-
NAC(NAM-ATAF1/2-CUC2)是植物中最大的转录因子家族之一,成员众多,功能多样[1]。NAC转录因子参与植物多个生物学过程,如植物茎尖分生组织的形成[2]、植物次生生长[3]、组织愈合过程中的细胞增殖[4]、生物和非生物胁迫响应[5-6]、植物开花时间调节[7-8]、叶片和花瓣衰老[9]和果实成熟时颜色变化等[10]。花瓣细胞的伸长和扩展也受到NAC转录因子的调控。花瓣的扩展是引起植物花开放的重要原因[11-13],主要受花瓣细胞的膨压变化、细胞壁代谢变化等影响[14]。研究发现:NAC转录因子NAP可能通过抑制拟南芥Arabidopsis thaliana花瓣细胞扩张而影响花器官生长,且在花发育细胞分裂向细胞扩展过渡阶段发挥重要作用[15]。RhNAC3可以作为上游调控因子结合具有调节渗透压功能的基因从而影响月季Rosa hybrida花瓣细胞扩张[16]。桂花Osmanthus fragrans是中国传统的十大名花之一,花是其最重要的观赏器官。桂花花的开放受到温度、湿度等环境因子的影响,其具体的分子机制至今尚未阐明。本研究通过桂花NAC家族基因鉴定和筛选,预测它们的理化性质,运用荧光定量PCR分析花开放进程中的表达模式,筛选出参与桂花花开放调控的关键NAC转录因子成员,为明确其在花开放中的功能作用奠定基础。
-
以桂花品种‘堰虹桂’O. fragrans‘Yanhonggui’盆栽植株为材料,种植于浙江农林大学桂花资源圃。选取株龄相同、生长一致的健康植株,分别于起始期(S1)以及花开放不同阶段:圆珠期(S2)、顶壳期(S3)、铃梗期(S4)、初开期(S5)、盛花期(S6)进行取样[17]。每个时期样品采集3份,取样时间均为10:00,液氮速冻后储藏于−80 ℃冰箱备用。
-
本研究从已构建的桂花‘堰虹桂’转录组数据库[18]中对NAC转录因子进一步进行NCBI-Blast(http://www.ncbi.nLm.nih.gov/BLAST/)比对分析。通过PlantTFDB以及Pfam查询候选基因中的NAC的结构域,并通过NCBI(https://www.ncbi.nlm.nih.gov/cdd)进行保守功能域预测,利用TBtools工具进行桂花NAC保守结构域可视化分析,TBtools-ORF finder分析开放阅读框[19],并通过NCBI-Blast获得开放阅读框完整的桂花OfNAC序列。运用ExPASy在线软件(http://web.expasy.org/protparam/)预测编码蛋白质分子量、理论电点、酸碱性等。
-
用DNAMAN对已筛选的桂花OfNAC转录因子氨基酸序列进行比对,分析其保守亚结构域;在线分析软件MEME 5.1.0分析NAC的保守基序(http://meme-suite.org/tools/meme),对桂花OfNAC蛋白质二级结构以及跨膜结构分别采用在线软件SOPMA和TMHMM Server V.2.0进行分析。用在线分析软件Wolfp sort Genscript(https://www.genscript.com/wolf-psort.html?src=leftbar)进行亚细胞定位预测。
-
从PlantTFDB中获取拟南芥、水稻Oryza sativa的NAC序列,使用MEGA 6.0软件构建系统发育树,采用邻近法(neighbor-joining),1 000 次Bootstrap抽样。
-
RNA提取采用UltraClean Polysaccharide and Phenol Plant RNA Purification Kit试剂盒(诺禾致源公司,中国天津),方法参照说明书。使用核酸分析仪(IMPLEN公司,德国)对RNA的纯度与浓度进行检测,并用质量分数1%的琼脂糖凝胶电泳检测RNA的完整性。依据说明书使用PrimeScript™RT Master Mix Perfect Real Time(TaKaRa,中国大连)试剂盒对桂花不同花开放时期的cDNA进行合成。利用Primer Premier 5.0对22个OfNAC基因进行定量特异性引物设计,桂花OfACT作为内参基因[20],引物序列如表1所示。荧光定量反应体系如下:SYBR Premix Ex Taq 10 μL,cDNA 2 μL,上游引物和下游引物(10 μmol·L−1)各8 μL,双蒸水(ddH2O)补足至20 μL,每个样品设置3次生物学重复。反应程序为:95 ℃预变性30 s,95 ℃变性5 s,60 ℃复性30 s,共40个循环;然后以95 ℃持续5 s,60 ℃持续1 min,95 ℃持续15 s作为溶解曲线分析程序。循环阈值由Light Cycler 480软件自动计算。最后根据
${2^{{\rm{ - }}\Delta \Delta {C_{\rm{t}}}}} $ 法计算目的基因的相对表达量。表 1 桂花OfNAC转录因子RT-PCR特异性引物
Table 1. Specific primers for RT-PCR of OfNAC in O. fragrans
基因名称 用于荧光定量的引物序列(5′→3′) 基因名称 用于荧光定量的引物序列(5′→3′) OfNAC100-1 F: TGAACAAGATTGAGCCTTGGG OfNAC104 F: TGCATTTTACATTGGTGAAGATGTC R: CCTTTCCTGTGGCTTTCCAG R: GCTCGTACACTTGACACACCA OfNAC53 F: AGATTGTGGGGATGAAGAAAA OfNAC92 F: TCCTAGTCGGAATGAAGAAAACTC R: CAACTCCATATCAGTAAGCCG R: ATGGCTTTCTAATCTGTATTCGTGC OfNTM1-9 F: GGTTGCTCTAATGCCCACTTC OfNAC72-1 F: GGAAAAGCCCCCAAAGGAAC R: CTGGTTCCGTAGCACGATACT R: CCCAATCATCCAACCTTGAGC OfNAC73 F: AGGCAAGGATGGCCAAATTC OfNAC72-2 F: ACGTAGGAAAAGCACCAAAAGG R: TTGTGCCATCTTGTTTCTCC R: AGCATCCAACCTTGCGCTTC OfNAC43 F: AGGCTACTGGTCGTGATAAAG OfNAC29-1 F: TTACAAGGGAAGGCCTCCAAAG R: GGGGTCATGAGTGTCGTCC R: TTGAGCCATTTTGCGTGTTAGG OfNAC91 F: TCTACAAAGGTCGTGCTCCG OfNAC29-2 F: CCCAAAGGGCGTCAAAACTG R: CCCTGACCAGGATAAGTGCC R: GCACACAATCATCCAACCTCA OfNAC50-X2 F: TGGCAAAGGGTATTGGAAAGC OfNAC71 F: CTATCGTGGAAGAGCACCACT R: TCGCCCACTATGGAAAACAAG R: TCCCTGAAATCTTGGGGTGTC OfNAC50 F: AAGCAACTGGAAAGGATCGC OfNAC2 F: TTGGGAATAAAGAAGGCTCTGGTG R: AATTCTGCATCGCAAAGCCTG R: ACACAACACCCAATCATCAAGCCTC OfNAC21/22 F: GAAGGGAAGCCTGGTTGGAAT OfNAC100-2 F: TCAGAGGAAAAATCCTCGTCGG R: CCCAATCCTCCTTGACAGATG R: TTTGGGAGGTTGTGGATCGAG OfNAC56 F: TCTATGGTGGAAAGCCTCCT OfNAC32 F: AAGCCTTGGTTTTCTATGCCG R: CATCAAGCCTTAAAGAGCCC R: AAGCTGTTGTTCTTGTTTCGA OfNAC17-X2 F: TCCTGTTGGGGTGAAGAAGA OfACT F: CCCAAGGCAAACAGAGAAAAAAT R: ATAGTCATCCTGTGCATCCTGC R: ACCCCATCACCAGAATCAAGAA OfNAC17 F: TGGTCTTCCATAAAGGTCGTGC R: TTGTACAGAGCATAGCAATCCCGTG -
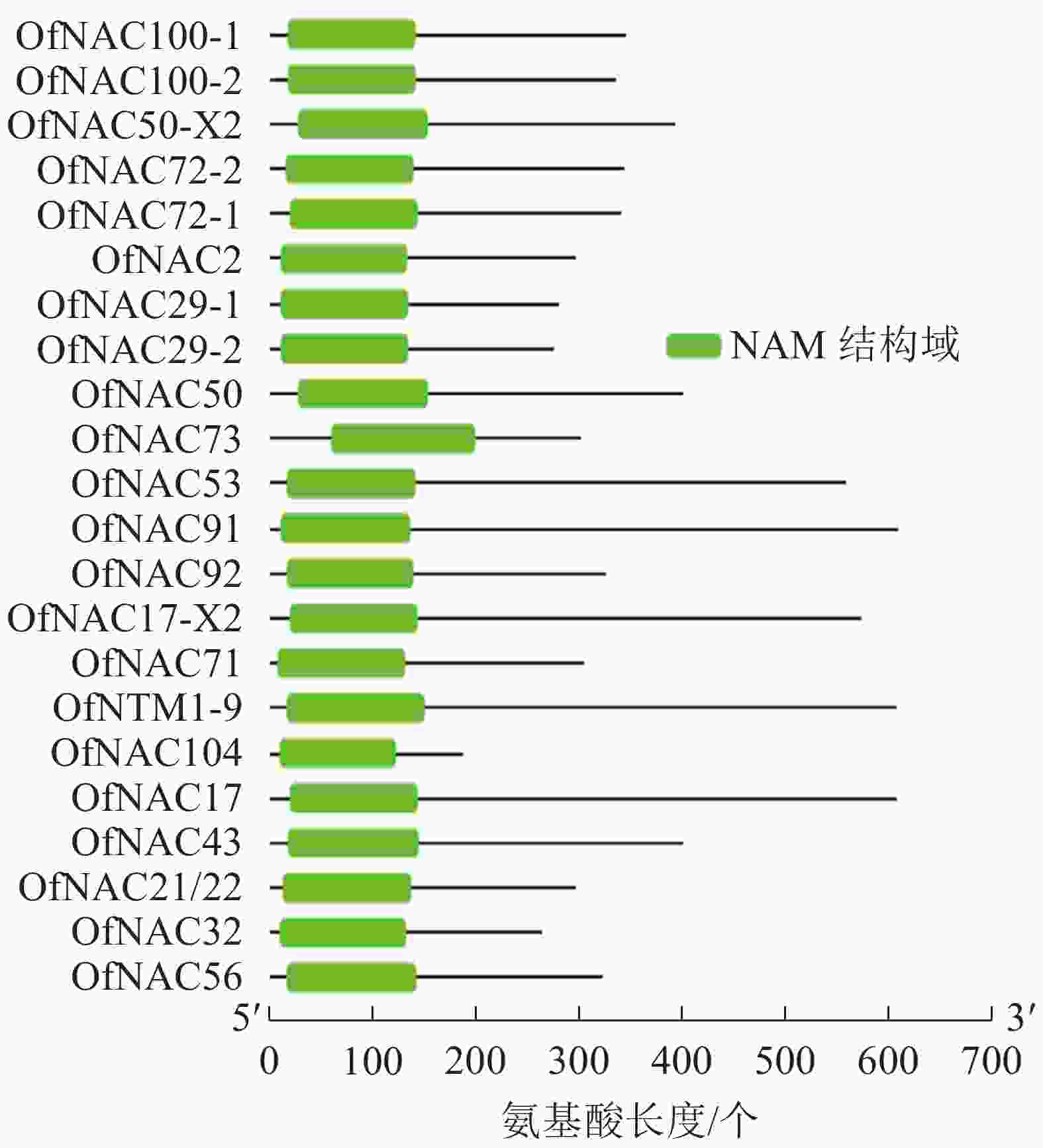
通过对桂花‘堰虹桂’转录组数据分析,并结合NCBI-Blast筛选,对其中22个桂花OfNAC基因进行分析。结果显示:它们均含有NAC家族蛋白特有的NAM保守功能域(图1)。通过ExPASy对22条桂花OfNAC转录因子进行理化性质分析(表2),结果表明:22条桂花OfNAC基因的氨基酸(OfNAC104~OfNAC91)长度为186~609个;蛋白质(OfNAC104~OfNAC17)相对分子量为21 498.77~68 408.12;等电点为4.59~9.33,其中OfNAC100-1、OfNAC73、OfNAC56、OfNAC72-1、OfNAC72-2、OfNAC29-1、OfNAC29-2、OfNAC32等8个蛋白质等电点大于7,偏碱性,其余14个OfNAC等电点小于7,偏酸性。OfNAC的不稳定系数为29.27~52.83,OfNAC53、OfNAC73、OfNAC56、OfNAC92、OfNAC29-2等5个蛋白质不稳定系数低于40,为稳定蛋白质,其余17个不稳定系数均高于40,为不稳定蛋白质。脂溶性指数为53.37(OfNAC71)~77.70(OfNAC17-X2),总平均疏水值为−0.816~−0.530,蛋白质的平均疏水指数均小于0,表明OfNAC均为亲水性蛋白质(表2)。
表 2 桂花OfNAC蛋白质理化性质及二级结构分析
Table 2. Physicochemical properties and secondary structure analysis of OfNAC of O. fragrans
序列名称 氨基酸长度/个 相对分子量 等电点 碱性氨基酸 酸性氨基酸 不稳定系数 脂溶性指数 总平均疏水值 亚细胞定位 位置 预测值/% OfNAC100-1 345 38 986.11 8.69 40 36 40.06 61.01 −0.574 细胞核 61.54 OfNAC53 558 62 807.47 4.60 51 88 38.87 69.37 −0.564 内质网 42.42 OfNTM1-9 606 68 053.70 5.63 71 86 51.49 62.43 −0.725 细胞核 76.92 OfNAC73 300 33 768.95 8.79 39 34 38.21 69.47 −0.754 细胞核 76.92 OfNAC43 399 45 513.66 5.77 43 55 46.31 63.26 −0.752 细胞核 69.23 OfNAC91 609 67 741.52 4.98 63 86 52.83 71.07 −0.593 细胞核 76.92 OfNAC50-X2 389 43 765.29 5.29 46 58 47.83 69.43 −0.629 叶绿体 58.33 OfNAC50 399 44 952.52 5.35 46 61 50.45 67.69 −0.652 叶绿体 46.15 OfNAC21/22 296 33 613.15 6.54 33 34 51.26 66.52 −0.570 细胞核 71.43 OfNAC56 322 35 946.51 8.62 37 34 39.48 64.81 −0.725 细胞核 92.30 OfNAC17-X2 573 64 828.71 4.87 63 91 40.60 77.70 −0.530 细胞核 53.85 OfNAC17 606 68 408.12 4.85 60 97 46.79 74.62 −0.571 细胞核 30.77 OfNAC104 186 21 498.77 4.59 19 31 46.96 68.60 −0.695 细胞核 84.61 OfNAC92 325 36 813.71 6.47 40 42 29.27 71.35 −0.579 细胞核 38.46 OfNAC72-1 339 38 289.93 8.64 40 37 40.14 64.96 −0.671 细胞核 84.61 OfNAC72-2 342 38 848.46 8.64 42 39 40.04 61.64 −0.762 细胞核 100 OfNAC29-1 280 32 538.56 7.71 36 35 42.96 61.96 −0.816 细胞核 52.50 OfNAC29-2 275 31 330.36 9.33 35 26 33.81 61.35 −0.741 细胞核 100 OfNAC71 303 34 864.61 5.42 33 43 51.89 53.37 −0.795 细胞核 85.71 OfNAC2 296 34 249.55 6.09 35 38 50.01 66.22 −0.735 细胞核 69.23 OfNAC100-2 334 37 778.80 6.51 38 40 43.10 67.96 −0.540 细胞核 85.71 OfNAC32 263 30 253.45 8.45 38 35 43.91 69.32 −0.635 细胞核 61.54 说明:不稳定系数大于40为不稳定序列,小于40为稳定序列 -
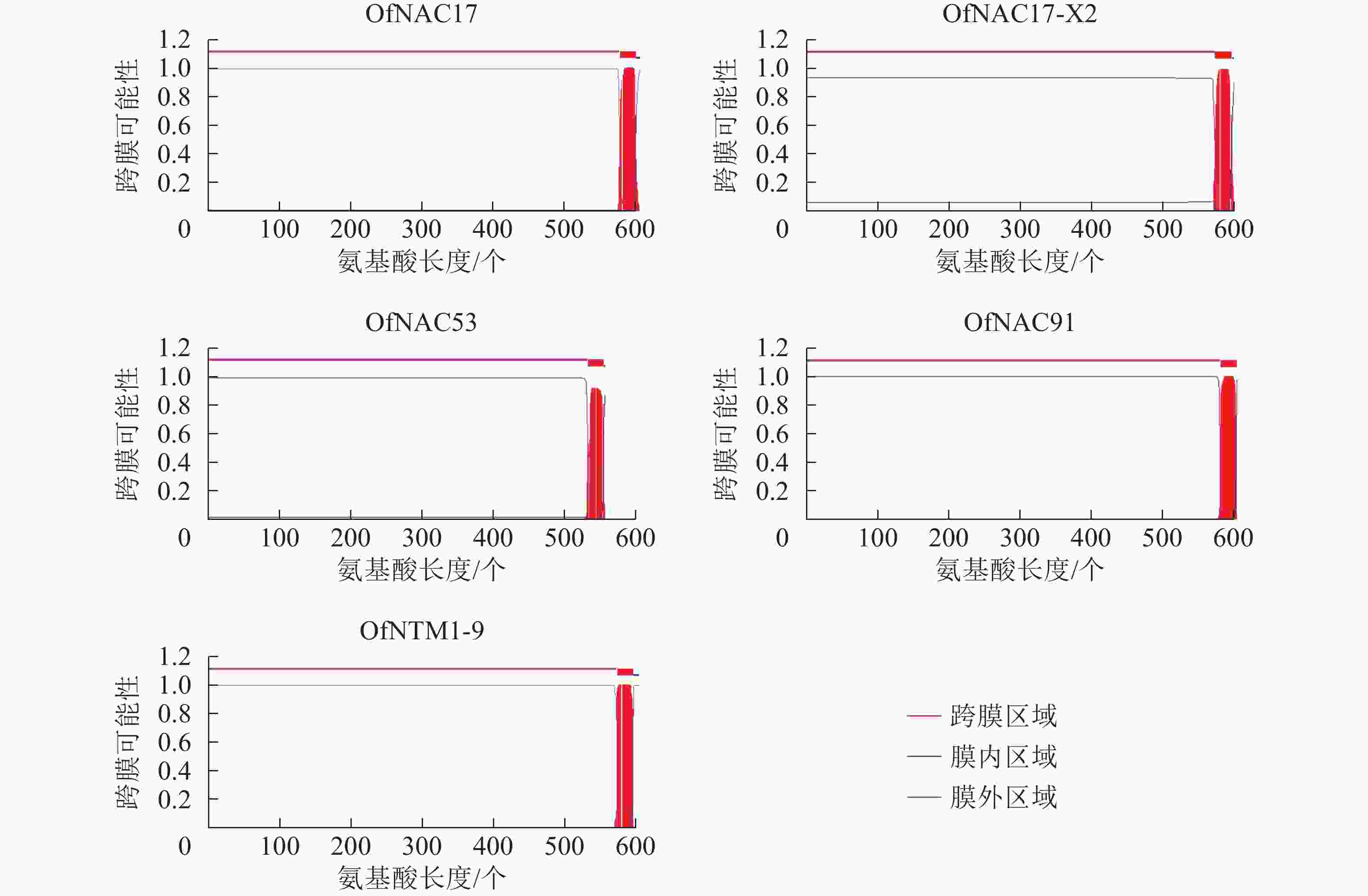
使用DNAMAN软件对桂花OfNAC转录因子的氨基酸序列进行比对,结果显示(图2):22个OfNAC的氨基酸序列均具有保守的亚结构域(A~E)。对这5个亚结构域进行分析发现:A区较为保守的氨基酸是甘氨酸(G)、苯丙氨酸(F)、脯氨酸(P)、谷氨酸(E);B区较为保守的氨基酸是脯氨酸(P);C区较为保守的氨基酸是苯丙氨酸(F)、精氨酸(R)、色氨酸(W)、甘氨酸(G);D区较为保守的氨基酸是甘氨酸(G)、赖氨酸(K)、色氨酸(W);E区没有完全稳定的氨基酸。各保守亚域的保守性由强到弱依次为C、A、D、B、E。对所有OfNAC的氨基酸序列进行保守基序分析发现(图3):大多数OfNAC均含有较保守的元件,除OfNAC73只有2个保守元件(元件1、元件2)外,其余OfNAC均含有4个保守元件(元件1、元件3、元件4、元件5)。另外,22条OfNAC转录因子中元件7、元件9和元件10出现在一些OfNAC蛋白质的C端,且对应的序列亲缘关系较近。桂花OfNAC蛋白质二级结构分析表明:OfNAC蛋白质的二级结构包括α-螺旋、β-折叠、延伸链和无规则卷曲4种结构,且不同结构占比从大到小依次表现为无规则卷曲、α-螺旋、延伸链、β-折叠(表3)。OfNAC32的无规则卷曲占比最高,为67.30%。此外,OfNAC73、OfNAC92、OfNAC29-2结构中α-螺旋占比均小于延伸链。跨膜结构预测分析表明(图4):OfNAC17、OfNAC17-X2、OfNAC53、OfNAC91、OfNTM1-9均在C端有1个跨膜结构。亚细胞定位预测分析表明:OfNAC53定位于内质网,OfNAC50-X2和OfNAC50定位于叶绿体,其余OfNAC蛋白质均定位在细胞核中。
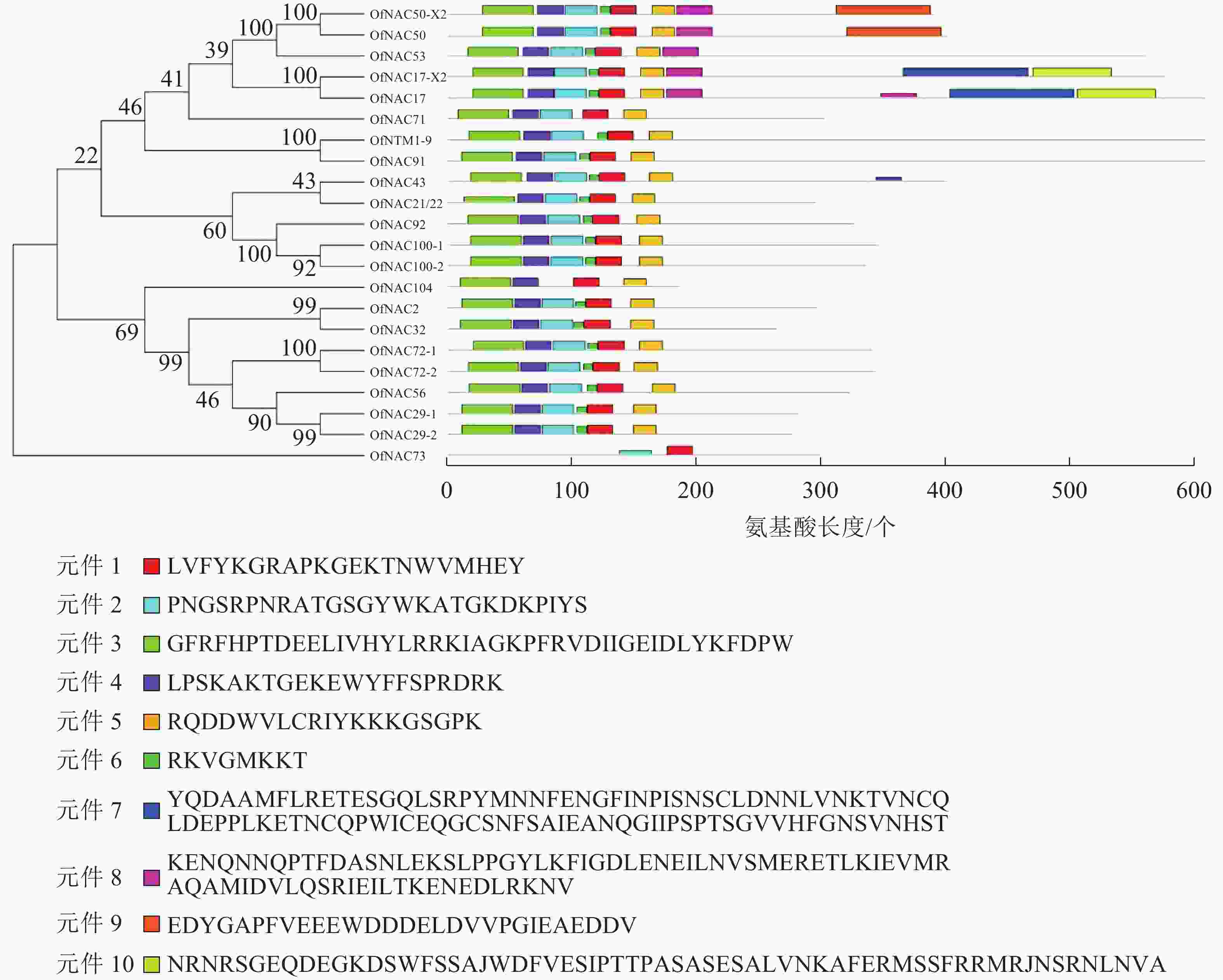
图 3 桂花OfNAC转录因子进化分析及保守基序分析
Figure 3. Evolution analysis of 22 OfNAC transcription factors and analysis of conserved motif
表 3 桂花OfNAC蛋白质理化性质及二级结构分析
Table 3. Secondary structure analysis of OfNAC of O. fragrans
序列名称 二级结构占比 序列名称 二级结构占比 α-螺旋/% β-螺旋/% 延伸/% 无规则卷曲/% α-螺旋/% β-螺旋/% 延伸/% 无规则卷曲/% OfNAC100-1 15.07 3.19 14.78 66.96 OfNAC17 24.59 2.81 12.71 59.90 OfNAC53 21.33 3.76 14.52 60.39 OfNAC104 18.82 3.76 15.59 61.83 OfNTM1-9 23.27 3.14 9.74 63.86 OfNAC92 14.15 3.69 15.69 66.46 OfNAC73 13.00 3.67 20.00 63.33 OfNAC72-1 17.99 3.54 16.22 62.24 OfNAC43 21.55 4.26 14.29 59.90 OfNAC72-2 16.96 3.51 16.08 63.45 OfNAC91 20.36 2.63 10.51 66.50 OfNAC29-1 3.21 3.57 17.86 55.36 OfNAC50-X2 31.11 5.91 12.34 50.64 OfNAC29-2 15.64 4.36 16.73 63.27 OfNAC50 27.82 5.01 10.53 56.64 OfNAC71 16.17 4.95 14.52 64.36 OfNAC21/22 17.23 3.04 15.20 64.53 OfNAC2 21.62 4.05 11.82 62.50 OfNAC56 16.15 3.11 15.84 64.91 OfNAC100-2 16.47 3.59 14.07 65.87 OfNAC17-X2 25.83 2.97 12.91 58.29 OfNAC32 17.11 2.66 12.93 67.30 -
为了进一步研究OfNAC的进化关系,对桂花、拟南芥和水稻的NAC蛋白构建系统进化树。结果表明:22个桂花OfNAC与拟南芥和水稻的NAC蛋白质共划分为11个亚族(图5),除a、b、c、e、h亚族外,其他亚族均有OfNAC分布。其中i亚族桂花NAC蛋白质较多,有8个OfNAC蛋白质。
-
为了研究OfNAC在桂花花开放进程中的表达模式,对其进行荧光定量表达分析(图6)。OfNAC100-2、OfNAC43、OfNAC73相对表达量在铃梗期(S4)出现表达高峰,在此之后相对表达量大幅度降低,其中OfNAC43只在铃梗期(S4)相对表达量最高,其余时期相对表达量很低;OfNAC56从铃梗期开始呈现上升趋势,并在盛花期(S6)到达顶峰;OfNAC71、OfNAC29-1、OfNAC21/22从起始期(S1)缓慢上升,在顶壳期 (S3)到达最高,随后急剧下降;OfNAC29-2在圆珠期(S2)相对表达量达到最大,之后在铃梗期(S4)相对表达达到最低。OfNAC在花开放不同阶段的表达情况说明:OfNAC100-2、OfNAC43、OfNAC73、OfNAC71、OfNAC29-1、OfNAC21/22、OfNAC29-2可能参与花开放的调控,其中又以OfNAC43和OfNAC29-2关联最为紧密。
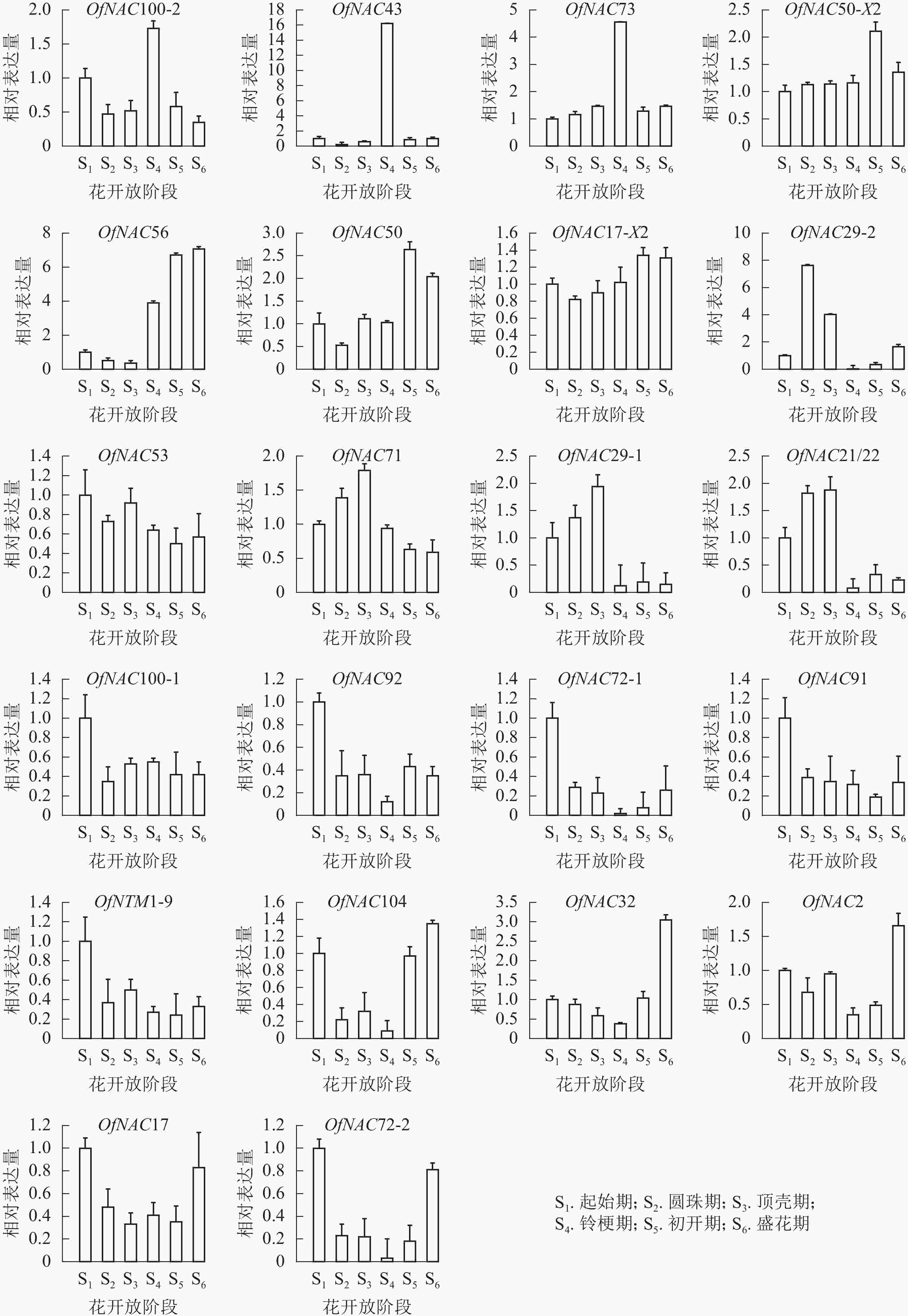
图 6 22个OfNAC基因在不同花开放时期的表达结果
Figure 6. Expression results of 22 OfNAC genes in different flower opening periods
OfNAC50-X2、OfNAC50、OfNAC17-X2整体呈现上升趋势,并在初开期(S5)达到最高;OfNAC100-1、OfNAC92、OfNAC72-1、OfNAC91、OfNTM1-9、OfNAC53在起始期(S1)表达量较高,整体呈下降趋势;OfNAC104、OfNAC32、OfNAC2、OfNAC17、OfNAC72-2均呈现先下调后上升的表达趋势,到盛花期(S6)相对表达量较高。这说明,这些基因成员可能不参与桂花花开放的调控。
-
NAC转录因子最初在矮牵牛Petunia hybrida中发现[21]。在模式植物拟南芥、水稻以及园艺作物中的研究较多。在拟南芥、水稻、黄瓜Cucumis sativus、紫花苜蓿Medicago sativa中分别有151、117、91、113个NAC基因被鉴定[22-24]。目前关于桂花OfNAC转录因子的报道较少。通过桂花OfNAC转录因子的鉴定与分析,可为进一步了解桂花OfNAC的生物学功能及其在花开放过程中的作用奠定基础。
本研究通过转录组测序鉴定出22条开放阅读框完整的OfNAC转录因子,对它们进行跨膜结构预测发现:OfNAC17、OfNAC17-X2、OfNAC53、OfNAC91、OfNTM1-9这5个转录因子均在C端有1个跨膜结构,推测它们可能是NAC膜结合转录因子。大多数OfNAC蛋白质预测定位在细胞核中的可能性较高,因此推测,OfNAC转录因子可能大多依旧在细胞核内发挥功能。目前,已经在模式植物中发现NAC膜结合转录因子,如拟南芥和水稻NAC基因组中已发现分别有18和6个膜结合NAC转录因子[25-26],其中拟南芥膜结合转录因子NTM1调节细胞分裂素信号传导而参与细胞分裂;NTL8调节拟南芥开花时间,过表达转基因植株出现晚花现象;ANAC089被确定为另一个膜结合转录因子,也有调控拟南芥开花时间的功能[27-28]。由此推测,桂花OfNAC17、OfNAC17-X2、OfNAC53、OfNAC91、OfNTM1-9可能也具有调控开花时间的功能。
桂花22个OfNAC与拟南芥和水稻NAC蛋白质共同划分为11个亚族。根据亲缘关系推测相关OfNAC基因的功能,如OfNAC100-1、OfNAC100-2与拟南芥At5G6140.1、At5G7680.1、At5G39610.1、At3G29035.1亲缘性较高,其中At5G39610.1(ANAC92/ATNAC2)存在于部分或完全开放的花的花器官中,且参与叶和花的衰老过程[29]。在月季中,RhNAC100是At5G39610.1(ANAC92/ATNAC2)的同源基因,乙烯通过微调microRNA164-RhNAC100模块来调节细胞的扩张从而影响月季花瓣的扩展[30],OfNAC100-1和OfNAC100-2与拟南芥At5G39610.1亲缘关系较近,所以推测OfNAC100-1与OfNAC100-2可能有调节花瓣扩展的相关功能。OfNTM1-9与At1G33060.1同源性较高,且At1G33060.1是膜结合转录因子,在拟南芥各个器官均有表达,影响叶片衰老[25],推测OfNTM1-9可能作为NAC膜结合转录因子参与细胞衰老。
花开放的主要原因是花瓣细胞的扩展[11-13],对OfNAC基因在桂花花开放时期的荧光定量表达分析表明:OfNAC在桂花花开放过程中发挥重要作用,且不同的OfNAC在此过程中表达模式有差异。OfNAC71、OfNAC29-1、OfNAC21/22在顶壳期相对表达到达最大。罗云等[31]发现:OfEXPA2和OfEXLA1在桂花花开放的顶壳期相对表达急剧增加并到达顶峰,推测此时期的高丰度表达是后续花开放的基础。OfNAC71、OfNAC29-1、OfNAC21/22与OfEXPA2和OfEXLA1表达模式相似,推测它们参与花开放进程。OfEXPA4在花芽萌发期至圆珠期表达量较低,而顶壳期开始急剧上升,铃梗期表达量达到最大。罗云等[31]推测:OfEXPA4是参与花瓣扩展的关键基因,OfNAC100-2、OfNAC43、OfNAC73相对表达量也在铃梗期出现峰值,推测它们亦作为关键基因参与桂花花瓣扩展。对月季的研究发现:RhNAC2和RhEXPA4参与月季花瓣细胞扩展,且RhEXPA4的表达可能受到RhNAC2的调控[32],拟南芥中NAC25和NAC1L被确定是EXPA2(EXPANSIN2)的上游调控因子[33]。以此推测,NAC转录因子可能与EXP的启动子结合,是EXP的上游调控因子,则OfNAC29-2在圆珠期相对表达最大,可能是作为OfEXPA2和OfEXLA1调控因子调节OfEXP影响花瓣细胞扩张从而为后续花瓣展开奠定基础,所以OfNAC100-2、OfNAC43、OfNAC73、OfNAC71、OfNAC29-1、OfNAC21/22、OfNAC29-2可能是参与桂花花开放的关键基因。由于OfNAC43、OfNAC29-2相对表达差异明显,推测它们与花开放关系更为紧密,但这些关键基因在桂花开花中的具体作用还需要进一步研究。
-
本研究从桂花品种‘堰虹桂’转录组数据中筛选得到22条OfNAC序列,生物信息学分析结果表明:22条OfNAC转录因子氨基酸序列含有5个保守的亚结构域(A~E),其保守性由强到弱依次为C、A、D、B、E;亚细胞定位及跨膜结构预测表明:OfNAC17、OfNAC17-X2、OfNAC53、OfNAC91、OfNTM1-9是膜结合转录因子,且大多数 OfNAC 定位在细胞核。实时荧光定量PCR分析表明:在桂花花开放进程中,22个OfNAC基因表达模式不同,其中OfNAC100-2、OfNAC43、OfNAC73相对表达量在铃梗期(S4)达到顶峰,在此之后相对表达降低。OfNAC43在铃梗期骤然升高,并且在此时期相对表达最大;OfNAC71、OfNAC29-1、OfNAC21/22从起始期呈缓慢上升趋势,在顶壳期到达最高,随后整体呈现下降趋势;OfNAC29-2在圆珠期相对表达量陡然上升,在铃梗期相对表达最低,推测OfNAC100-2、OfNAC43、OfNAC73、OfNAC71、OfNAC29-1、OfNAC21/22、OfNAC29-2等成员极有可能参与调控桂花的花开放。
Identification and expression analysis of OfNAC transcription factors in Osmanthus fragrans during flower opening stage
-
摘要:
目的 研究OfNAC基因对桂花Osmanthus fragrans花开放的调控作用。 方法 从桂花品种‘堰虹桂’O. fragrans ‘Yanhonggui’转录组数据中,筛选获得相关OfNAC基因序列,分析预测其理化性质和结构,运用实时荧光定量PCR技术分析花开放过程的表达特性。 结果 筛选得到22条OfNAC序列。生物信息学分析发现:22条OfNAC转录因子均含有NAM结构域,氨基酸序列含有5个保守的亚结构域(A~E),其保守性由强到弱依次为C、A、D、B、E;二级结构中不同结构的占比由大到小表现为无规则卷曲、α-螺旋、延伸链、β-折叠;亚细胞定位及跨膜结构预测表明:OfNAC17、OfNAC17-X2、OfNAC53、OfNAC91、OfNTM1-9是膜结合转录因子,且大多数OfNAC定位在细胞核。在桂花花开放进程中,OfNAC100-2、OfNAC43、OfNAC73相对表达量在铃梗期(S4)到达顶峰,在此之后相对表达降低;OfNAC43在铃梗期(S4)骤然升高,并且在此时期相对表达最大;OfNAC71、OfNAC29-1、OfNAC21/22从起始期(S1)呈缓慢上升趋势,在顶壳期 (S3) 到达最高,随后整体呈现下降趋势;OfNAC29-2在圆珠期(S2)相对表达量陡然上升,在铃梗期(S4)相对表达最低。 结论 推测OfNAC100-2、OfNAC43、OfNAC73、OfNAC71、OfNAC29-1、OfNAC21/22、OfNAC29-2等成员极有可能参与调控桂花的花开放。图6表3参33 Abstract:Objective This study aims to investigate the regulation of OfNAC genes on flower opening of Osmanthus fragrans. Method From the transcriptome data of O. fragrans ‘Yanhonggui’, the related OfNAC g enes were screened and analyzed to predict the physicochemical properties and structure. Real-time fluorescence quantitative PCR was used to analyze the expression characteristics of the flower opening process. Result 22 OfNAC sequences were screened from transcriptome. Bioinformatics analysis showed that all of the 22 OfNAC transcription factors contained NAM domains, and the amino acid sequence contained 5 conserved subdomains (A−E). The sequence of conservatism ranging from strong to weak was C, A, D, B, and E. The proportion of different structures in the secondary structure from large to small was random coils, α-helices, extended strands, and β-sheets. Subcellular localization and transmembrane structure prediction showed that OfNAC17, OfNAC17-X2, OfNAC53, OfNAC91, OfNTM1-9 were membrane-bound transcription factors, and most OfNACs were located in the nucleus. During the O. fragrans flowering process, the relative expression levels of OfNAC100-2, OfNAC43 and OfNAC73 reached the peak at the boll stem stage (S4), and then decreased. The relative expression of OfNAC43 was the highest at the boll stem stage (S4). The relative expression of OfNAC71, OfNAC29-1, and OfNAC21/22 increased slowly from the initial stage (S1), reached the highest at the apical shell stage, and then decreased as a whole. The relative expression of OfNAC29-2 increased sharply at the bead stage (S2) and reached the lowest at the boll stem stage (S4). Conclusion It is speculated that OfNAC100-2, OfNAC43, OfNAC73, OfNAC71, OfNAC29-1, OfNAC21/22, OfNAC29-2 may be involved in the regulation of O. fragrans flower opening. [Ch, 6 fig. 3 tab. 33 ref.] -
Key words:
- botany /
- Osmanthus fragrans /
- flower opening /
- NAC transcription factor /
- expression analysis
-
表 1 桂花OfNAC转录因子RT-PCR特异性引物
Table 1. Specific primers for RT-PCR of OfNAC in O. fragrans
基因名称 用于荧光定量的引物序列(5′→3′) 基因名称 用于荧光定量的引物序列(5′→3′) OfNAC100-1 F: TGAACAAGATTGAGCCTTGGG OfNAC104 F: TGCATTTTACATTGGTGAAGATGTC R: CCTTTCCTGTGGCTTTCCAG R: GCTCGTACACTTGACACACCA OfNAC53 F: AGATTGTGGGGATGAAGAAAA OfNAC92 F: TCCTAGTCGGAATGAAGAAAACTC R: CAACTCCATATCAGTAAGCCG R: ATGGCTTTCTAATCTGTATTCGTGC OfNTM1-9 F: GGTTGCTCTAATGCCCACTTC OfNAC72-1 F: GGAAAAGCCCCCAAAGGAAC R: CTGGTTCCGTAGCACGATACT R: CCCAATCATCCAACCTTGAGC OfNAC73 F: AGGCAAGGATGGCCAAATTC OfNAC72-2 F: ACGTAGGAAAAGCACCAAAAGG R: TTGTGCCATCTTGTTTCTCC R: AGCATCCAACCTTGCGCTTC OfNAC43 F: AGGCTACTGGTCGTGATAAAG OfNAC29-1 F: TTACAAGGGAAGGCCTCCAAAG R: GGGGTCATGAGTGTCGTCC R: TTGAGCCATTTTGCGTGTTAGG OfNAC91 F: TCTACAAAGGTCGTGCTCCG OfNAC29-2 F: CCCAAAGGGCGTCAAAACTG R: CCCTGACCAGGATAAGTGCC R: GCACACAATCATCCAACCTCA OfNAC50-X2 F: TGGCAAAGGGTATTGGAAAGC OfNAC71 F: CTATCGTGGAAGAGCACCACT R: TCGCCCACTATGGAAAACAAG R: TCCCTGAAATCTTGGGGTGTC OfNAC50 F: AAGCAACTGGAAAGGATCGC OfNAC2 F: TTGGGAATAAAGAAGGCTCTGGTG R: AATTCTGCATCGCAAAGCCTG R: ACACAACACCCAATCATCAAGCCTC OfNAC21/22 F: GAAGGGAAGCCTGGTTGGAAT OfNAC100-2 F: TCAGAGGAAAAATCCTCGTCGG R: CCCAATCCTCCTTGACAGATG R: TTTGGGAGGTTGTGGATCGAG OfNAC56 F: TCTATGGTGGAAAGCCTCCT OfNAC32 F: AAGCCTTGGTTTTCTATGCCG R: CATCAAGCCTTAAAGAGCCC R: AAGCTGTTGTTCTTGTTTCGA OfNAC17-X2 F: TCCTGTTGGGGTGAAGAAGA OfACT F: CCCAAGGCAAACAGAGAAAAAAT R: ATAGTCATCCTGTGCATCCTGC R: ACCCCATCACCAGAATCAAGAA OfNAC17 F: TGGTCTTCCATAAAGGTCGTGC R: TTGTACAGAGCATAGCAATCCCGTG 表 2 桂花OfNAC蛋白质理化性质及二级结构分析
Table 2. Physicochemical properties and secondary structure analysis of OfNAC of O. fragrans
序列名称 氨基酸长度/个 相对分子量 等电点 碱性氨基酸 酸性氨基酸 不稳定系数 脂溶性指数 总平均疏水值 亚细胞定位 位置 预测值/% OfNAC100-1 345 38 986.11 8.69 40 36 40.06 61.01 −0.574 细胞核 61.54 OfNAC53 558 62 807.47 4.60 51 88 38.87 69.37 −0.564 内质网 42.42 OfNTM1-9 606 68 053.70 5.63 71 86 51.49 62.43 −0.725 细胞核 76.92 OfNAC73 300 33 768.95 8.79 39 34 38.21 69.47 −0.754 细胞核 76.92 OfNAC43 399 45 513.66 5.77 43 55 46.31 63.26 −0.752 细胞核 69.23 OfNAC91 609 67 741.52 4.98 63 86 52.83 71.07 −0.593 细胞核 76.92 OfNAC50-X2 389 43 765.29 5.29 46 58 47.83 69.43 −0.629 叶绿体 58.33 OfNAC50 399 44 952.52 5.35 46 61 50.45 67.69 −0.652 叶绿体 46.15 OfNAC21/22 296 33 613.15 6.54 33 34 51.26 66.52 −0.570 细胞核 71.43 OfNAC56 322 35 946.51 8.62 37 34 39.48 64.81 −0.725 细胞核 92.30 OfNAC17-X2 573 64 828.71 4.87 63 91 40.60 77.70 −0.530 细胞核 53.85 OfNAC17 606 68 408.12 4.85 60 97 46.79 74.62 −0.571 细胞核 30.77 OfNAC104 186 21 498.77 4.59 19 31 46.96 68.60 −0.695 细胞核 84.61 OfNAC92 325 36 813.71 6.47 40 42 29.27 71.35 −0.579 细胞核 38.46 OfNAC72-1 339 38 289.93 8.64 40 37 40.14 64.96 −0.671 细胞核 84.61 OfNAC72-2 342 38 848.46 8.64 42 39 40.04 61.64 −0.762 细胞核 100 OfNAC29-1 280 32 538.56 7.71 36 35 42.96 61.96 −0.816 细胞核 52.50 OfNAC29-2 275 31 330.36 9.33 35 26 33.81 61.35 −0.741 细胞核 100 OfNAC71 303 34 864.61 5.42 33 43 51.89 53.37 −0.795 细胞核 85.71 OfNAC2 296 34 249.55 6.09 35 38 50.01 66.22 −0.735 细胞核 69.23 OfNAC100-2 334 37 778.80 6.51 38 40 43.10 67.96 −0.540 细胞核 85.71 OfNAC32 263 30 253.45 8.45 38 35 43.91 69.32 −0.635 细胞核 61.54 说明:不稳定系数大于40为不稳定序列,小于40为稳定序列 表 3 桂花OfNAC蛋白质理化性质及二级结构分析
Table 3. Secondary structure analysis of OfNAC of O. fragrans
序列名称 二级结构占比 序列名称 二级结构占比 α-螺旋/% β-螺旋/% 延伸/% 无规则卷曲/% α-螺旋/% β-螺旋/% 延伸/% 无规则卷曲/% OfNAC100-1 15.07 3.19 14.78 66.96 OfNAC17 24.59 2.81 12.71 59.90 OfNAC53 21.33 3.76 14.52 60.39 OfNAC104 18.82 3.76 15.59 61.83 OfNTM1-9 23.27 3.14 9.74 63.86 OfNAC92 14.15 3.69 15.69 66.46 OfNAC73 13.00 3.67 20.00 63.33 OfNAC72-1 17.99 3.54 16.22 62.24 OfNAC43 21.55 4.26 14.29 59.90 OfNAC72-2 16.96 3.51 16.08 63.45 OfNAC91 20.36 2.63 10.51 66.50 OfNAC29-1 3.21 3.57 17.86 55.36 OfNAC50-X2 31.11 5.91 12.34 50.64 OfNAC29-2 15.64 4.36 16.73 63.27 OfNAC50 27.82 5.01 10.53 56.64 OfNAC71 16.17 4.95 14.52 64.36 OfNAC21/22 17.23 3.04 15.20 64.53 OfNAC2 21.62 4.05 11.82 62.50 OfNAC56 16.15 3.11 15.84 64.91 OfNAC100-2 16.47 3.59 14.07 65.87 OfNAC17-X2 25.83 2.97 12.91 58.29 OfNAC32 17.11 2.66 12.93 67.30 -
[1] OLSEN A N, ERNST H A, LEGGIO L L, et al. NAC transcription factors: structurally distinct, functionally diverse [J]. Trends Plant Sci, 2005, 10(2): 79 − 87. [2] HIBARA K, TAKADA S, TASAKA M. CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation [J]. Plant J, 2004, 36(5): 687 − 696. [3] ZHANG Qian, LUO Fang, ZHONG Yu, et al. Modulation of NAC transcription factor NST1 activity by XYLEM NAC DOMAIN1 regulates secondary cell wall formation in Arabidopsis [J]. J Exp Bot, 2019, 71(4): 1449 − 1458. [4] PITAKSARINGKARN W, MATSUOKA K, ASAHINA M, et al. XTH20 and XTH19 regulated by ANAC071 under auxin flow are involved in cell proliferation in incised Arabidopsis inflorescence stems [J]. Plant J, 2014, 80(4): 604 − 614. [5] KANEDA T, TAGA Y, TAKAI R, et al. The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death [J]. EMBO J, 2009, 28(7): 926 − 936. [6] JEONG J S, KIM Y S, BAEK K H, et al. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions [J]. Plant Physiol, 2010, 153(1): 185 − 197. [7] CHEN Xu, LU Songchong, WANG Yaofeng, et al. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice [J]. Plant J, 2015, 82(2): 302 − 314. [8] LIU Yuanlong, KE Lili, WU Guizhi, et al. miR3954 is a trigger of phasiRNAs that affects flowering time in citrus [J]. Plant J, 2017, 92(2): 263 − 275. [9] TRUPKINSA, ASTIGUETA F, BAIGORRIA A H, et al. Identification and expression analysis of NAC transcription factors potentially involved in leaf and petal senescence in Petunia hybrid[J]. Plant Science, 2019, 287: 110195. doi: 10.1016/j.plantsci.2019.110195. [10] JIANG Guoxiang, LI Zhiwei, SONG Yunbo, et al. LcNAC13 physically interacts with LcR1MYB1 to coregulate anthocyanin biosynthesis-related genes during litchi fruit ripening[J]. Biomolecules, 2019, 9(4): 135. doi: 10.3390/biom9040135. [11] van DOORN W G, van MEETEREN U. Flower opening and closure: a review [J]. J Exp Bot, 2003, 54(389): 1801 − 1812. [12] van DOORN W G, KAMDEE C. Flower opening and closure: an update [J]. J Exp Bot, 2014, 65(20): 5749 − 5757. [13] IRISH V F. The Arabidopsis petal: a model for plant organogenesis [J]. Trends Plant Sci, 2008, 13(8): 430 − 436. [14] ZONIA L, MUNNIK T. Life under pressure: hydrostatic pressure in cell growth and function [J]. Trends Plant Sci, 2007, 12(3): 90 − 97. [15] SABLOWSKI R, MEYEROWITZ E M. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA [J]. Cell, 1998, 92(1): 93 − 103. [16] JIANG Xinqiang, ZHANG Changqing, LÜ Peitao, et al. RhNAC3, a stress-associated NAC transcription factor, has a role in dehydration tolerance through regulating osmotic stress-related genes in rose petals [J]. Plant Biotechnol J, 2014, 12(1): 38 − 48. [17] 王英, 张超, 付建新, 等. 桂花花芽分化和花开放研究进展[J]. 浙江农林大学学报, 2016, 33(2): 340 − 347. WANG Ying, ZHANG Chao, FU Jianxin, et al. Progress on flower bud differentiation and flower opening in Osmanthus fragrans [J]. J Zhejiang A&F Univ, 2016, 33(2): 340 − 347. [18] ZHANG Chao, WANG Yiguang, FU Jianxin, et al. Transcriptomic analysis and carotenogenic gene expression related to petal coloration in Osmanthus fragrans‘Yanhong Gui’ [J]. Trees-Struct Funct, 2016, 30(4): 1207 − 1223. [19] CHEN Chengjie, CHEN Hao, ZHANG Yi, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194 − 1202. [20] 付建新, 张超, 王艺光, 等. 桂花组织基因表达中荧光定量PCR内参基因的筛选[J]. 浙江农林大学学报, 2016, 33(5): 727 − 733. FU Jianxin, ZHANG Chao, WANG Yiguang, et al. Reference gene selection for quantitativereal-time polymerase chain reaction(qRT-PCR) normalization in the gene expression of sweet osmanthus tissues [J]. J Zhejiang A&F Univ, 2016, 33(5): 727 − 733. [21] SOUER E, van HOUWELINGEN A, KLOOS D, et al. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries [J]. Cell, 1997, 85(2): 159 − 170. [22] NURUZZAMAN M, MANIMEKALAI R, AKHTER S, et al. Genome-wide analysis of NAC transcription factor family in rice [J]. Gene, 2010, 465(1/2): 30 − 44. [23] LIU Xingwang, WANG Ting, BARTHOLOMEW E, et al. Comprehensive analysis of NAC transcription factors and their expression during fruit spine development in cucumber (Cucumis sativus L.)[J]. Hortic Res, 2018, 5(1): 31. doi: 10.1038/s41438-018-0036-z. [24] MIN Xueyang, JIN Xiaoyu, ZHANG Zhengshe, et al. Genome-wide identification of NAC transcription factor family and functional analysis of the abiotic stress-responsive genes in Medicago sativa L. [J]. J Plant Growth Regul, 2020, 39(1): 324 − 337. [25] KIM S M, KIM S G, KIM Y S, et al. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation [J]. Nucl Acids Res, 2007, 35(1): 203 − 213. [26] FANG Yujie, YOU Jun, XIE Kabin, et al. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice [J]. Mol Genet Genomics, 2008, 280(6): 547 − 563. [27] KIM S G, LEE A K, YOON H K, et al. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination [J]. Plant J, 2008, 55(1): 77 − 88. [28] 李建琴, 张娟, 王学臣, 等. 膜系留转录因子ANAC089在拟南芥开花诱导过程中起负调控作用[J]. 中国科学: 生命科学, 2010, 40(5): 408 − 417. LI Jianqin, ZHANG Juan, WANG Xuechen, et al. A membrane-tethered transcription factor ANAC089 negatively regulates floral initiation in Arabidopsis thaliana [J]. Sci Sin Vitae, 2010, 40(5): 408 − 417. [29] BALAZADEH S, SIDDIQUI H, ALLU A D, et al. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence [J]. Plant J, 2010, 62(2): 250 − 264. [30] PEI Haixia, MA Nan, TIAN Ji, et al. An NAC transcription factor controls ethylene-regulated cell expansion in flower petals [J]. Plant Physiol, 2013, 163(2): 775 − 791. [31] 罗云, 张超, 付建新, 等. 桂花扩展蛋白基因家族的鉴定和表达分析[J]. 农业生物技术学报, 2017, 25(8): 1289 − 1299. LUO Yun, ZHANG Chao, FU Jianxin, et al. Identification and expression analysis of expansin gene family in Osmanthus fragrans [J]. J Agric Biotechnol, 2017, 25(8): 1289 − 1299. [32] DAI Fanwei, ZHANG Changqing, JIANG Xinqiang, et al. RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals [J]. Plant Physiol, 2012, 160(4): 2064 − 2082. [33] SÁNCHEZ-MONTESINO R, BOUZA-MORCILLO L, MARQUEZ J, et al. A regulatory module controlling GA-mediated endosperm cell expansion is critical for seed germination in Arabidopsis [J]. Mol Plant, 2019, 12(1): 71 − 85. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.20200474






 下载:
下载: