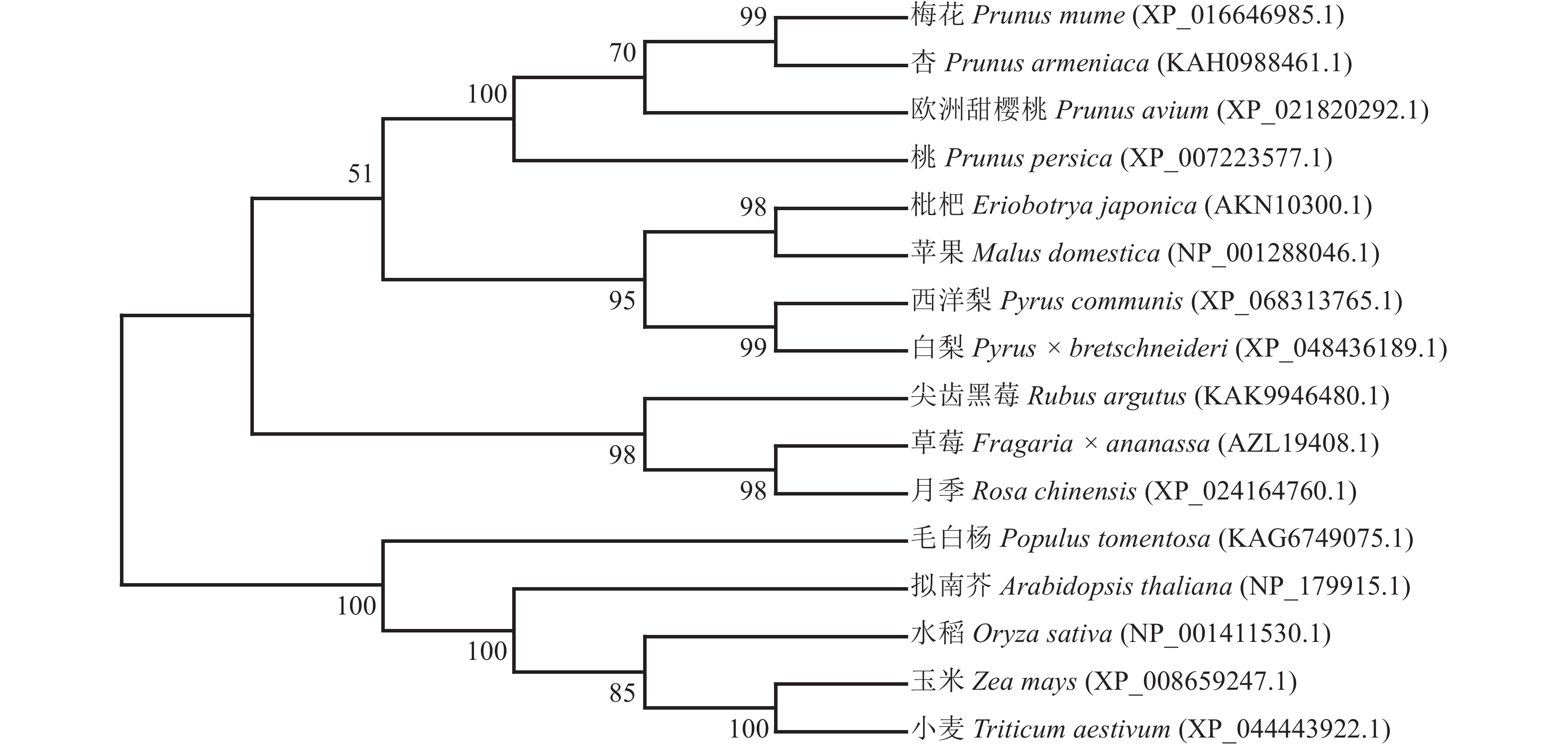
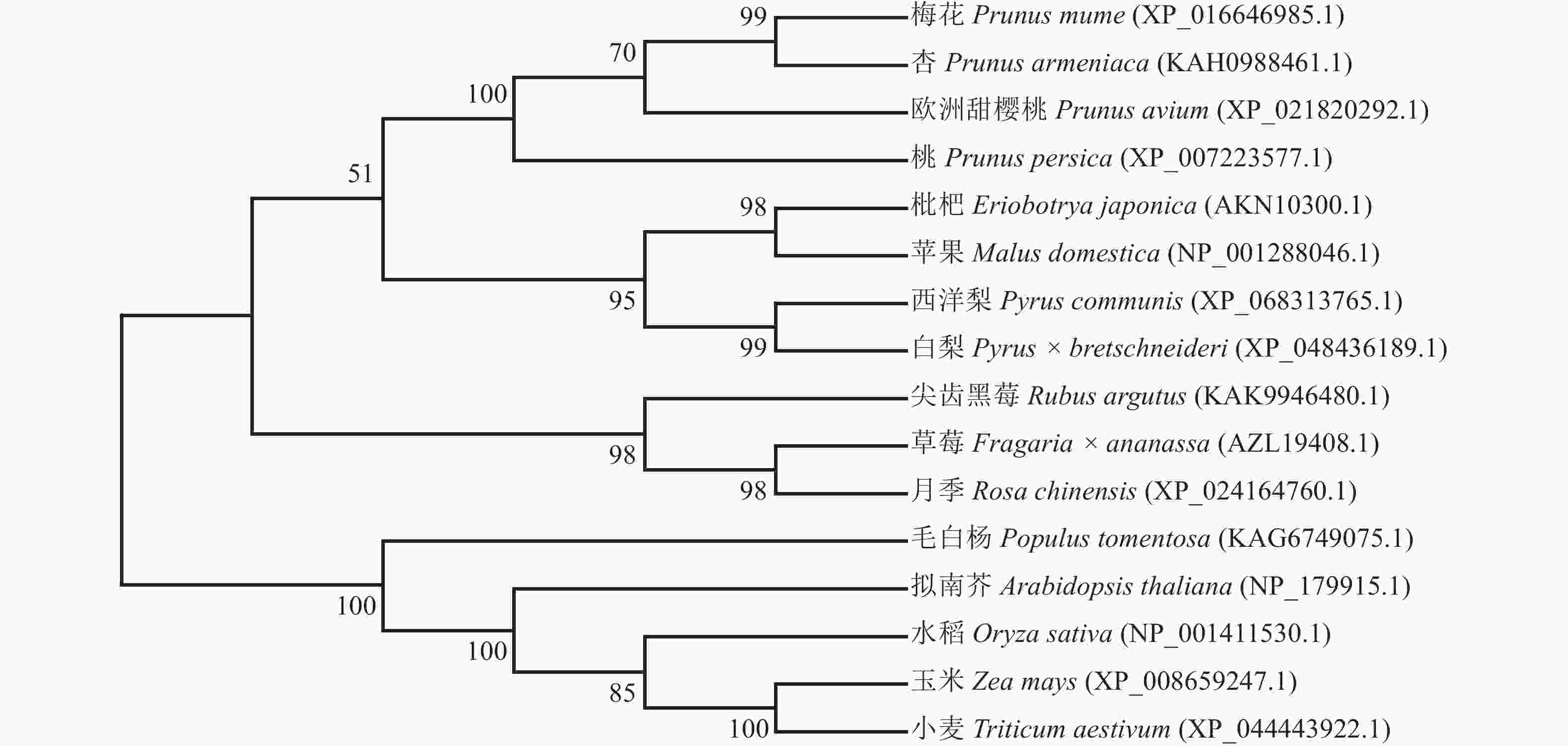
利用MEGA 7.0软件,采用邻接法构建PmERF011同源蛋白的多物种进化树,结果显示:PmERF011与杏ERF011处于进化树的同一分支上,两者亲缘关系最为接近,次之的是欧洲甜樱桃和桃,证实了蔷薇科李属内该基因的高度保守性;而与其他蔷薇科属(如草莓属Fragaria、梨属Pyrus、苹果属Malus等)及非蔷薇科物种(拟南芥、水稻、毛白杨等)的ERF011蛋白亲缘关系较远,反映了该基因在长期进化过程中可能产生的科属特异性分化(图4)。
-
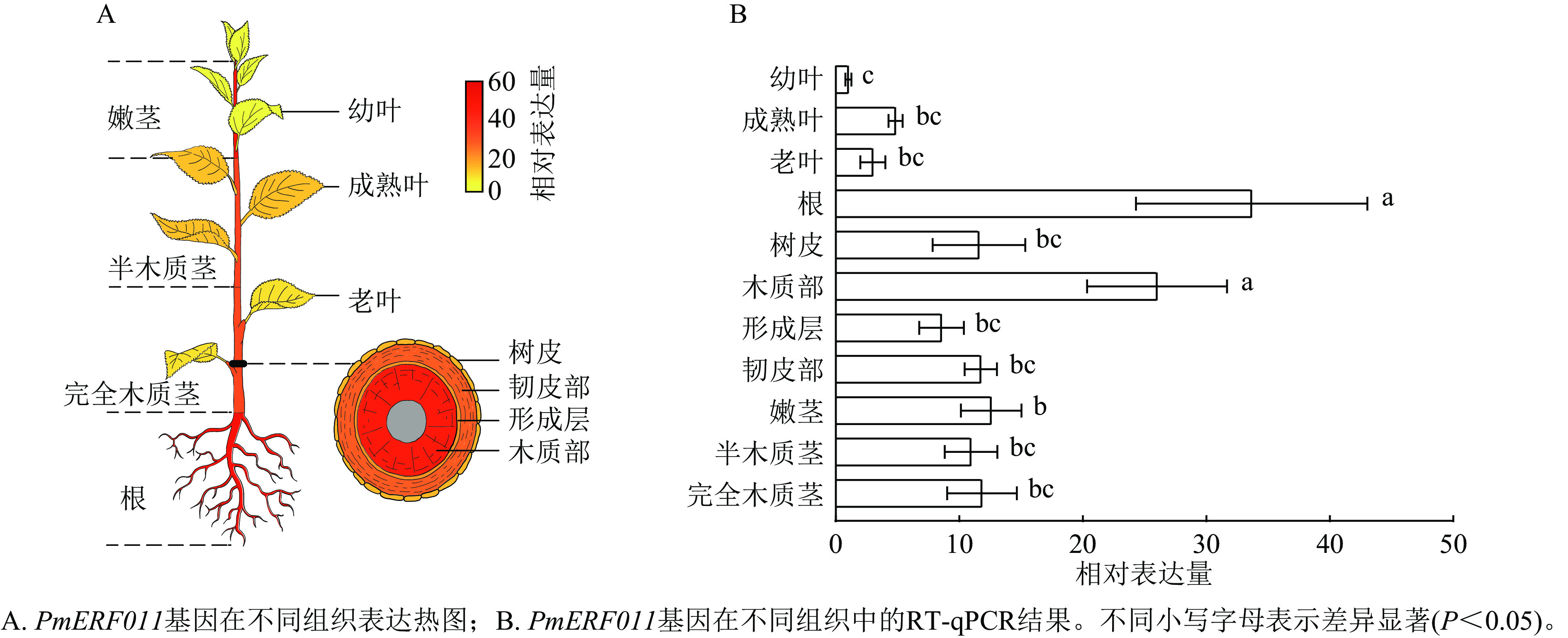
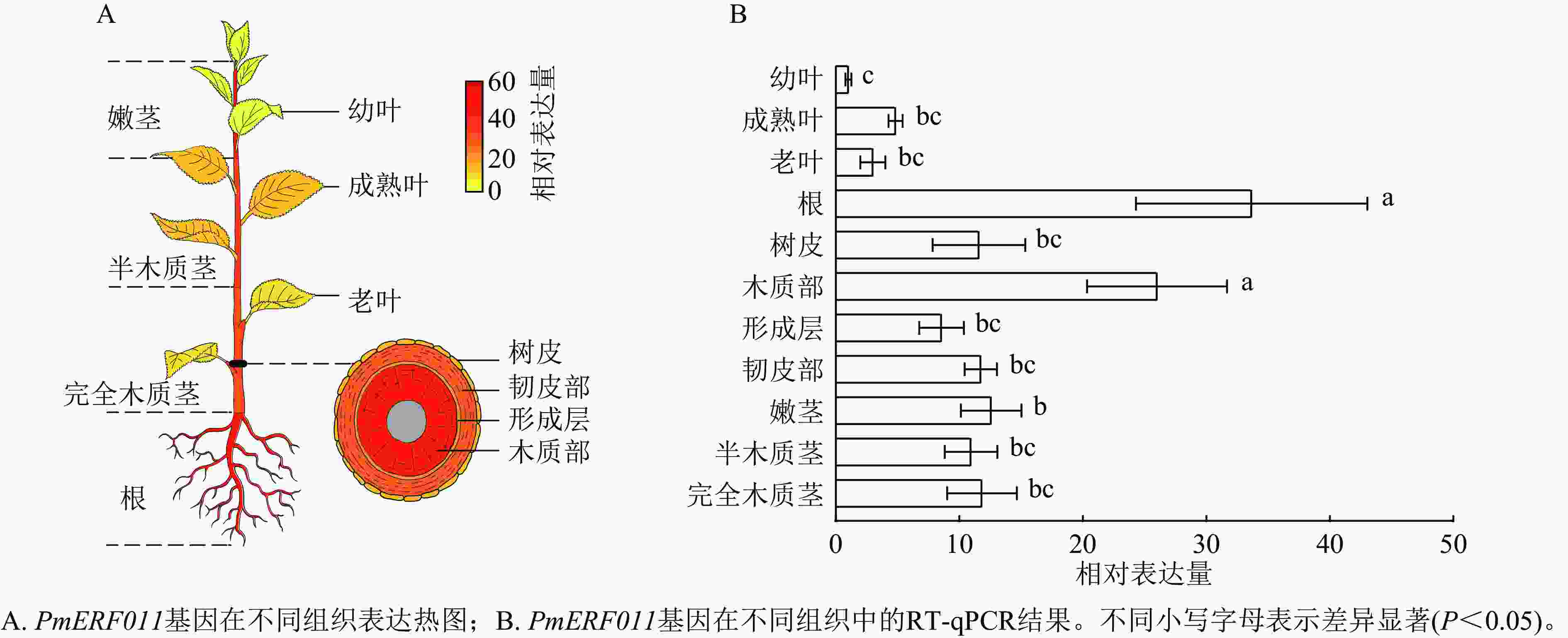
转录组数据表达热图和RT-qPCR检测结果显示:PmERF011基因在幼叶、成熟叶、老叶、根、树皮、木质部、形成层、韧皮部、嫩茎、半木质茎、完全木质茎11种组织中均有表达。值得注意的是,PmERF011的表达水平存在显著的组织差异:在根和茎木质部中表达量最高,在嫩茎和茎韧皮部表达量次之,而在叶片中表达量最低(图5)。这种表达模式说明PmERF011主要参与根部发育和茎次生生长相关的调控过程。
-
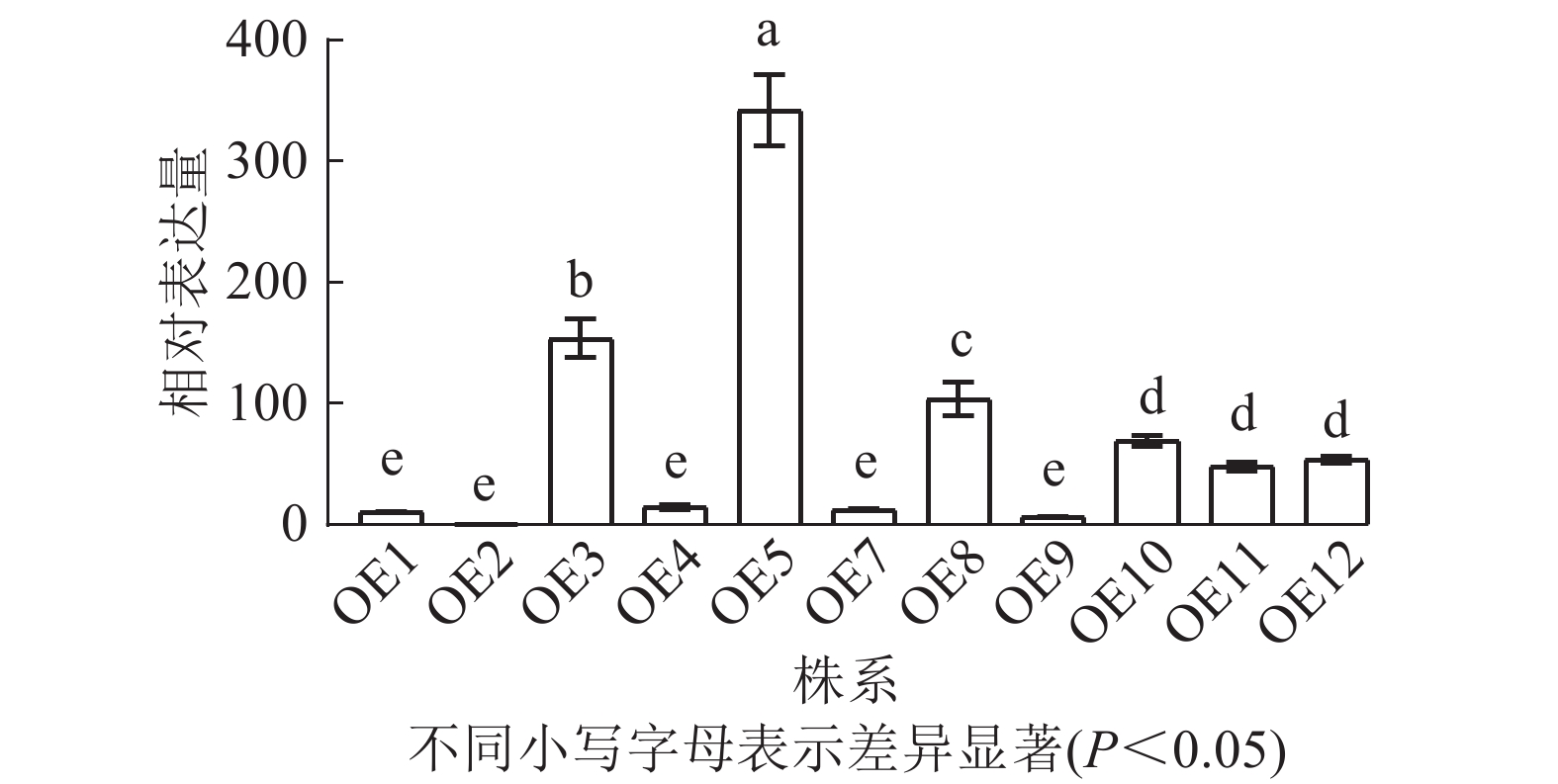
去除假阳性苗,对获得的11个转基因拟南芥过表达株系进行RT-qPCR检测,分析PmERF011基因的表达水平。结果表明:不同转基因株系间表达水平存在显著差异,其中OE2表达量最低,OE5、OE3、OE8表达量较高(图6)。最终筛选出OE3、OE5和OE8这3个高表达株系用于后续表型观察。
-
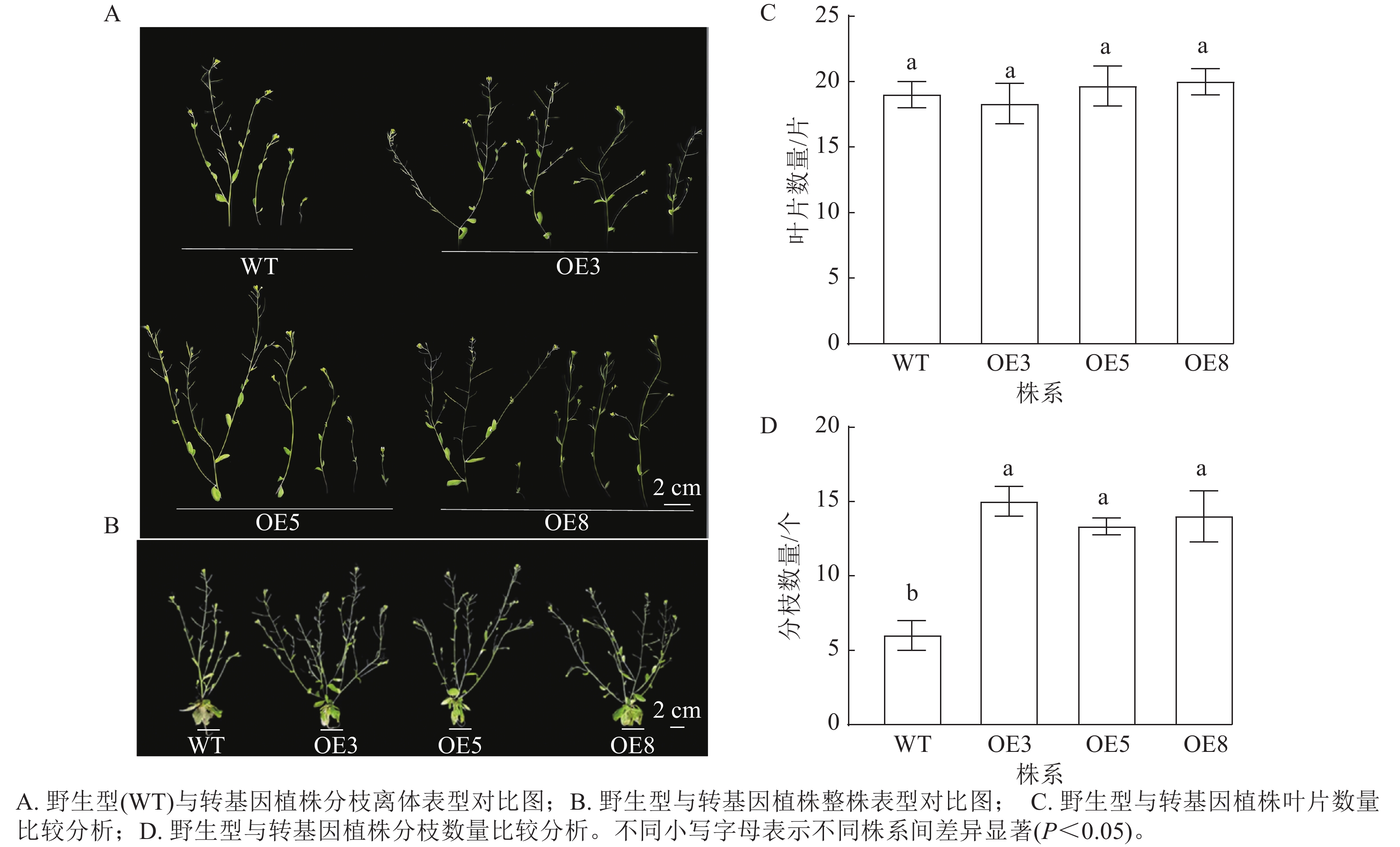
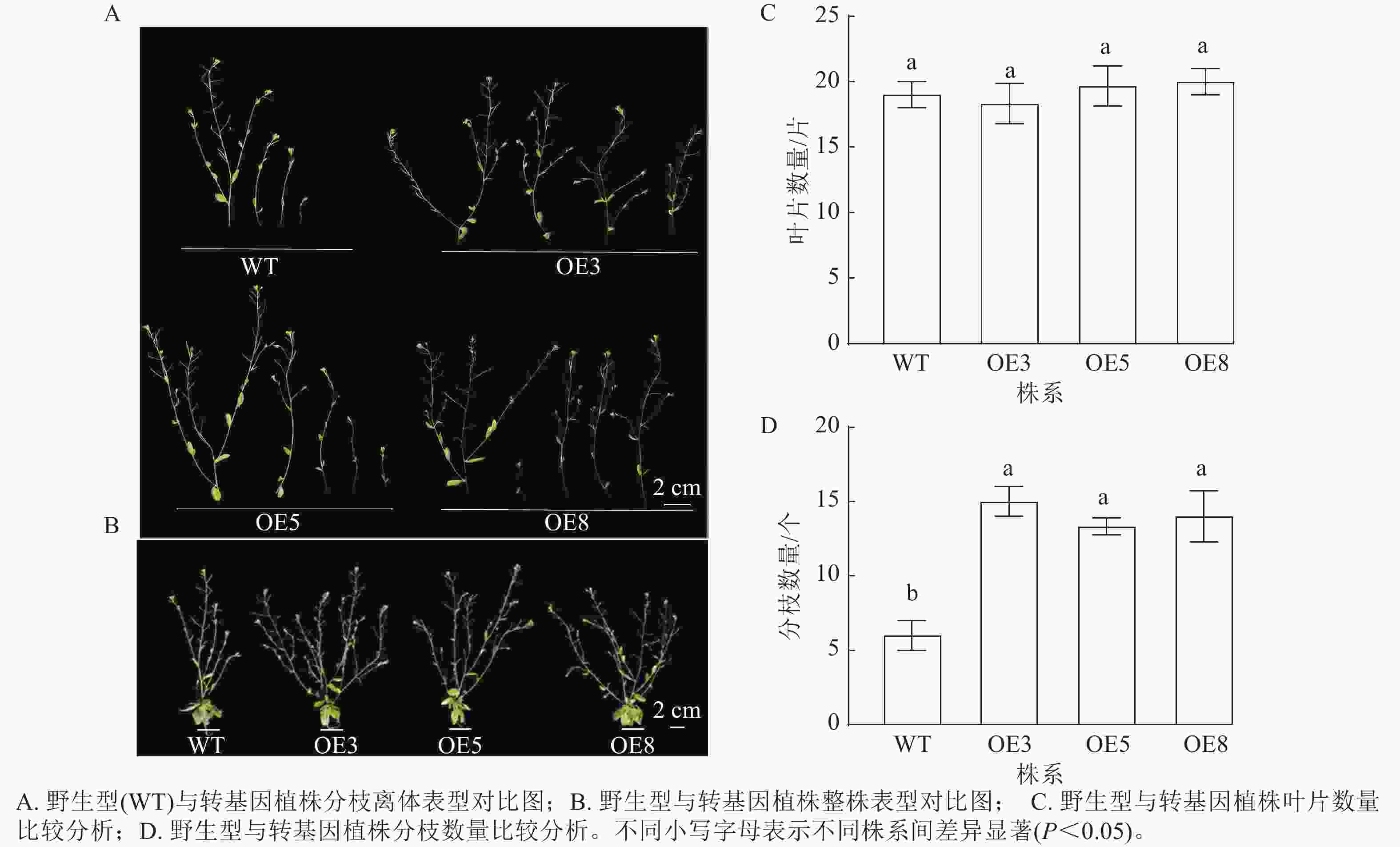
选择表达量高的T2代3个株系(OE3、OE5和OE8)进行播种并观察表型。拟南芥幼苗移栽40 d后,野生型和过表达植株均进入花期。对比野生型和PmERF011转基因植株的表型性状,发现转基因植株的分枝数量有所增加(图7 A、B)。统计结果表明:野生型和转基因株系拟南芥的叶片数量相近,无显著差异(图7 C);野生型拟南芥的平均分枝数为6.00个,3个PmERF011过表达株系的平均分枝数量分别为15.00、13.33、14.00个,与野生型相比差异显著(P<0.05,图7 D)。
-
植物的侧枝在形态建成中具有核心作用,其发育调控直接影响株型结构及生物量积累。然而,木本植物分枝的分子基础仍不清晰,尤其是AP2/ERF转录因子在其中的功能尚未明确。本研究以梅花为对象,通过整合生物信息学分析、组织特异性表达模式及异源转基因功能验证,明确了PmERF011基因序列特征及其在分枝调控中的作用,为梅花株型改良提供理论依据。结果表明:PmERF011是一个具有典型AP2/ERF结构域的转录因子,与蔷薇科近缘物种桃、杏的ERF011蛋白高度保守,推测它在木本植物中可能具有功能保守性,而与其他科属物种(如拟南芥、水稻)的序列差异则反映出存在物种特异性调控机制。
组织特异性表达结果显示:PmERF011在根和木质部中表达量最高,嫩茎和韧皮部次之,叶片中表达量最低,暗示AP2/ERF家族成员可能参与根系激素合成及维管组织分化。例如,拟南芥ESR1的过表达促进细胞分裂素合成,诱导芽再生[42];在水稻中,SNORKELs基因是水稻茎间伸长的关键调节因子,乙烯在植物中积累并诱导该基因表达[43]。菊花Chrysanthemum × morifolium去除顶芽后,其顶端优势释放,腋芽生长素减少,细胞分裂素生物合成加速,进而诱导CmERF053表达,促进侧枝发育[44]。此外,根、茎组织的高表达暗示其可能直接调控分生组织活性,促进维管组织的分化。值得注意的是,PmERF011在叶片中表达量极低,这可能与其功能偏好性相关:叶片作为光合器官,其发育可能受其他转录因子(如KNOX[45]或TCP[46]家族)主导,而PmERF011更倾向于通过维管组织与分生组织调控株型结构。启动子顺式元件分析进一步支持这一推论,其中存在茉莉酸甲酯响应元件及生长素响应元件,暗示PmERF011可能通过整合激素信号协调分枝进程。
转基因功能验证结果显示:PmERF011过表达显著增加拟南芥分枝数量,尤其是侧枝数量,但其对叶片数无显著影响,这一表型与欧洲油菜ERF114通过重塑顶端生长素梯度促进分枝的机制相似[37]。不过,PmERF011调控分枝的具体分子机制尚未明确,PmERF011是否直接抑制分枝抑制因子(如BRC1)或激活分生组织标记基因(如STM)仍需进一步验证。
-
本研究成功克隆了梅花AP2/ERF转录因子家族成员PmERF011,并揭示了PmERF011在植物分枝调控中的作用。生物信息学分析证实:PmERF011编码一个典型的亲水性核蛋白并在蔷薇科李属植物(如桃、杏)中高度保守。组织特异性表达模式分析表明:PmERF011主要在根和茎木质部中高表达。拟南芥异源过表达结果显示:转基因植株分枝数量显著增加,证实PmERF011具有促进植物侧枝形成的能力。
Cloning and functional validation of the transcription factor PmERF011 in Prunus mume
doi: 10.11833/j.issn.2095-0756.20250366
- Received Date: 2025-07-05
- Accepted Date: 2025-09-02
- Rev Recd Date: 2025-08-31
- Available Online: 2026-01-27
- Publish Date: 2026-02-20
-
Key words:
- AP2/ERF /
- PmERF011 /
- Prunus mume /
- plant architecture /
- transcription factor
Abstract:
| Citation: | WU Jinyue, LI Xue, WANG Yuexin, et al. Cloning and functional validation of the transcription factor PmERF011 in Prunus mume[J]. Journal of Zhejiang A&F University, 2026, 43(1): 76−85 doi: 10.11833/j.issn.2095-0756.20250366 |









 DownLoad:
DownLoad: