-
黄薇Heimia myrtifolia是千屈菜科Lythraceae黄薇属Heimia落叶丛生灌木,原产于巴西,南亚、东亚地区有零星分布,在中国的上海、浙江、广西等地也有引种栽培[1]。黄薇喜温暖湿润且阳光充沛的环境,不耐干旱,夏季开花,花色金黄,花量丰富,可作为园林绿篱、花坛花境背景应用,点缀和美化植物景观,还可以净化城市水质,具有较高的园林价值和应用前景[2]。目前,国内外主要在黄薇的分类和应用[3]、叶内酚类化合物[4]、类黄酮组分[5]、非生物胁迫的抵御机制[6−8]、叶绿体基因组分析[9]等方面进行了研究,但关于黄薇的分子生物学方面的研究则十分匮乏。
实时荧光定量PCR (RT-qPCR)具有灵敏度高、重复性好、特异性强及高通量等优点,近年来已成为研究基因表达分析的常用方法[10],在分子生物学领域,RT-qPCR对深入研究植物基因表达调控机制、发现新基因、细胞因子表达分析、预测基因功能、医学诊断以及食品药品检测等至关重要[11]。在实际的应用过程中,为了确保RT-qPCR结果的准确性,需要引用一个表达量稳定的内参基因作为参照,而内参基因的稳定性也会影响RT-qPCR结果的准确性[12−13]。因此,筛选到适宜的内参基因是进行RT-qPCR表达分析时的重要前提[14]。目前暂未见黄薇内参基因筛选的相关研究,这严重制约了对黄薇目标基因的定量分析。因此,本研究基于黄薇转录组数据,对不同组织及不同干旱胁迫下黄薇的内参基因进行研究,为提高不同组织及不同干旱胁迫下基因RT-qPCR分析的准确性提供依据。
-
材料取自浙江农林大学风景园林与建筑学院人工气候室内生长健壮、长势一致的黄薇1年生扦插苗。采样时选取黄薇盛花期的根、茎、叶、花,以及模拟自然条件下的对照组 (土壤相对含水量为65%~75%,ck)和4个干旱胁迫处理组 (T1、T2、T3、T4土壤相对含水量分别为45%~60%、30%~45%、15%~30%、10%~15%)的成熟叶片。采样后立即将样品置于液氮中速冻,并保存在−80 ℃冰箱备用。
-
样品总RNA的提取按诺唯赞公司的RNA isolater Total RNA Extraction Reagent (RC401-01) 试剂盒使用方法进行。利用质量分数为1.2%的琼脂糖凝胶电泳检测RNA的完整性,使用紫外分光光度计测定浓度。总RNA纯度D(260)/D(280)为1.8~2.4,D(260)/D(230)为1.5~2.4。使用Reverse Transcriptase M-MLV试剂(Takara)合成cDNA的第1链,反转录程序为:37 ℃,15 min (反转录过程);85 ℃,5 s (反转录酶的失活过程),4 ℃冰箱保存。最后将cDNA产物于−20 ℃储存备用。
-
基于黄薇转录组数据(序列号:PRJNA804698),选取9个基因作为候选内参基因:延伸因子1α蛋白基因(elongationfactors-1α, EF-1α)、α-微管蛋白基因(α-tubulin, TUA)、亲环蛋白基因(cyclophilin, CYP)、甘油醛-3-磷酸脱氢酶(glyceraldehyde-3-phosphat dehydrogenase, GAPDH)、18S核糖体RNA(18S ribosomal RNA, 18S RNA)、泛素化酶基因(ubiquitin-conjugating enzyme, UBC)、β-维管蛋白基因(tubulin, TUB)、肌动蛋白基因(actin, ACT)和伴侣蛋白基因(chaperone protein, DNA J)。基于转录组数据,选择与纤维素合成和干旱胁迫相关的差异表达基因CSLD、SOD基因,验证内参基因对基因表达量的影响。利用Primer 5软件,设计候选内参基因的荧光定量PCR引物,并由杭州有康生物技术有限公司合成。
-
以所有试样的cDNA为模板依次稀释5个浓度梯度(5−1、5−2、5−3、5−4、5−5),设3次重复,制作9个内参基因的标准曲线。反转录产物cDNA稀释10倍,RT-qPCR反应使用诺唯赞公司出品的SYBR®Primix Ex TaqTM试剂盒(Q712)。荧光定量反应在Roche LightCycler 480Ⅱ荧光定量仪上进行,反应体系为20 μL:2×Taq Pro Universal SYBR qPCR Master Mix荧光染料10 μL、cDNA模板2 μL、上下游引物各0.8 μL、ddH2O 6.4 μL。PCR扩增反应程序为:预变性95 ℃,10 s;PCR反应95 ℃,10 s,60 ℃,30 s,共40个循环。采集溶解曲线荧光信号进行分析,反应体系为:从60 ℃到95 ℃,15 s;60 ℃,60 s;95 ℃,15 s。
-
使用Excel 2016统计基因的Ct值,用Origin 9.1制图。采用geNorm[15]、NormFinder[16]、BestKeeper[17]软件以及在线分析软件RefFinder ,综合分析不同情况下最稳定的内参基因。
-
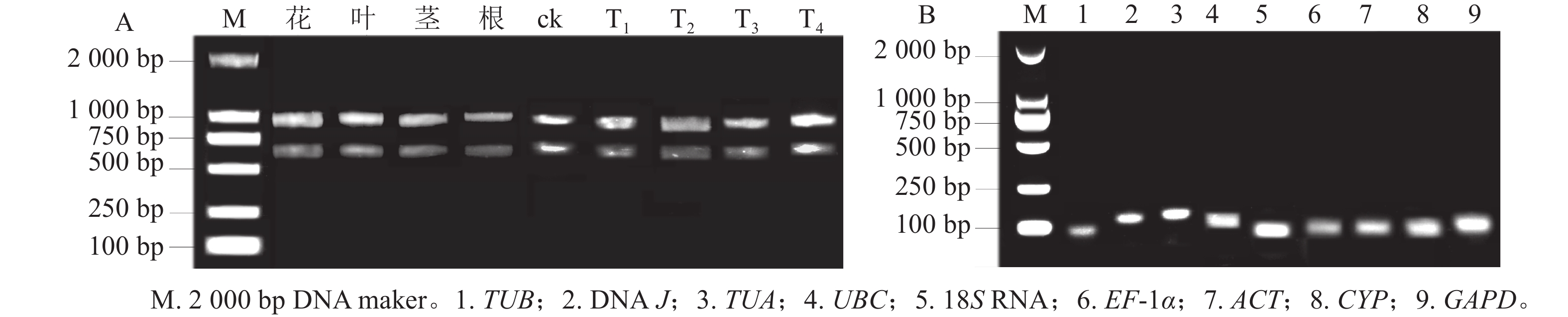
所得RNA产物D(260)/D(280)为1.8~2.2,D(260)/D(230)为1.8~2.4,电泳条带清晰(图1A),说明提取的总RNA质量良好,无明显降解,满足后续实验要求。9个候选内参基因琼脂糖凝胶电泳检测后出现与预期大小一致的单一条带(图1B),说明引物不存在二聚体,可进行特异性扩增。根据结果绘制标准曲线并计算斜率,如表1显示:候选基因的线性相关系数R2>0.990,引物扩增效率为95.6%~109.7%。综上,候选内参基因的扩增效率、产物特异性都符合RT-qPCR的条件,可用于后续的分析。
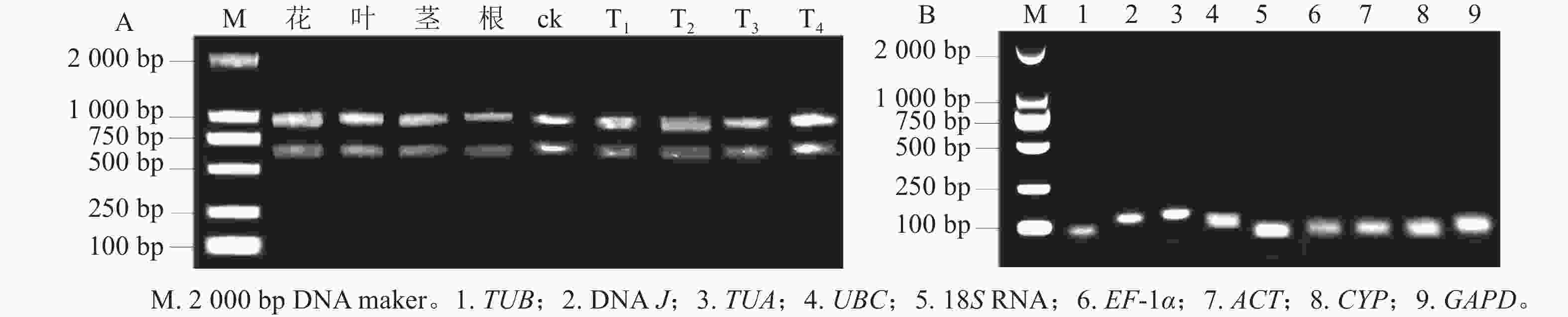
图 1 黄薇总RNA琼脂糖凝胶电泳分析(A)和RT-qPCR扩增产物特异性(B)
Figure 1. Agarose gel electrophoresis analysis of total RNA (A) and specificity of products amplified by RT-qPCR (B) in H. myrtifolia
表 1 黄薇候选内参基因引物信息
Table 1. Primer information of candidate reference gene in H. myrtifolia
基因 正向引物序列(5′→3′) 反向引物序列(5′→3′) 扩增大小/bp 退火温度/℃ 扩增效率/% R2 EF-1α TGGTTTTGAGGCTGGTATCTCC TTTGCTTGACACCAAGGGTGA 80 56.5 95.6 0.998 TUA TCTCTGCCTTGACCGAGTGA ACCACCAACGGCACTGAAAA 82 56.5 96.6 0.998 CYP ACCCCGACTCGTCCTACAAG TCGGTGTTCCGCTCCAAATG 130 58.0 106.8 0.999 GAPDH AGAAGGTCGTCATTTCTGCCC TGGTTGTGCAGCTAGCGTTG 114 57.5 108.4 0.999 18S RNA CAGGGCCTAGGATTTCGTCC GCCTTCAATCTTAGTCGTGGC 113 58.5 100.5 0.992 UBC GACCTGATGACACTCCCTGG TCACAGTTGGTGGTTTGTTCG 87 57.5 99.1 0.999 TUB GGGTGCTGAGCTTATTGATGC TGAGCAATGTCCCCATGCCT 131 57.5 96.8 0.996 ACT AGGGAATGCCTTTTGATTGATCC AAACATAAGCTCCACTGCCCTC 102 57 109.7 0.999 DNA J CGGAGCTATCACCCCGATG CGGCCTCACCATACCTGTCA 127 59.5 100.2 0.997 CSLD TACCTTGTCCCTTTCGGCG TCAGCGTCCTCATCCCGATA 149 57.4 95.1 0.997 SOD GTTGACGCAAGACGAGGGA CCGTTGGTCGTGTCACCAT 108 57.3 96.0 0.997 -
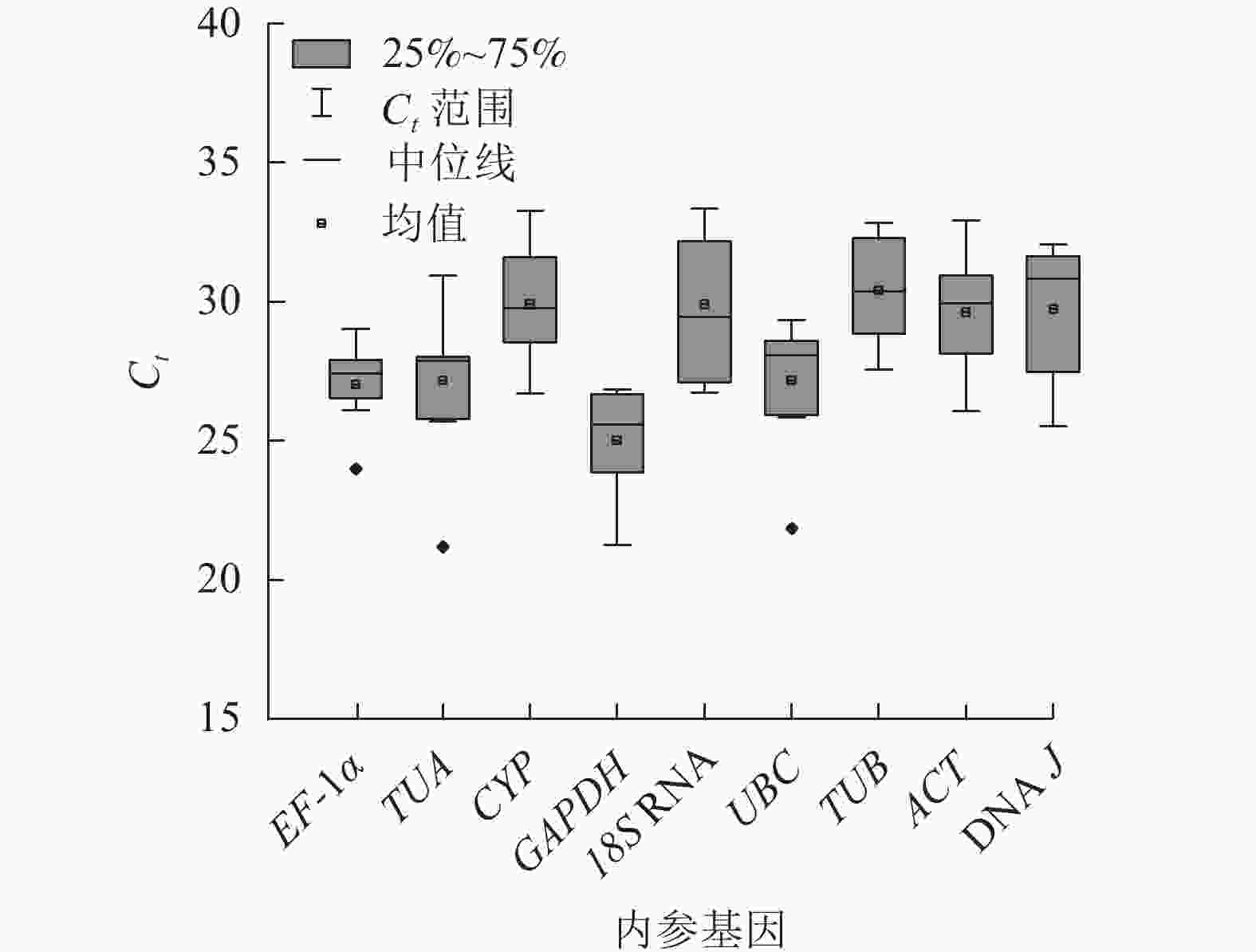
根据Ct表示不同条件下基因的相对表达量,Ct越小,表达丰度越高[18]。所有样品中,9个候选内参基因的Ct为21.12~33.74 (图2),其中平均表达量最高的基因为GAPDH,Ct平均值为25.01;18S RNA的Ct平均为29.90,其表达量最低。EF-1α基因的表达水平变化最小,Ct为23.97~29.06;18S RNA的Ct为26.64~33.47,表达变化最大。从Ct的大小和变化情况来看,GAPDH基因的表达相对稳定且丰度最高,可初步评价候选内参基因的表达水平和稳定性。
-
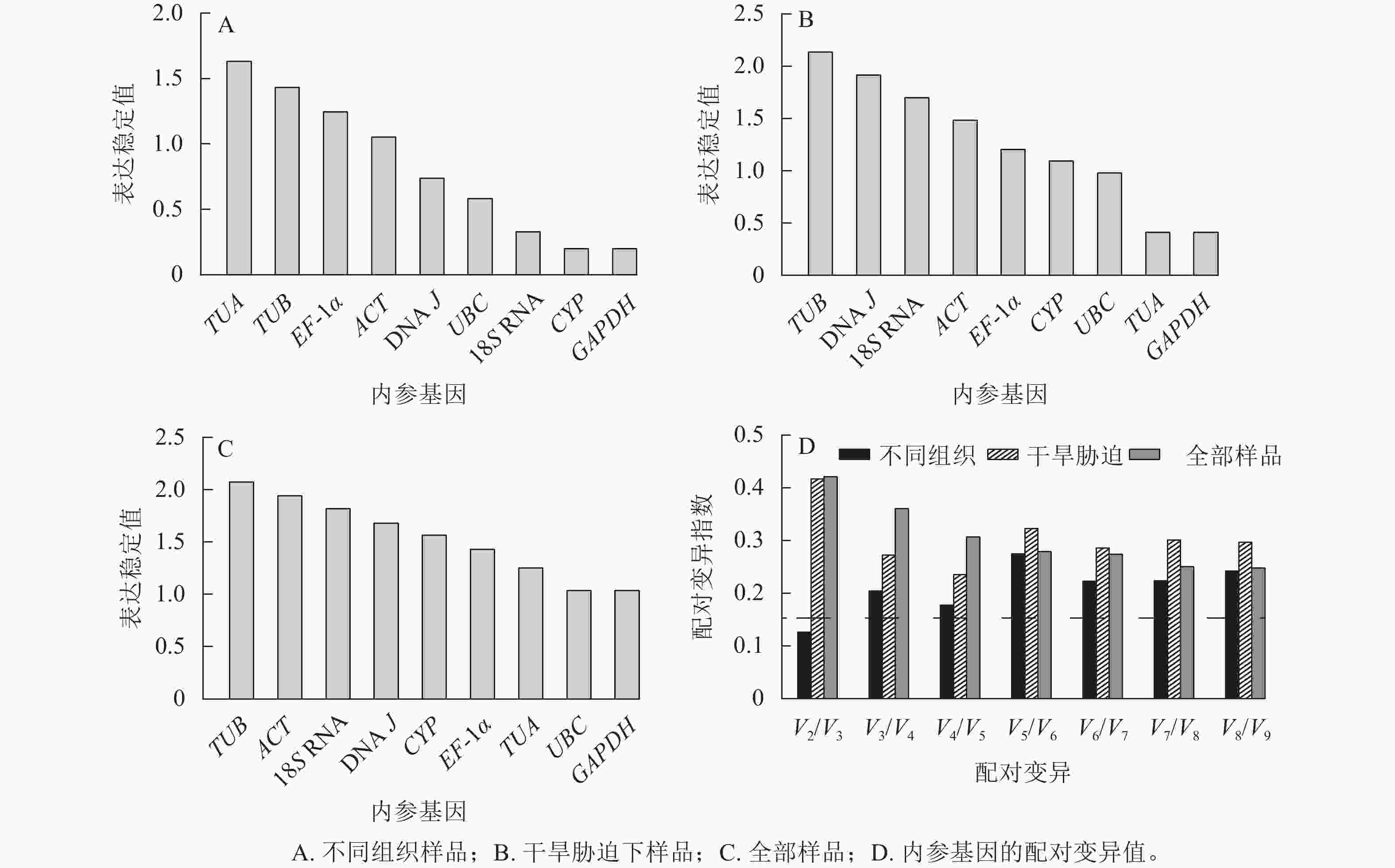
geNorm软件根据平均稳定值M来判定结果,M越小,基因表达越稳定,反之越不稳定[19]。使用2−ΔΔCt法计算内参基因的表达量:在黄薇不同组织中,候选内参基因的表达稳定性从高到低依次为CYP=GAPDH、18S RNA、UBC、DNA J、ACT、EF-1α、TUB、TUA (图3A)。在全部样品和干旱胁迫中,黄薇GAPDH和UBC基因最稳定,TUB基因最不稳定(图3B和图3C)。geNorm软件还可以根据成对变异值,分析最适的内参基因数目,软件默认的配对差异值(Vn/Vn+1,V为配对变异指数,n为可使RT-qPCR结果准确的最少基因数目)为0.15[20]。当Vn/Vn+1>0.15,需要加入第n+1基因;当Vn/Vn+1<0.15,不需要引入新的基因。黄薇在不同组织和干旱胁迫下(图3D),除不同组织中V2/V3<0.15外,其余情况均大于0.15。该现象可能是因为在不同组织和胁迫处理下,黄薇基因的表达量变化差异大,但软件说明书上也提出Vn/Vn+1最大值设定为0.15并非总是必须的[21]。
-
NormFinder软件按照2−ΔΔCt法计算稳定性,其中表达稳定值越小,候选内参基因越稳定[22]。结果表明(表2):在不同组织中,GAPDH和CYP基因表达最稳定,TUA基因最不稳定。在不同干旱胁迫处理下,GAPDH基因是最稳定的内参基因,TUB基因最不稳定。在黄薇的全部样品中,最稳定的内参基因是GAPDH基因,这与geNorm软件得出的结果相同。
表 2 NormFinder分析内参基因的表达稳定值
Table 2. Expression stability values of nine candidate reference genes calculated by the NormFinder
基因 不同组织 干旱胁迫 全部样品 表达稳定值 排序 表达稳定值 排序 表达稳定值 排序 GAPDH 0.069 2 0.142 1 0.359 1 UBC 0.593 5 0.452 3 0.578 2 EF-1α 0.904 6 0.800 5 0.862 3 TUA 1.483 9 0.239 2 0.910 4 CYP 0.069 1 0.611 4 0.911 5 DNA J 0.579 4 1.479 6 1.094 6 18S RNA 0.494 3 1.542 8 1.366 7 ACT 1.095 7 1.507 7 1.390 8 TUB 1.294 8 1.816 9 1.505 9 -
BestKeeper软件以内参基因的标准差(SD)和变异系数(CV)为基准,其数值越小越稳定[23]。从表3可见:在不同组织中,GAPDH基因的CV±SD为2.65 ± 0.69,排名第一,最稳定;在干旱胁迫和全部样品中,EF-1α基因的CV±SD分别为1.97±0.53和2.58±0.71,表达最稳定。综合不同条件下的分析结果,EF-1α基因被鉴定为最稳定的内参基因。
表 3 BestKeeper分析内参基因的表达稳定值
Table 3. Expression stability values of nine candidate reference genes calculated by the BestKeeper
基因 不同组织 干旱胁迫 全部样品 CV±SD 排序 CV±SD 排序 CV±SD 排序 EF-1α 3.46 ± 0.95 2 1.97 ± 0.53 1 2.58 ± 0.71 1 GAPDH 2.65 ± 0.69 1 3.78 ± 0.94 4 3.59 ± 0.91 2 UBC 3.73 ± 1.04 4 4.32 ± 1.18 5 4.21 ± 1.16 3 TUB 3.47 ± 1.06 3 6.14 ± 1.86 9 4.98 ± 1.51 4 TUA 6.65 ± 1.89 8 3.52 ± 0.95 3 5.26 ± 1.45 5 ACT 5.03 ± 1.46 5 6.11 ± 1.84 8 5.72 ± 1.69 6 CYP 6.36 ± 1.99 6 3.34 ± 0.96 2 6.11 ± 1.83 7 DNA J 7.31 ± 2.18 9 6.11 ± 1.81 7 6.81 ± 2.03 8 18S RNA 6.57 ± 2.05 7 5.68 ± 1.64 6 7.61 ± 2.28 9 -
上述3种软件的结果存在一定的差异,使用在线分析软件RefFinder,整合上述分析结果并进行综合分析。由表4显示:不同组织中,最稳定的内参基因是GAPDH和CYP,最不稳定的内参基因是TUA;在干旱胁迫下,最稳定的内参基因是GAPDH和TUA,最不稳定的内参基因是TUB。在全部样本中,最稳定的内参基因为GAPDH,最不稳定的内参基因为18S RNA。
表 4 RefFinder分析内参基因的表达稳定值
Table 4. Expression stability of candidate reference genes ranked by RefFinder
排序 不同组织 干旱胁迫 全部样品 基因 表达稳
定值基因 表达稳
定值基因 表达稳
定值1 GAPDH 1.41 GAPDH 1.19 GAPDH 1.19 2 CYP 2.11 TUA 1.86 UBC 2.11 3 18S RNA 3.57 EF-1α 3.34 EF-1α 2.45 4 UBC 4.76 UBC 3.41 TUA 4.28 5 TUB 4.76 CYP 4.00 CYP 5.23 6 EF-1α 5.05 ACT 6.70 DNA J 6.45 7 ACT 5.12 DNA J 6.96 ACT 6.73 8 DNA J 5.44 18S RNA 7.20 TUB 6.84 9 TUA 9.00 TUB 9.00 18S RNA 7.45 -
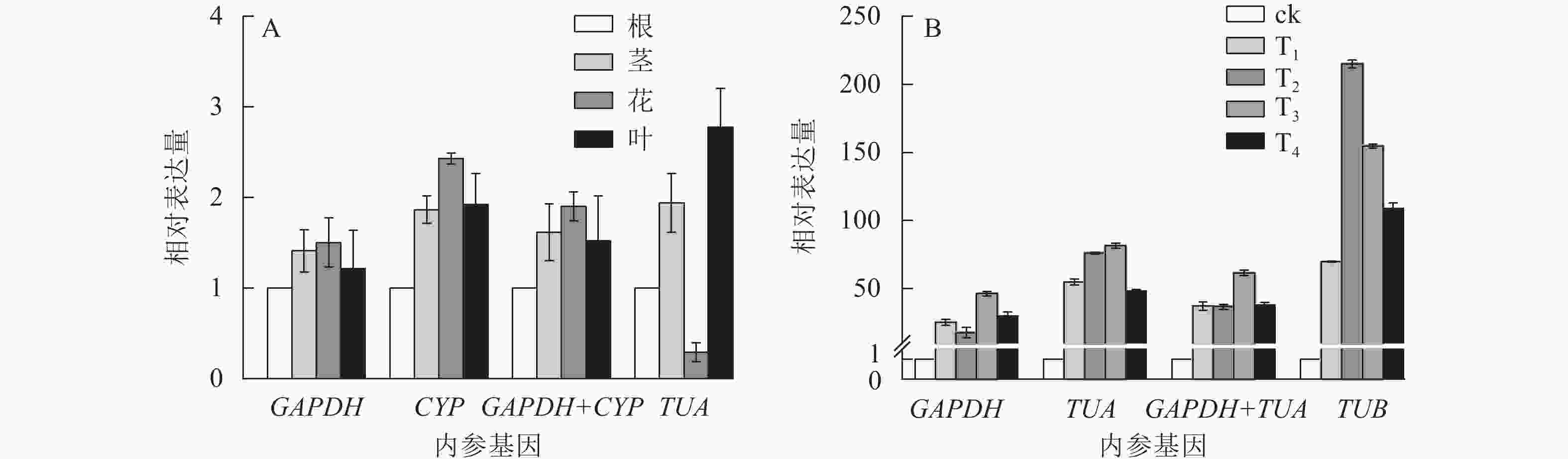
为验证候选内参基因的稳定性,采用RT-qPCR技术,根据筛选的稳定内参基因,对干旱胁迫下的靶基因CSLD和SOD的表达模式进行验证。CSLD基因是一种纤维素合成酶的结构蛋白,在茎中的表达量一般较高[24]。SOD基因属于抗氧化酶功能基因,在植物遭遇干旱胁迫时上调表达以抵御外界伤害[25]。从图4可见:在不同组织中,GAPDH和CYP基因在单独和组合校正下,CSLD均在茎中表达量最高,在其他组织中表达变化不大。但使用TUA作内参基因时,CSLD在茎中的表达量最低,在叶片中表达却最高。干旱胁迫下,使用GAPDH和基因组合GAPDH+TUA时,SOD基因在T3处理表达量最高,表达趋势大致相同;而利用TUB基因进行验证时,SOD基因在T2处理表达量最高,与其他结果差异较大。以上结果进一步验证了GAPDH基因作为内参基因的稳定性与可靠性。
-
RT-qPCR技术是一种分析基因表达量的研究工具,而使用相对定量法检测的可靠性,极大依赖内参基因的稳定性[26]。理想的内参基因在不同条件下,基因的表达变化较小,常用的内参基因一般都是维持植物生命活动必须的看家基因[27]。本研究基于黄薇转录组数据,筛选了9个常用内参基因(EF-1α、TUA、CYP、GAPDH、18S RNA、UBC、TUB、ACT、DNA J),其中,GAPDH在不同组织和干旱胁迫下都较稳定,最不稳定的是18S RNA。geNorm和NormFinder结果中,得出的最适内参基因组合为GAPDH + UBC,BestKeeper得出最佳基因为EF-1α,但GAPDH的稳定值排名综合也较靠前。3个筛选软件得出的结果基本一致,但也存在一定的差异,这可能是由于算法的不同所导致的[28]。与本研究结果不同的是,张海洋等[29]探究了菠菜Spinacia oleracea在不同胁迫处理下稳定的内参基因,3个软件的结果一致,在氯化钠和高温胁迫下,G6PD的表达稳定性最好,聚乙二醇胁迫下ELF1B的表达稳定性最好。王蕊等[30]研究了不同发育时期大豆Glycine max在不同组织、不同非生物胁迫下稳定表达的最适内参基因,4个软件在全部组织及全部胁迫中综合分析结果均一致,在全部组织中最适内参基因为ACT,全部胁迫中最适内参基因为EF-1α。
非生物胁迫会改变植物正常的代谢过程,筛选在胁迫处理下稳定表达的内参基因,有利于后续开展目的基因的鉴定,以便进一步了解目的基因的功能[31]。目前已有大量研究筛选不同非生物胁迫下的最佳内参基因。如紫鸭跖草Tradescantia pallida在不同浓度的铜离子胁迫下,最佳内参基因组合为18S RNA + TUB + UBI[32]。马铃薯Solanum tuberosum在干旱和渗透胁迫条件下,EF-1α和sec3基因表达最稳定[33]。上述结果与本研究的结果存在一定差异,说明不同植物在不同非生物胁迫下最适内参基因也在变化,特定条件下的最适内参基因仍需通过试验筛选。
Screening and validation of reference genes in Heimia myrtifolia in different tissues and under drought stress
-
摘要:
目的 为黄薇Heimia myrtifolia不同组织及不同干旱胁迫下基因表达分析筛选最适内参基因。 方法 选取黄薇盛花期的根、茎、叶、花,以及5种不同干旱处理的叶片作为实验材料,借助RT-qPCR技术对黄薇转录组数据筛选的9个候选内参基因进行分析,并利用软件geNorm、BestKeeper、NormFinder和RefFinder综合评价候选基因的表达稳定性。最后选取2个与胁迫相关的基因CSLD和SOD,对所选内参基因进行验证。 结果 geNorm、BestKeeper和NormFinder分析得出的候选内参基因排序存在一定差异。利用RefFinder对上述3个软件的结果综合分析得出:在不同组织中,最稳定的内参基因为GAPDH,最不稳定的内参基因为TUA;在干旱胁迫中,最稳定的内参基因为GAPDH,最不稳定的内参基因为TUB。在全部样本中,最稳定的内参基因为GAPDH,最不稳定的内参基因为18S RNA。对不同组织和干旱胁迫下的CSLD和SOD基因表达模式进行验证表明:上述2个基因与筛选所得内参基因的表达量和变化趋势均较为一致。 结论 在不同组织和干旱处理后,GAPDH是最适合黄薇基因表达的内参基因。图4表4参33 Abstract:Objective The objective is to study the gene expression of Heimia myrtifolia in different tissues and under different drought stress. Method The root, stem, leaf, and flower of H. myrtifolia at full flowering stage and 5 leaves under different drought treatments were selected as experimental materials, and 9 candidate internal reference genes screened from the transcriptome data were analyzed by RT-qPCR. The software geNorm, BestKeeper, NormFinder, and RefFinder were used to comprehensively evaluate the expression stability of candidate reference genes. Finally, 2 stress-related genes CSLD and SOD were selected to verify the selected internal reference genes. Result GeNorm, BestKeeper, and NormFinder showed some differences in the ranking of candidate internal parameters. By using the online software RefFinder to comprehensively analyze the results of the above 3 software, it was concluded that GAPDH was the most stable internal reference gene and TUA was the most unstable in different tissues. In drought stress, GAPDH was the most stable internal reference gene and TUB was the least stable. In all samples, the most stable reference gene was GAPDH, and the most unstable one was 18S RNA. The expression patterns of CSLD and SOD genes in different tissues and under drought stress were verified, which showed that the expression levels and change trends of the above 2 genes were consistent with those of the selected internal reference genes. Conclusion GAPDH is the best internal reference gene for gene expression of H. myrtifolia in different tissues and under drought treatment. [Ch, 4 fig. 4 tab. 33 ref.] -
表 1 黄薇候选内参基因引物信息
Table 1. Primer information of candidate reference gene in H. myrtifolia
基因 正向引物序列(5′→3′) 反向引物序列(5′→3′) 扩增大小/bp 退火温度/℃ 扩增效率/% R2 EF-1α TGGTTTTGAGGCTGGTATCTCC TTTGCTTGACACCAAGGGTGA 80 56.5 95.6 0.998 TUA TCTCTGCCTTGACCGAGTGA ACCACCAACGGCACTGAAAA 82 56.5 96.6 0.998 CYP ACCCCGACTCGTCCTACAAG TCGGTGTTCCGCTCCAAATG 130 58.0 106.8 0.999 GAPDH AGAAGGTCGTCATTTCTGCCC TGGTTGTGCAGCTAGCGTTG 114 57.5 108.4 0.999 18S RNA CAGGGCCTAGGATTTCGTCC GCCTTCAATCTTAGTCGTGGC 113 58.5 100.5 0.992 UBC GACCTGATGACACTCCCTGG TCACAGTTGGTGGTTTGTTCG 87 57.5 99.1 0.999 TUB GGGTGCTGAGCTTATTGATGC TGAGCAATGTCCCCATGCCT 131 57.5 96.8 0.996 ACT AGGGAATGCCTTTTGATTGATCC AAACATAAGCTCCACTGCCCTC 102 57 109.7 0.999 DNA J CGGAGCTATCACCCCGATG CGGCCTCACCATACCTGTCA 127 59.5 100.2 0.997 CSLD TACCTTGTCCCTTTCGGCG TCAGCGTCCTCATCCCGATA 149 57.4 95.1 0.997 SOD GTTGACGCAAGACGAGGGA CCGTTGGTCGTGTCACCAT 108 57.3 96.0 0.997 表 2 NormFinder分析内参基因的表达稳定值
Table 2. Expression stability values of nine candidate reference genes calculated by the NormFinder
基因 不同组织 干旱胁迫 全部样品 表达稳定值 排序 表达稳定值 排序 表达稳定值 排序 GAPDH 0.069 2 0.142 1 0.359 1 UBC 0.593 5 0.452 3 0.578 2 EF-1α 0.904 6 0.800 5 0.862 3 TUA 1.483 9 0.239 2 0.910 4 CYP 0.069 1 0.611 4 0.911 5 DNA J 0.579 4 1.479 6 1.094 6 18S RNA 0.494 3 1.542 8 1.366 7 ACT 1.095 7 1.507 7 1.390 8 TUB 1.294 8 1.816 9 1.505 9 表 3 BestKeeper分析内参基因的表达稳定值
Table 3. Expression stability values of nine candidate reference genes calculated by the BestKeeper
基因 不同组织 干旱胁迫 全部样品 CV±SD 排序 CV±SD 排序 CV±SD 排序 EF-1α 3.46 ± 0.95 2 1.97 ± 0.53 1 2.58 ± 0.71 1 GAPDH 2.65 ± 0.69 1 3.78 ± 0.94 4 3.59 ± 0.91 2 UBC 3.73 ± 1.04 4 4.32 ± 1.18 5 4.21 ± 1.16 3 TUB 3.47 ± 1.06 3 6.14 ± 1.86 9 4.98 ± 1.51 4 TUA 6.65 ± 1.89 8 3.52 ± 0.95 3 5.26 ± 1.45 5 ACT 5.03 ± 1.46 5 6.11 ± 1.84 8 5.72 ± 1.69 6 CYP 6.36 ± 1.99 6 3.34 ± 0.96 2 6.11 ± 1.83 7 DNA J 7.31 ± 2.18 9 6.11 ± 1.81 7 6.81 ± 2.03 8 18S RNA 6.57 ± 2.05 7 5.68 ± 1.64 6 7.61 ± 2.28 9 表 4 RefFinder分析内参基因的表达稳定值
Table 4. Expression stability of candidate reference genes ranked by RefFinder
排序 不同组织 干旱胁迫 全部样品 基因 表达稳
定值基因 表达稳
定值基因 表达稳
定值1 GAPDH 1.41 GAPDH 1.19 GAPDH 1.19 2 CYP 2.11 TUA 1.86 UBC 2.11 3 18S RNA 3.57 EF-1α 3.34 EF-1α 2.45 4 UBC 4.76 UBC 3.41 TUA 4.28 5 TUB 4.76 CYP 4.00 CYP 5.23 6 EF-1α 5.05 ACT 6.70 DNA J 6.45 7 ACT 5.12 DNA J 6.96 ACT 6.73 8 DNA J 5.44 18S RNA 7.20 TUB 6.84 9 TUA 9.00 TUB 9.00 18S RNA 7.45 -
[1] 方文培. 中国植物志: 第52卷第2分册[M]. 北京: 科学出版社, 1983. FANG Wenpei. Flora of China: Vol. 52, No. 2[M]. Beijing: Science Press, 1983. [2] LOURTEIG A. Legitimacy of Heimia myrtifolia Chamisso et Schlechtendal (Lythraceae) [J]. Taxon, 1989, 38(2): 279 − 280. [3] CLEMENS S. Frequent oligolecty characterizing a diverse bee-plant community in a xerophytic bushland of subtropical Brazil [J]. Studies on Neotropical Fauna and Environment, 1998, 33(1): 46 − 59. [4] AYOUB N, SINGAB A N, ELNAGGAR M, et al. Investigation of phenolic leaf extract of Heimia myrtifolia (Lythraceae): pharmacological properties (stimulation of mineralization of Saos-2 osteosarcoma cells) and identification of polyphenols [J]. Drug Discovery Today, 2010, 4(5): 341 − 348. [5] 林启芳, 刘婷婷, 刘洁茹, 等. 紫薇属与黄薇属植物花瓣类黄酮组成及含量分析[J]. 园艺学报, 2021, 48(10): 1956 − 1968. LIN Qifang, LIU Tingting, LIU Jieru, et al. Flavonoids composition and content in petals of Lagerstroemia and Heimia species and cultivars [J]. Acta Horticulturae Sinica, 2021, 48(10): 1956 − 1968. [6] 郑钢, 顾翠花, 王杰, 等. 干旱胁迫对黄薇光合特性和若干生理生化指标的影响[J]. 浙江农业学报, 2021, 33(9): 1650 − 1659. ZHENG Gang, GU Cuihua, WANG Jie, et al. Effects of drought stress on photosynthetic characteristics and several physiological and biochemical indexes of Heimia myrtifolia Cham. et Schlechtend [J]. Acta Agriculturae Zhejiangensis, 2021, 33(9): 1650 − 1659. [7] 顾帆, 季梦成, 顾翠花, 等. 高温干旱胁迫对黄薇抗氧化防御系统的影响[J]. 浙江农林大学学报, 2019, 36(5): 894 − 901. GU Fan, JI Mengcheng, GU Cuihua, et al. Heat and drought stress with an antioxidant defense system in Heimia myrtifolia [J]. Journal of Zhejiang A&F University, 2019, 36(5): 894 − 901. [8] 徐涛, 张柯岩, 顾翠花. 盐胁迫对黄薇若干生理生化指标的影响[J/OL]. 分子植物育种, 2022[2022-04-28]. https://kns.cnki.net/kcms/detail/46.1068.S.20220425.1310.008.html. XU Tao, ZHANG Keyan, GU Cuihua. Effects of salt stress on several physiological and biochemical indexes of Heimia myrtifolia [J/OL]. Molecular Plant Breeding, 2022-04-26[2022-04-28]. https://kns.cnki.net/kcms/detail/46.1068.S.20220425.1310.008.html. [9] GU Cuihua, DONG Bin, XU Liang, et al. The complete chloroplast genome of Heimia myrtifolia and comparative analysis within Myrtales [J/OL]. Molecules, 2018, 23(4): 846[2022-04-30]. doi:10.3390/molecules23040846. [10] WANG Hao, CAI Qizhong, LIU Lu, et al. Reference gene screening for real-time quantitative PCR in Polygonum multiflorum [J]. China Journal of Chinese Materia Medica, 2021, 46: 80 − 85. [11] LUO Meng, GAO Zhen, LI Hui, et al. Selection of reference genes for miRNA qRT-PCR under abiotic stress in grapevine [J/OL]. Scientific Reports, 2018, 8(1): 4444[2022-04-30]. doi:10.1038/s41598-018-22743-6. [12] FU Jianxin, WANG Yi, HUANG He, et al. Reference gene selection for RT-qPCR analysis of Chrysanthemum lavandulifolium during its flowering stages [J]. Molecular Breeding, 2013, 31(1): 205 − 215. [13] SUN Huapeng, LI Fang, RUAN Qinmei, et al. Identification and validation of reference genes for quantitative real-time PCR studies in Hedera helix L. [J]. Plant Physiology and Biochemistry, 2016, 108: 286 − 294. [14] KUMAR D, DAS P K, SARMAH B K. Reference gene validation for normalization of RT-qPCR assay associated with germination and survival of rice under hypoxic condition [J]. Journal of Applied Genetics, 2018, 59(4): 419 − 430. [15] VANDESOMPELE J, PRETER K D, PATTYN F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes [J/OL]. Genome Biology, 2002, 3(7): 0034.1[2022-04-28]. doi:10.1186/gb-2002-3-7-research0034. [16] ANDERSEN C L, JENSEN J L, ØRNTOFT T F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets [J]. Cancer Research, 2004, 64(15): 5245 − 5250. [17] PFAFFL M W, TICHOPAD A, PRGOMET C, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations [J]. Biotechnology Letters, 2004, 26(6): 509 − 515. [18] 崔运启, 朱再标, 郭巧生, 等. 垂盆草实时荧光定量PCR内参基因筛选[J/OL]. 分子植物育种, 2022-03-18[2022-04-30]. https://kns.cnki.net/kcms/detail/46.1068.S.20220317.1731.024.html. CUI Yunqi, ZHU Zaibiao, GUO Qiaosheng, et al. Screening of internal reference genes by quantitative real-time PCR in Sedum sarmentosum [J/OL]. Molecular Plant Breeding, 2022-03-18[2022-04-30]. https://kns.cnki.net/kcms/detail/46.1068.S.20220317.1731.024.html. [19] 干思宸, 师悦, 梁立军. 山麦冬果实花青素生物合成中内参基因的筛选与验证[J]. 浙江农林大学学报, 2022, 39(2): 307 − 317. GAN Sichen, SHI Yue, LIANG Lijun. Selection and validation of reference genes for anthocyanin biosynthesis in Liriope spicata fruits [J]. Journal of Zhejiang A&F University, 2022, 39(2): 307 − 317. [20] 李桥, 王淑安, 王鹏, 等. 铁线莲属萼片荧光定量PCR内参基因的筛选和评价[J/OL]. 分子植物育种, 2022-04-13[2022-04-30]. https://kns.cnki.net/kcms/detail/46.1068.s.20220410.2211.014.html. LI Qiao, WANG Shu’an, WANG Peng, et al. Selection and evaluation of reference genes for quantitative real-time PCR in sepals of different Clematis Varieties[J/OL]. Molecular Plant Breeding, 2022-04-13[2022-04-30]. https://kns.cnki.net/kcms/detail/46.1068.s.20220410.2211.014.html. [21] 章丽珍, 韩晓云, 吴菁华, 等. 甜瓜实时荧光定量PCR分析中内参基因的筛选[J]. 福建农业学报, 2020, 35(11): 1179 − 1187. ZHANG Lizhen, HAN Xiaoyun, WU Jinghua, et al. Reference gene selection for RT-qPCR analysis on Cucumis melo [J]. Fujian Journal of Agricultural Sciences, 2020, 35(11): 1179 − 1187. [22] 杨婷, 薛珍珍, 李娜, 等. 铁十字秋海棠斑叶发育过程内参基因筛选及验证[J]. 园艺学报, 2021, 48(11): 2251 − 2261. YANG Ting, XUE Zhenzhen, LI Na, et al. Reference genes selection and validation in Begonia masoniana leaves of different developmental stages [J]. Acta Horticulturae Sinica, 2021, 48(11): 2251 − 2261. [23] 钱猛, 杨娜, 朱昌华, 等. 绿豆实时荧光定量PCR内参基因的筛选与验证[J]. 植物生理学报, 2021, 57(11): 2203 − 2212. QIAN Meng, YANG Na, ZHU Changhua, et al. Selection and validation of reference genes for real-time fluorescence quantitative PCR in mung beans [J]. Plant Physiology Journal, 2021, 57(11): 2203 − 2212. [24] 奚航献. 铁皮石斛葡甘聚糖生物合成途径关键催化酶类纤维素合成酶CslD的挖掘与功能分析[D]. 杭州: 浙江农林大学, 2021. XI Hangxian. Discovery and Functional Analysis of Cellulose Synthase D, a Key Catalytic Enzyme in Glucomannan Biosynthesis Pathway in Dendrobium candidum [D]. Hangzhou: Zhejiang A&F University, 2021. [25] 朱冉冉, 吉雪花, 张中荣, 等. 辣椒超氧化物歧化酶基因家族的生物信息学分析[J]. 石河子大学学报(自然科学版), 2020, 38(6): 712 − 717. ZHU Ranran, JI Xuehua, ZHANG Zhongrong, et al. Bioinformatics analysis of Capsicum superoxide dismutase gene family [J]. Journal of Shihezi University (Natural Science), 2020, 38(6): 712 − 717. [26] ZHAO Zeying, ZHOU Hanwen, NIE Zhongnan, et al. Appropriate reference genes for RT-qPCR normalization in various organs of Anemone flaccida Fr. Schmidt at different growing stages [J/OL]. Genes, 2021, 12(3): 459[2022-04-25]. doi:10.3390/genes12030459. [27] TONG Zhaoguo, GAO Zhihong, WANG Fei, et al. Selection of reliable reference genes for gene expression studies in peach using real-time PCR [J/OL]. BMC Molecular Biology, 2009, 10(1): 71[2022-04-25]. doi:10.1186/1471-2199-10-71. [28] WARD D S, JUTTA D W, ROSWITHA W, et al. Reference gene validation for RT-qPCR, a note on different available software packages [J/OL]. PLoS One, 2015, 10(3): e0122515[2022-04-25]. doi: 10.1371/journal.pone.0122515. [29] 张海洋, 付娆, 李茹霞, 等. 菠菜非生物胁迫下实时荧光定量PCR分析中内参基因的选择[J]. 山东农业科学, 2020, 52(5): 21 − 25. ZHANG Haiyang, FU Rao, LI Ruxia, et al. Reference gene selection for real-time quantitative PCR in spinach treated with abiotic stresses [J]. Shandong Agricultural Sciences, 2020, 52(5): 21 − 25. [30] 王蕊, 胡绍旺, 刘金凤, 等. 大豆不同发育时期及非生物胁迫下实时荧光定量PCR内参基因筛选[J/OL]. 吉林农业大学学报, 2021-06-03[2022-04-30]. https://kns.cnki.net/kcms/detail/22.1100.S.20210602.1200.006.html. WANG Rui, HU Shaowang, LIU Jinfeng, et al. Screening of reference genes under abiotic stress and different development stages of soybean by real-time fluorescence quantitative PCR [J/OL]. Journal of Jilin Agricultural University, 2021-06-03[2022-04-30]. https://kns.cnki.net/kcms/detail/22.1100.S.20210602.1200.006.html. [31] HE Meijing, CUI Shunli, YANG Xinlei, et al. Selection of suitable reference genes for abiotic stress-responsive gene expression studies in peanut by real-time quantitative PCR [J]. Electronic Journal of Biotechnology, 2017, 28: 76 − 86. [32] 杨坤, 黄超, 卢山, 等. 铜胁迫下紫鸭跖草根组织实时定量PCR内参基因的选择[J]. 植物生理学报, 2021, 57(1): 195 − 204. YANG Kun, HUANG Chao, LU Shan, et al. Reference gene selection for quantitative real-time PCR in purple setcreasea (Setcreasea purpurea) root tissue under copper stress [J]. Plant Physiology Journal, 2021, 57(1): 195 − 204. [33] TANG Xun, ZHANG Ning, SI Huaijun, et al. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress [J/OL]. Plant Methods, 2017, 13(1): 85[2022-04-30]. doi:10.1186/s13007-017-0238-7. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.20220342






 下载:
下载: