-
甘油脂的从头生物合成途径(de novo glycerolipid biosynthesis)是细胞中最基本的代谢过程。3-磷酸甘油酰基转移酶(glycerol-3-phosphate acyltransferase, GPAT)催化甘油脂从头合成的初始步骤,生成的溶血磷脂酸(LPA)在LPA酰基转移酶(lysophosphatidic acid acyltransferase, LPAAT)的作用下转化为磷脂酸(PA)[1−6]。PA是调节真核生物中多种细胞过程的重要信号分子,也是极性甘油磷脂与中性三酰甘油(TAG)的生物合成前体。在磷酸酶的催化下PA转化为二酰甘油,后者可经脂酰辅酶A-依赖型二酰甘油酰基转移酶(acyl-CoA: diacylglycerol acyltransferease, DGAT)和(或)磷脂-依赖型二酰甘油酰基转移酶(phospholipid-dependent diacylglycerol acyltransferase, PDAT)作用生成TAG[7−10]。TAG合成能力是油料作物的关键性状,同时与人类肥胖症等疾病密切相关。因而,运用遗传或化学遗传方法操控TAG生物合成,提高油料作物含油量,降低与肥胖症相关联的人类疾病,具有重要实践意义[11−16]。
脂酰基转移酶在TAG生物合成过程中发挥重要作用,但目前对于这类酶的结构与功能的内在关系知之甚少,参与第1步酰化反应的GPAT亦不例外,仅有少量关于其结构与功能关系的报道[17−18]。已知GPAT、LPAAT、磷酸二羟丙酮酰基转移酶(dihydroxyacetone-phosphate acyltransferase, DHAPAT)等脂酰基转移酶均含4个高度保守的结构域,结构域Ⅰ的组氨酸(H)、天冬氨酸(D),结构域Ⅲ的甘氨酸(G)和结构域Ⅳ的脯氨酸(P)是GPAT催化所必需的;而结构域Ⅱ的精氨酸(R)与结构域Ⅲ的谷氨酸(E)在结合底物3-磷酸甘油中起作用[18−20]。迄今为止,对于保守结构域外的其他氨基酸残基在酰基转移酶活性调控中的作用,及与保守结构域中的氨基酸残基存在的潜在互作关系的了解非常有限。
GPAT9位于植物细胞的内质网,参与膜脂和TAG的生物合成,其功能缺失会导致种子发育异常、油脂合成减弱[20−22]。本实验室前期的酵母遗传互补研究也显示:油菜Brassica napus BnGPAT9的异源表达能够恢复酵母条件致死型双敲除突变体(ZAFU1)因GPAT酶活性缺失引起的生长缺陷。然而,拟南芥Arabidopsis thaliana AtGPAT9却不具备这种互补能力[23−25],尽管AtGPAT9与BnGPAT9的进化关系密切[26−27],两者氨基酸序列的一致性高达 94.1%,且两者在4个酰基转移酶保守结构域的氨基酸残基完全一致。因此,可以假设保守结构域之外的某些氨基酸残基对GPAT9的活性起着重要的调节作用。本研究充分利用AtGPAT9和BnGPAT9在酵母异源系统中表现出的不同性质,并结合定点突变与酵母遗传互补技术,剖析单个和多个氨基酸残基改变对GPAT9酶活性的影响,鉴定新的关键活性位点,以深化对脂酰基转移酶结构与功能内在关系的认知,为酰基转移酶的分子改造与结构优化、真核生物中TAG合成途径的改良以及全新TAG从头合成途径的构建提供理论基础。
-
通过Vector NTI 11.5.4软件对AtGPAT9 (Genebank 登录号: ACT32031.1)和BnGPAT9 (Genebank 登录号: ANV28166.1)的氨基酸序列进行比对。使用TMHMM 2.0和Protter进行跨膜结构域和蛋白质拓扑异构模型预测[28-29]。由I-TASSER预测三维结构[30]。
-
将AtGPAT9和BnGPAT9编码序列分别通过BamH I/XhoⅠ和BamH I/EcoR I双酶切位点克隆至pMD19-T载体,得到新的质粒pMD19-T-AtGPAT9和pMD19-T-BnGPAT9;以之为模板,对AtGPAT9和BnGPAT9进行定点突变。具体方法如下:利用包含突变位点的引物(表1),PCR扩增整个质粒;质粒DNA经Dpn I消化与纯化后,转入大肠埃希菌Escherichia coli并进行测序分析;序列正确的质粒经BamH I和Xho Ⅰ双酶切后,连接到经相同酶切处理的pYES2-yADH1-Kan V2酵母表达载体,并对产生的重组质粒再次进行DNA测序分析,以确保突变位点的正确性。需要说明的是,选择pMD19-T质粒作为定点突变过程中的中间质粒,而非直接在pYES2-yADH1-Kan V2质粒上进行GPAT9基因的定点突变,是因为后者DNA长度(6 998 bp)是前者(2 660 bp)的2.6倍,采取这样的策略可以降低因PCR扩增时间延长导致潜在的错误碱基出现频率。
表 1 拟南芥AtGPAT9和油菜BnGPAT9定点突变所用的引物序列
Table 1. Sequences of the primers used for site-directed mutagenesis of A. thaliana AtGPAT9 and B. napus BnGPAT9
突变位点 引物序列 (5′→3′) 突变位点 引物序列 (5′→3′) BnGPAT9(R40S) AGCCTCGTGGCAAGCTCAGCCTGCGTGATTTGCTAGACAT AtGPAT9(N119H) TTTCATTGTTTATCCCTGTACACGCGTTGCTGAAAGGTCAAG BnGPAT9(W85Y) TCTACTTGTTTCCTTTATACTGCTGTGGTGTTGTTGTTAG AtGPAT9(D230N) TTGTAGCAAAAAAGTTAAGGAACCATGTCCAAGGAGCTGAC BnGPAT9(C87F) TTGTTTCCTTTATGGTGCTTTGGTGTTGTTGTTAGATACT AtGPAT9(A235T) TAAGGGACCATGTCCAAGGAACTGACAGTAATCCTCTTCTC BnGPAT9(I102F) TTCTCTTTCCCTTGAGGTGCTTCACTTTAGCTTTTGGATG AtGPAT9(S237N) ACCATGTCCAAGGAGCTGACAATAATCCTCTTCTCATATTTCC BnGPAT9(F109I) CATCACTTTAGCTTTTGGATGGATTATTTTCCTTTCAACG AtGPAT9(D230N/A235T) TTGTAGCAAAAAAGTTAAGGAACCATGTCCAAGGAACTGAC BnGPAT9(T114L) TGGTTTATTTTCCTTTCATTGTTTATCCCTGTACACTCTC AtGPAT9(A235T/ S237N) TAAGGGACCATGTCCAAGGAACTGACAATAATCCTCTTCTCATATTTC BnGPAT9(H119N) TTCAACGTTTATCCCTGTAAATTCTCTCCTGAAAGGTCAG AtGPAT9(D230N/A235T/
S237N)TTGTAGCAAAAAAGTTAAGGAACCATGTCCAAGGAACTGAC BnGPAT9(N230D) GTAGCAAGAAAGTTAAGGGACCATGTTCAAGGAACTGACA AtGPAT9(G332A) CATAAGGCCCGGTGAAACAGCAATTGAATTTGCAGAGAGGG BnGPAT9(T235A) TAAGGAACCATGTTCAAGGAGCTGACAATAACCCTCTTCT AtGPAT9(L335H) GGTCAGAGACATGATATCTCATCGGGCGGGTCTCAAAAAGG BnGPAT9(N237S) CATGTTCAAGGAACTGACAGTAACCCTCTTCTTATATTTC AtGPAT9(P355S) TGAAGTATTCGAGACCAAGCTCCAAGCATAGTGAACGCAAG BnGPAT9(A322G) AAGGCCTGGTGAAACAGGAATTGAGTTTGCAGAGAGGGTC AtGPAT9(T10A) GTACGGCAGGGAGGCTCGTGGCTTCAAAATCCGAGCTTGAC AtGPAT9(S40R) ATGAACCTCGCGGCAAGCTCCGCCTGCGTGATTTGCTAGA AtGPAT9(S11A) CGGCAGGGAGGCTCGTGACTGCAAAATCCGAGCTTGACCTC AtGPAT9(Y85W) ATTTACTTATTCCCACTATGGTGCTTTGGGGTTGTTGTTAG AtGPAT9(S13A) GGAGGCTCGTGACTTCAAAAGCCGAGCTTGACCTCGATCAC AtGPAT9(F87C) CTTATTCCCACTATACTGCTGTGGGGTTGTTGTTAGATACT AtGPAT9(S28A) AACATCGAAGATTACCTTCCTGCTGGTTCTTCCATCAATGAAC AtGPAT9(F102I) TCCTCTTTCCCTTGAGGTGCATCACTTTAGCTTTTGGGTGG AtGPAT9(S30A) GAAGATTACCTTCCTTCTGGTGCTTCCATCAATGAACCTCGCG AtGPAT9(I109F) TCACTTTAGCTTTTGGGTGGTTTATTTTCCTTTCATTGTTT AtGPAT9(S31A) GATTACCTTCCTTCTGGTTCTGCCATCAATGAACCTCGCGGCA AtGPAT9(L114T) GGGTGGATTATTTTCCTTTCAACGTTTATCCCTGTAAATGCG -
条件致死型酵母双突变体ZAFU1[BY4742, gat1Δgat2Δ+(pGAL1::AtGPAT1 Leu2)] [25, 31],可在半乳糖的培养基上生长,但在葡萄糖培养基上丧失了生长能力。基于菌株ZAFU1建立的酵母遗传互补法对GPAT的鉴定具有很强的专一性[25, 31],本研究运用它鉴定不同氨基酸残基突变对GPAT9活性的影响。
使用基于醋酸锂的标准方法将重组酵母表达质粒导入菌株ZAFU1感受态细胞[25, 31],复苏4 h,分别涂布转化液于以葡萄糖(Glu)或半乳糖(Gal)为碳源,不含尿嘧啶、组氨酸和亮氨酸的培养基(SC-Ura-His-Leu)上, 30 ℃下培养3~5 d。为了准确地比较不同氨基酸残基突变对酶活性的影响,挑取在半乳糖培养基上生长的不同单菌落酵母进行浓度梯度稀释培养实验:从半乳糖培养基上随机挑选生长良好的单菌落至SC-Ura-His-Leu+Gal液体培养基,30 ℃振荡培养1~2 d至光密度[D(600)]为2.000 0~3.000 0,稀释菌液浓度至D(600)为1.000 0、0.200 0、0.040 0、0.008 0和0.001 6,取5 µL接种于SC-Ura-His-Leu+Glu和SC-Ura-His-Leu+Gal固体培养基上,30 ℃培养3~5 d。
-
30 ℃下,将表达不同GPAT9突变基因的ZAFU1细胞在SC-Ura-His-Leu+Gal液体培养基中培养至D(600)为3.000 0~4.000 0,稀释接种于SC-Ura-His-Leu+Glu液体培养基至D(600)为0.100 0,振荡培养并定时记录D(600)。
将平台生长期收获的细胞在真空冷冻干燥机中干燥,提取酵母总脂质[32],点样于硅胶板,通过薄层色谱法分离总脂质。喷洒质量浓度为0.05%樱草黄显色剂,在紫外灯下观察板上的脂质,从硅胶中提取TAG,并通过气相色谱法定量分析TAG含量[32]。
-
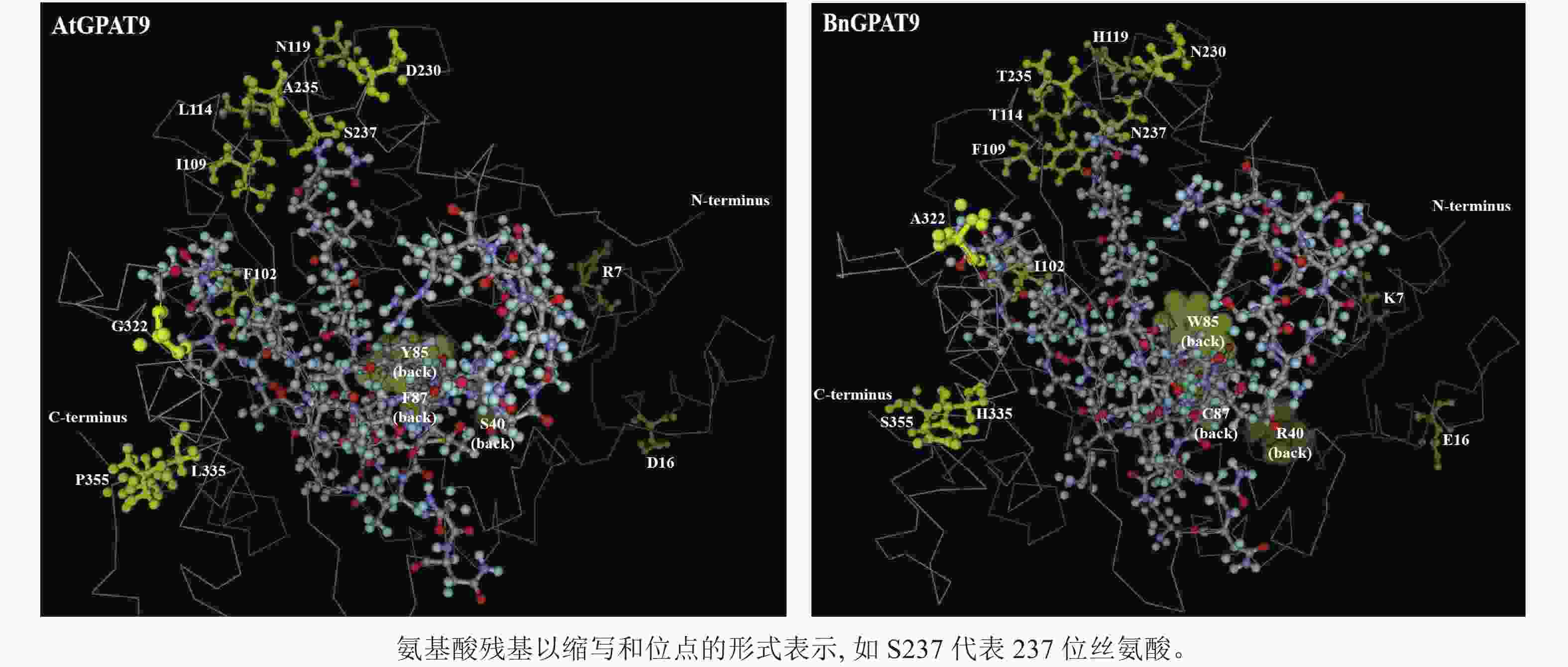
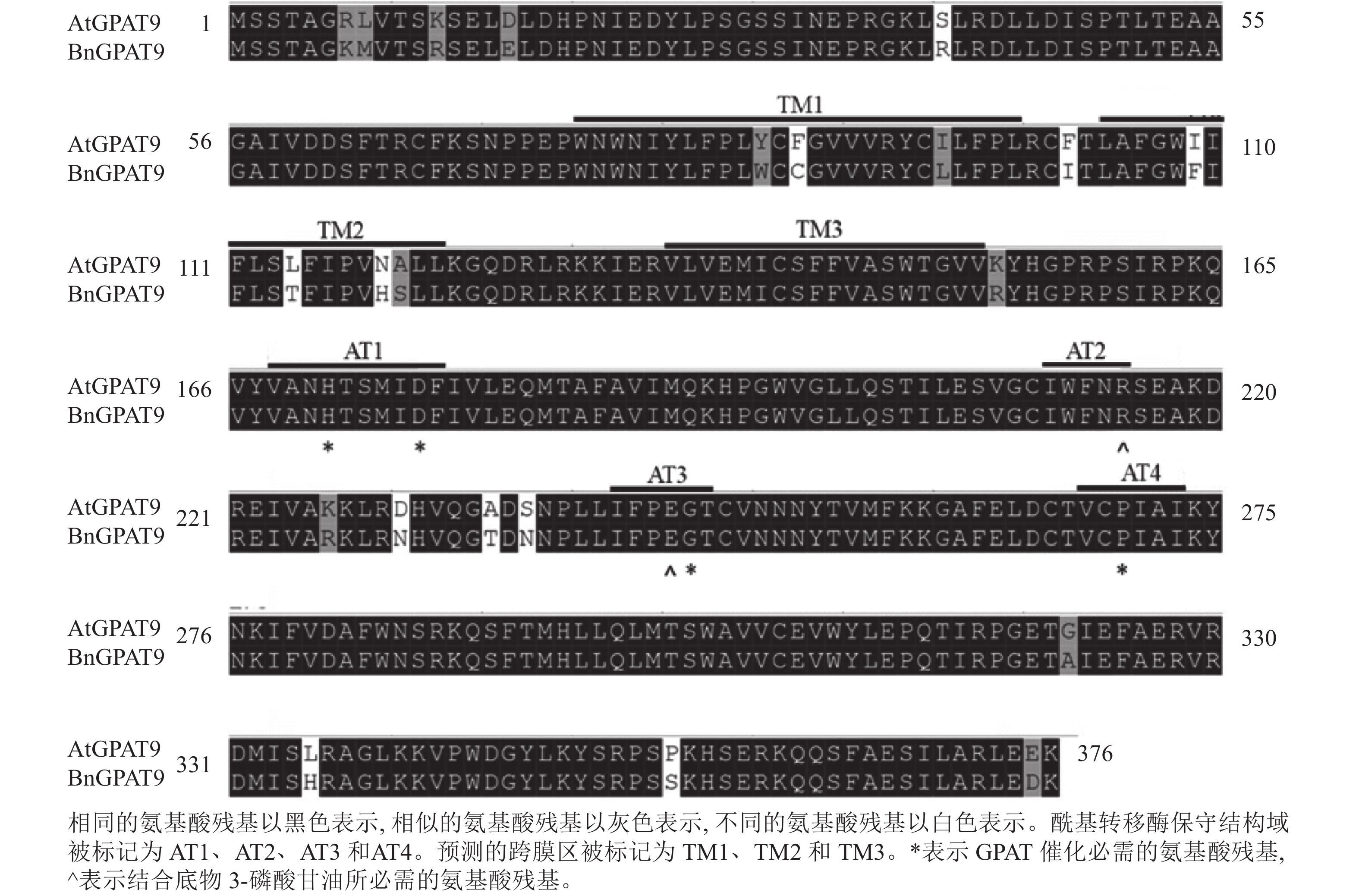
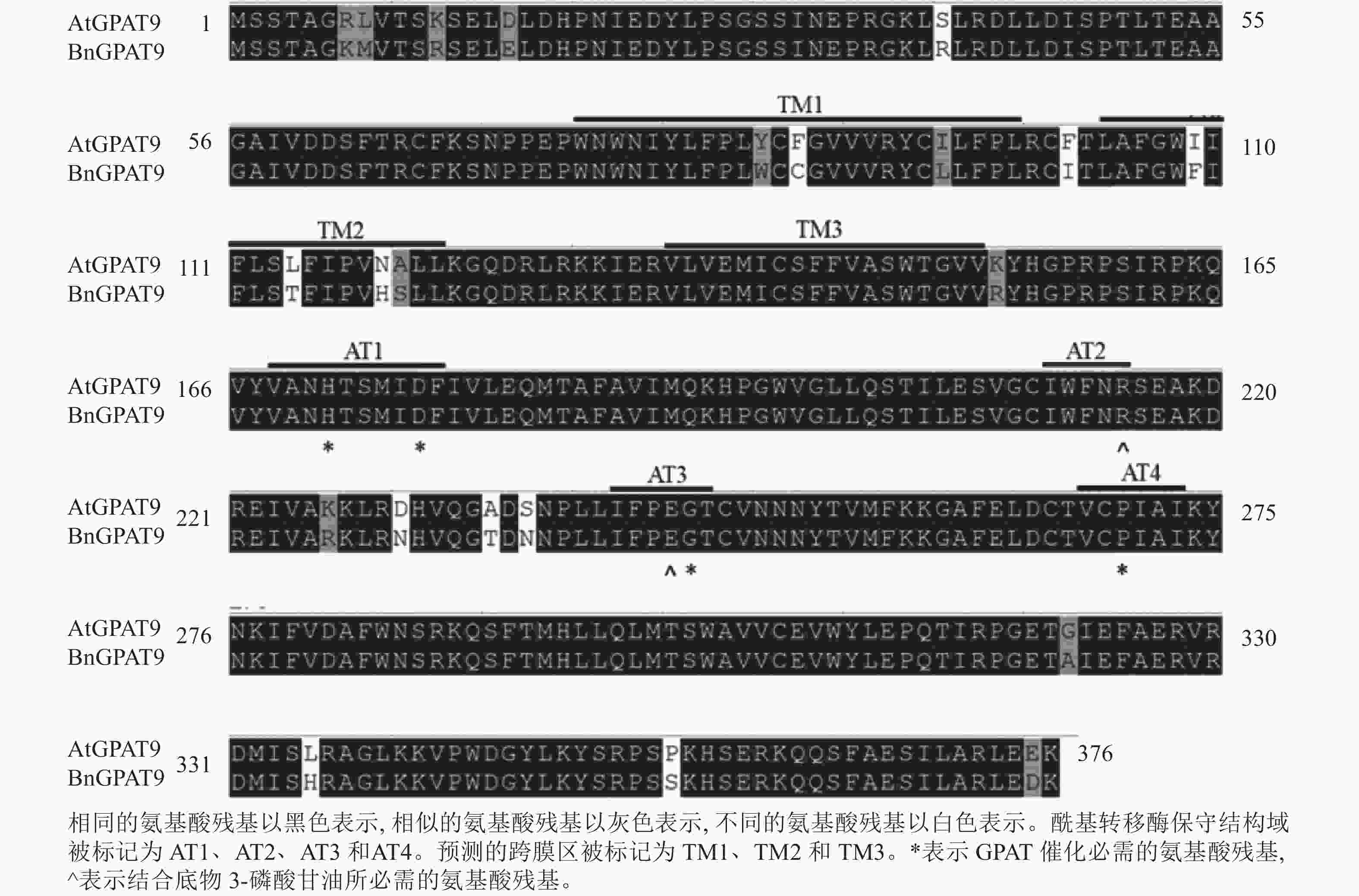
早期研究发现:在酵母中异源表达时,拟南芥AtGPAT9与油菜BnGPAT9表现出不同的活性[23-24]。为了找出潜在的关键活性调控位点,对AtGPAT9与BnGPAT9进行了序列比对。AtGPAT9和BnGPAT9均由376个氨基酸组成,两者序列一致性高达94.1%,4个保守的酰基转移酶结构域和C端内质网定位必需的疏水五肽结构域(−ILARL−)的氨基酸残基完全一致[18, 20],且在N端都含多个潜在的磷酸化位点,主要由丝氨酸和苏氨酸组成(图1)。它们之间共有22个不同的氨基酸残基,其中11个具有相似的性质。基于TMHMM和Protter的跨膜结构域预测显示:跨膜区结构具有较高相似性,但3个跨膜区域中有7个不同的氨基酸残基(图1)。另外,基于I-TASSER的三维结构预测发现:AtGPAT9和BnGPAT9在40、109、114、119、230、235、237和322位氨基酸残基的不同,可能会引起两者三维空间结构的差异,因此它们成为本研究重点剖析的位点(图2)。
-
为了明确哪些氨基酸位点对GPAT酶活性起着重要调控作用,据上述AtGPAT9和BnGPAT9中存在的氨基酸残基差异,运用定点突变技术对两者相应位置的氨基酸位点进行相互替代,即将AtGPAT9中的单个或多个氨基酸残基同时替换成与BnGPAT9中相应位置完全相同的氨基酸残基,反之亦然。本研究共构建了58种不同的GPAT9突变基因(表2)。
表 2 AtGPAT9和BnGPAT9中单和多位点氨基酸残基的定点突变
Table 2. Site-directed mutagenesis of amino acid residues at single and multiple sites in AtGPAT9 and BnGPAT9
单个氨基酸残基突变 AtGPAT9氨基酸残基突变组合 BnGPAT9 AtGPAT9 BnR40S** AtT10A AtF102I/S237N AtY85W/D230N BnW85Y AtS11A AtI109F/S237N AtY85W/A235T BnC87F** AtS13A AtD230N/A235T AtY85W/S237N** BnI102F** AtS28A AtD230N/S237N AtY85W/D230N/A235T BnF109I** AtS30A AtA235T/S237N AtY85W/D230N/S237N BnT114L*** AtS31A AtD230N/A235T/S237N AtY85W/A235T/S237N BnH119N AtS40R AtS237N/G322A AtY85W/D230N/A235T/S237N BnN230D*** AtY85W AtY85W/N119H*** AtS40R/Y85W/S237N** BnT235A** AtF87C AtY85W/L114T/N119H* AtN119H/D230N BnN237S* AtF102I AtY85W/N119H/S237N*** AtN119H/A235T BnA322G* AtI109F AtY85W/L114T/N119H/S237N*** AtN119H/S237N*** AtL114T AtY85W/N119H/D230N** AtN119H/D230N/A235T AtN119H* AtY85W/N119H/A235T** AtN119H/D230N/S237N** AtD230N AtN119H/A235T/S237N*** AtA235T AtN119H/D230N/A235T/S237N AtS237N AtG322A AtL335H AtP355S 说明:每种突变以物种的首字母缩写和突变前后的氨基酸残基缩写表示,如AtS40R/S237N代表AtGPAT9的40位由丝氨酸(S)变为精氨酸(R),237位由丝氨酸(S)变为天冬酰胺(N)。*代表基因的异源表达能够恢复酵母双突变体ZAFU1的生长缺陷;*数目代表恢复能力的大小,数目越多,能力越强。 -
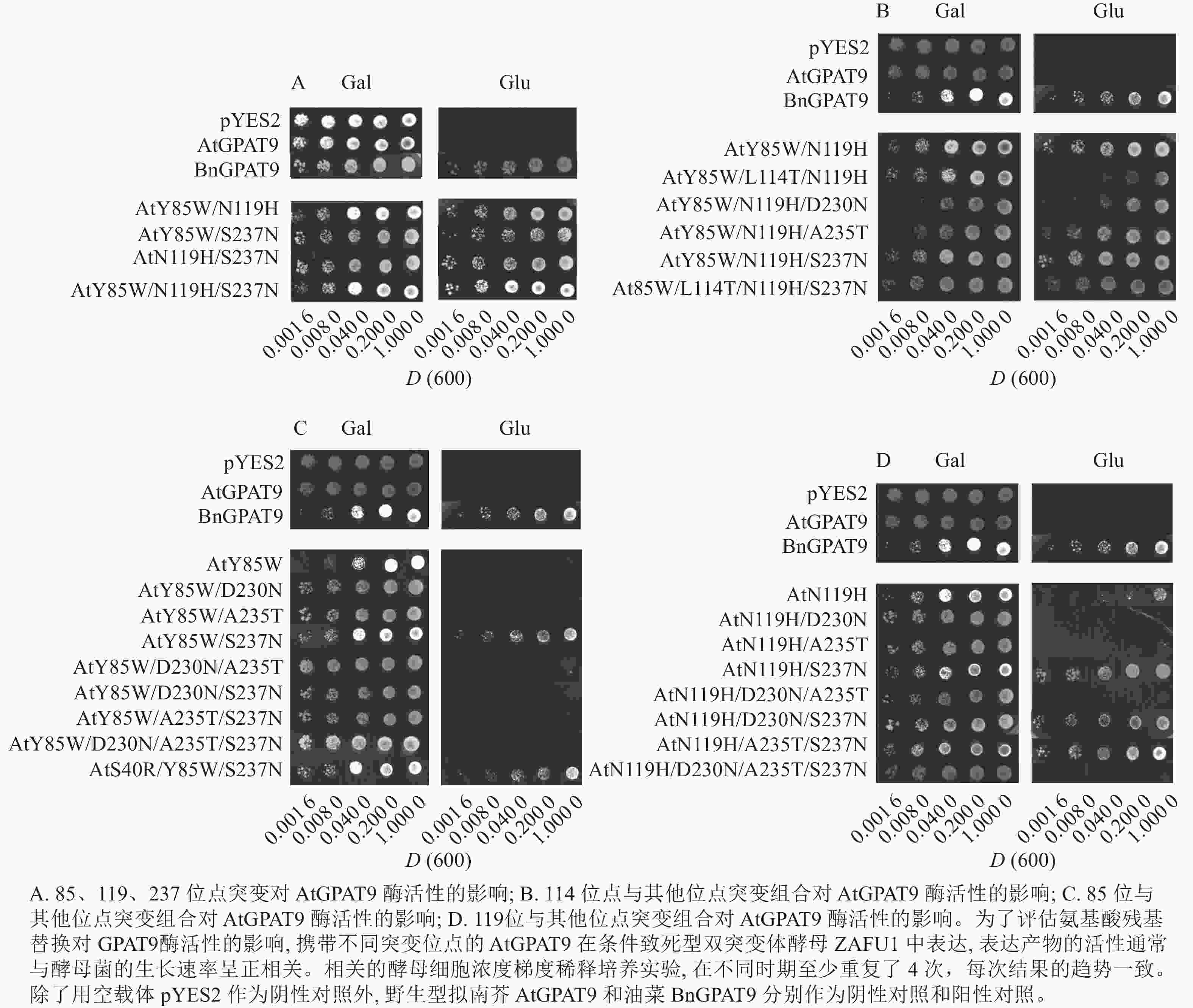
将不同GPAT9突变基因克隆至带有葡萄糖诱导启动子(ADH1)的质粒中,并以空载体与含野生型AtGPAT9的质粒为阴性对照,以含野生型BnGPAT9的质粒为阳性对照,将这些重组质粒导入到ZAFU1菌株中,测定不同GPAT9突变基因恢复ZAFU1在葡萄糖培养基上的生长缺陷能力。根据转化酵母细胞的生长速率,可以比较直观地评估不同突变位点对GPAT9酶活性的影响。结果显示:当ZAFU1突变体在葡萄糖培养基上培养时,W85Y或H119N的单位点替换导致BnGPAT9丧失对突变体生长缺陷的恢复能力(图3A),说明W85和H119是BnGPAT9正常功能所必需的。另外,与野生型BnGPAT9相比,含N237S或A322G突变位点的BnGPAT9对ZAFU1菌株生长的促进作用下降(图3A),表明这2个位点亦参与BnGPAT9活性的调节。相反,其他5个单位点替换(R40S、C87F、I102F、F109I、T235A)对BnGPAT9的活性不产生明显影响(图3A)。特别是N230D和T114L单位点替换增强了BnGPAT9活性,这种上调作用对于BnT114L而言尤为突出。如图4所示:与野生型BnGPAT9相比,含T114L突变位点的BnGPAT9在酵母突变体中的表达能促使细胞生长速率大幅度提高,表现为经2 d培养,表达BnGPAT9 (T114L)的菌落在葡萄糖培养基上生长的数量为野生型BnGPAT9的4倍,且每个单菌落表面积更大。
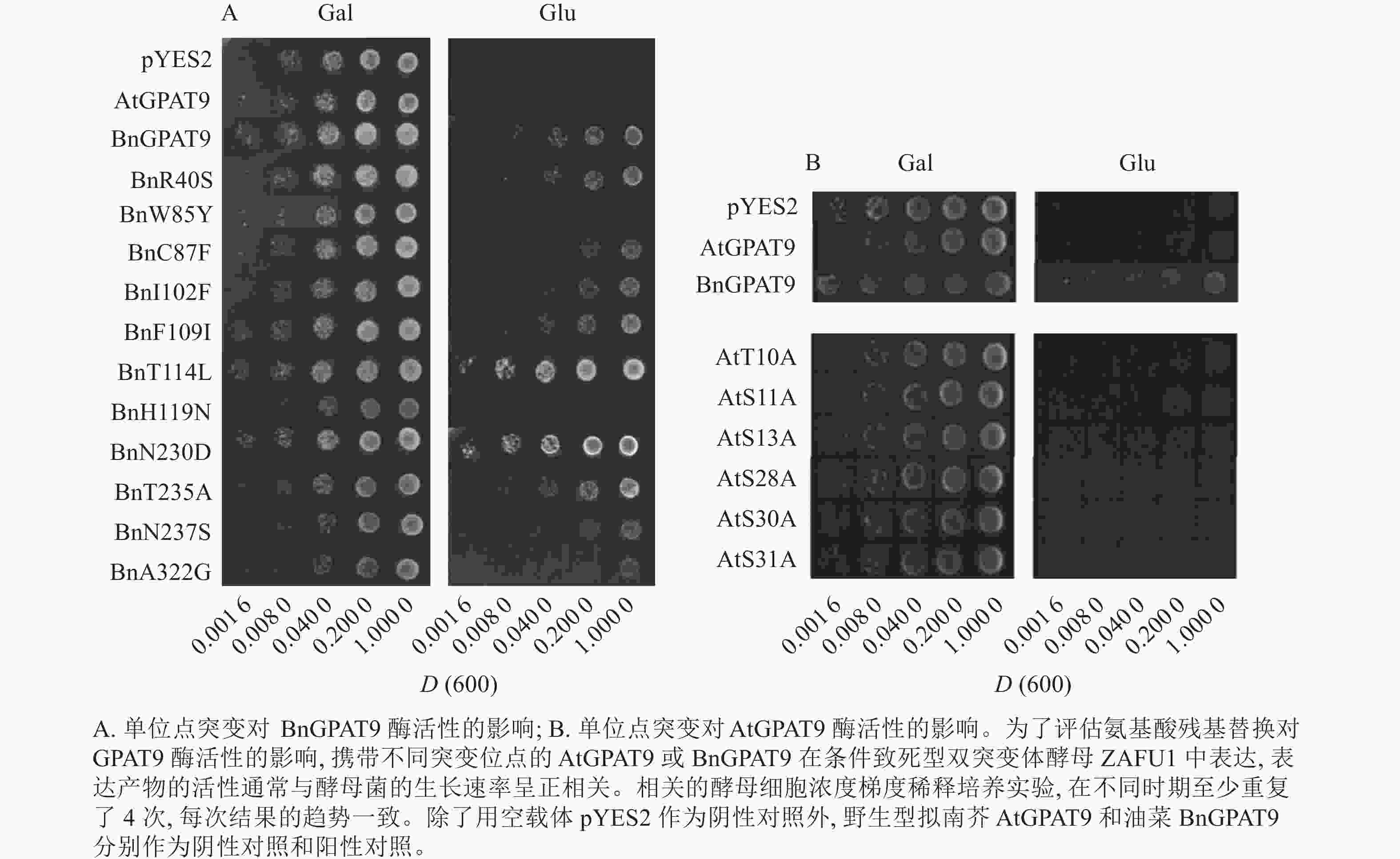
图 3 不同单位点突变对AtGPAT9和BnGPAT9酶活性的影响
Figure 3. Effects of different single mutations on AtGPAT9 and BnGPAT9 activities

图 4 亮氨酸替换114位的苏氨酸(T114L)对BnGPAT9活性的影响
Figure 4. Effects of the substitution of threonine for leucine at residue 114 (T114L) on BnGPAT9 activity
类似地,对19个不同的AtGPAT9突变基因进行了酵母遗传互补鉴定,其中的6个编码蛋白分别在N端的潜在磷酸化位点发生T10A、S11A、S13A、S28A、S30A和S31A 替换,另外13个分别发生了S40R、Y85W、F87C、F102I、I109F、L114T、N119H、D230N、A235T、S237N、G322A、L335H和P355S替换。结果显示:N端6个潜在磷酸化位点分别替换成中性氨基酸残基并不能改善AtGPAT9活性(图3B)。除了N119H,其他的单位点突变亦对AtGPAT9在酵母异源系统中的活性不产生可见影响(表2)。在酵母菌浓度梯度稀释培养实验中,N119H替换能使AtGPAT9恢复ZAFU1突变体在葡萄糖上的生长缺陷,但这种作用相对较弱(表2,图5D)。
-
基于相邻与非相邻氨基酸残基之间均可能对酶活性产生某种特定的互作效应的假设,根据AtGPAT9和BnGPAT9之间存在的氨基酸差异,进一步构建了28个含有2~4个氨基酸残基替换的AtGPAT9突变酶。与W85、H119和N237位点对BnGPAT9活性产生重要调节作用一致,同步替换AtGPAT9上2或3个相应位点的氨基酸残基(Y85W、N119H、S237N)均能大幅提高AtGPAT9酶活性,表现为携带Y85W/N119H、Y85W/S237N、N119H/S237N、Y85W/N119H/S237N突变的AtGPAT9均能互补菌株ZAFU1的生长缺陷(图5A);不过,单位点突变对AtGPAT9活性的影响十分有限(表2)。进一步调查发现:携带Y85W/N119H或Y85W/N119H/S237N突变组合的AtGPAT9比野生型BnGPAT9更能促进菌株ZAFU1的生长,且与携带T114L突变的BnGPAT9具有相近的作用效果(图3A,图5A)。
为了更好地剖析114位氨基酸性质对GAPT9活性的调节作用以及与其他氨基酸相互作用产生的效应,将双位点突变酶AtGPAT9 (Y85W/N119H)和三位点突变酶AtGPAT9 (Y85W/N119H/S237N)的114位亮氨酸(L114)替换为BnGPAT9相应位置存在的苏氨酸(T)。结果显示:生成的三位点突变酶AtGPAT9 (Y85W/L114T/N119H)的活性明显弱于AtGPAT9 (Y85W/N119H),但四位点突变酶AtGPAT9(Y85W/L114T/N119H/S237N)仍与AtGPAT9 (Y85W/N119H/S237N)的活性相当(图5B)。对这种现象的可能解释是,当2个潜在磷酸化位点即T114和S237同时出现于AtGPAT9 (Y85W/L114T/N119H) (其中含S237)或BnGPAT9 (N237S)(其中含T114)时,GPAT9的磷酸化程度可能加剧,从而抑制酶的活性。因此,推测植物GPAT9活性可能受磷酸化和非磷酸化机制调节,野生型GPAT9中存在的2个潜在磷酸化位点T114和S237可能对酰基转移酶活性产生负面效应。
进一步研究发现:230位氨基酸残基能与85、119位氨基酸残基产生互作效应而影响GPAT活性。虽然D230N本身或与A235T、S237N组合突变未能对AtGPAT9酶活性产生明显影响(表2),但当与N119H、Y85W/N119H或Y85W/S237N结合时, D230N突变能对AtGPAT9活性产生抑制作用。这是因为,与表达含N119H、Y85W/N119H或Y85W/S237N突变位点的酶的菌株相比,表达AtGPAT9 (N119H/D230N)、AtGPAT9 (Y85W/N119H/D230N)或AtGPAT9 (Y85W/D230N/S237N)的菌株ZAFU1在葡萄糖培养基上呈现生长速率明显减弱或不能生长的现象,暗示着D230N的替换不利于AtGPAT9活性(表2,图5B~D)。这一推测得到下述结果的支持,即N230D替换能提高BnGPAT9活性(图3A)。但是AtGPAT9(N119H/D230N/S237N)与AtGPAT9 (N119H/S237N)的活性相当,这说明D230N的负效应依赖于其他氨基酸的互作关系(表2,图5D)。
另外,N119H/D230N/S237N和N119H/A235T/S237N三突变组合均能提高AtGPAT9活性,使之具有拯救ZAFU1生长缺陷的能力,但在前者和后者中分别添加A235T和D230N得到的N119H/D230N/A235T/S237N四突变组合,能使相应蛋白丧失GPAT活性,如野生型AtGPAT9一样,无法恢复ZAFU1的生长缺陷(表2,图5D)。此外,无论是S237N和G322A单或双替换,均不能增强AtGPAT9在酵母中表达时的活性(表2),这与N237S或A322G的替换导致BnGPAT9活性下降现象不一致。由此可见,尽管在237位保留非磷酸化氨基酸(天冬酰胺,N)对BnGPAT9活性有利,但其效应取决于其他位置的氨基酸性质。
综上所述,GPAT9的酶活性受85、114、119、230、237和322位氨基酸残基性质的影响,它们之间存在互作效应;当这些位置的氨基酸残基分别为色氨酸(W)、亮氨酸(L)、组氨酸(H)、天冬氨酸(D)、天冬酰胺(N)和丙氨酸(A)时,酰基转移酶的活性较高。上述多位点突变增强AtGPT9酶活性的事实说明:运用分子设计能有效地改造和优化酰基转移酶的结构,并使之产生新的特性,这对将来人为操控甘油脂的从头合成十分有益。
-
为进一步探究氨基酸位点突变对GPAT9功能的影响,选取了3个活性程度不同的AtGPAT9突变酶,分别含N119H、Y85W/N119H 和Y85W/N119H/S237N突变,使其进行异源表达,测定其表达对酵母细胞中TAG合成的影响;阳性对照为野生型BnGPAT9。当相应酵母细胞的生长进入平台生长期时,提取总脂质,经薄层层析分离后,采用气相色谱法测定TAG含量。
需要指出的是,因不同突变酶活性的差异,相应酵母菌到达平台生长期所需的培养时间不一,且平台生长期时的细胞密度也不完全相同。譬如,表达BnGPAT9、AtGPAT9 (Y85W/N119H)和AtGPAT9 (Y85W/N119H/S237N)的酵母细胞在平台生长期时的细胞密度,以D(600)表示,分别为3.72、4.40、5.32 (图6)。总体而言,酶活性愈大,细胞生长越快、密度越高。
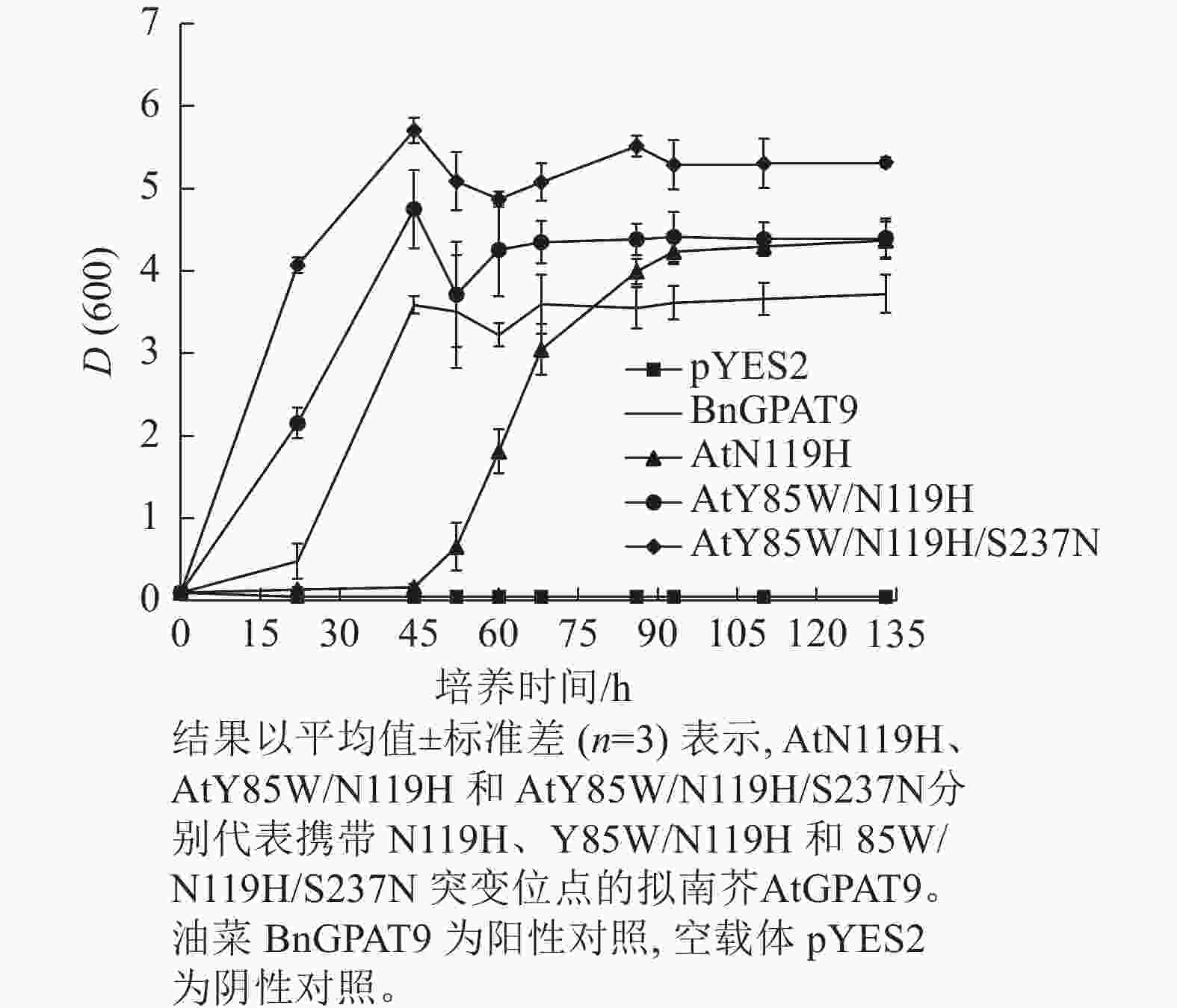
图 6 含不同突变位点的AtGPAT9表达对不同生长期酵母菌株ZAFU1的细胞密度影响
Figure 6. Effects of expression of AtGPAT9 bearing different mutation sites on cell density of the yeast strain ZAFU1 in different periods of growth
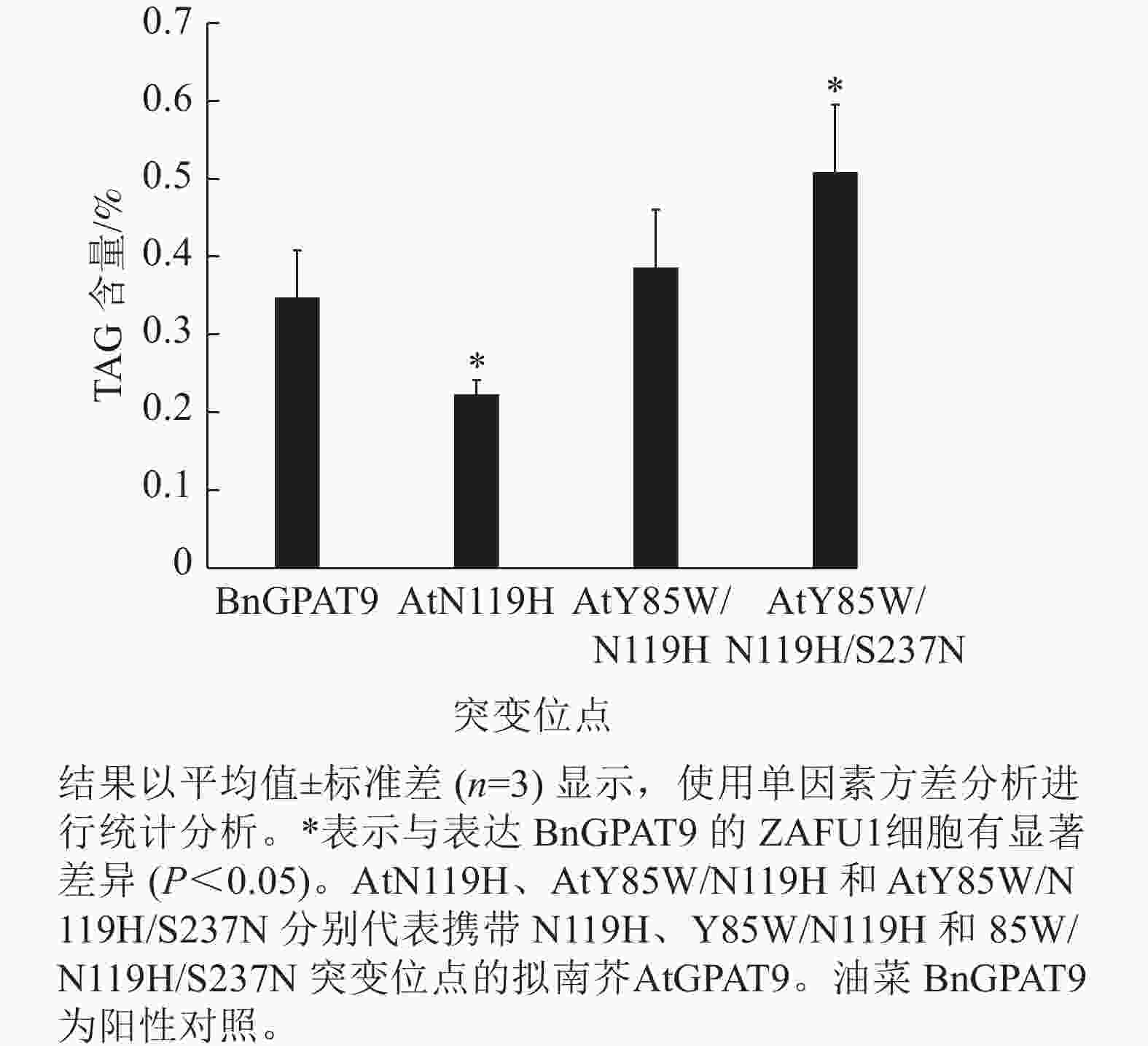
脂质分析显示:AtGPAT9 (Y85W/N119H/S237N)的表达使突变体酵母ZAFU1细胞中的TAG含量达0.51%,而BnGPAT9和AtGPAT9 (Y85W/N119H)的表达则分别产生0.35%和0.39%的含油量,前者比后两者分别增加了45.7%和 30.8% (图7)。与AtGPAT9 (N119H)具有相对较低的活性一致,表达此酶的酵母细胞,其TAG含量仅为0.22%,显著低于表达BnGPAT9的酵母细胞(0.35%)。以上结果表明:酵母细胞中TAG的合成能力受GPAT活性的调节,而GPAT活性大小与特定位置的氨基酸性质密切相关,因此通过分子设计优化GPAT9的氨基酸组成将有助于修饰细胞中TAG的合成能力。
-
提高油料作物油脂的合成对于保障食用油的供需平衡至关重要;相反,人类细胞中油脂合成能力的增强并非对健康有利,因为这会诱发肥胖症和心血管疾病等。目前人们试图运用遗传或化学遗传方法操控TAG的生物合成[11-16],但仍存在诸多因素影响,如人们对TAG合成途径中酶的结构与功能的内在关系知之甚少,这阻碍了酶结构的优化,并限制了基于翻译后修饰机制调控酶活性的技术开发。本研究充分利用GPAT专一的酵母遗传互补法[25, 31],鉴定控制植物GPAT9酶活性的关键氨基酸位点以及不同位点之间存在的互作效应,以深化对酰基转移酶结构与功能内在关系的认知,为将来运用合成生物学等手段有效操控真核生物中TAG的合成提供基础。
植物GPAT9与哺乳动物GPAT3结构相似,两者均参与极性膜脂和中性三酰甘油的生物合成[20-22]。尽管AtGPAT9和BnGPAT9序列相似性大于90%,它们在酵母异源表达时呈现的活性却相去甚远[23-24],这一特性有助于有效寻找调控酶活性的候选位点。在此基础上,本研究首次明确了酰基转移酶保守结构域外的6个氨基酸位点对植物GPAT酶活性的重要调节作用。
-
AtGPAT9的N端和C端均暴露于细胞质,意味着该蛋白应有偶数个跨膜结构域[20],然而这与生物信息学预测结果不一致,即AtGPAT9含3个潜在的跨膜结构域。对于这一现象的可能解释是,位于N端的几个脯氨酸残基可能会形成一个铰链状结构,使得疏水结构域Ⅰ不能跨膜,而是附着在内质网的表面[20]。基于85和119位氨基酸残基分别位于预测的第Ⅰ和Ⅱ个疏水结构域这一特点推测,将85位的疏水色氨酸替换成亲水的酪氨酸(Y)或将119位带正电荷的组氨酸替换成中性的天冬酰胺,可能会改变GPAT9在膜中的组装方式[33],这可能是构成AtGPAT9的酶活性低于BnGPAT9的原因之一。
-
本研究结果表明:尽管AtGPAT9的N端的6个磷酸化位点单独突变(T10A、S11A、S13A、S28A、S30A和 S31A)不能增强AtGPAT9在酵母异源表达时的活性,但T114L替换能增强BnGPAT9酶活性,而N237S替换则降低其活性。当114和237位氨基酸残基为非磷酸化氨基酸,即亮氨酸和天冬酰胺,而不是潜在的磷酸化位点苏氨酸(T)和丝氨酸(S)时,AtGPAT9和BnGPAT9突变酶保持较高活性。因此,推测GPAT9的活性受磷酸化机制调节,在114和237位点的磷酸化程度升高不利于维持酰基转移酶的活性。这种假设可以在某种程度上得到过去研究的支持。蛋白质磷酸化与非磷酸化修饰是酶活性的一种重要调节方式,已有研究报道哺乳动物线粒体GPAT (mtGPAT1)通过其C端S632和S639残基的磷酸化修饰调节其活性,酵母GPAT (Gat1p和Gat2p)的C端氨基酸残基发生磷酸化后也能使酶活性下调[34-35]。
-
当多个氨基酸残基同时突变时,它们之间的物理相互作用会导致蛋白质分子内的上位效应[36]。某些氨基酸残基突变组合可以产生协同作用,即产生正向上位效应,如Y85W、N119H和S237N任意突变组合均能增强AtGPAT9活性。相反,其他突变组合可能形成拮抗作用,导致负向上位效应,下调酶活性或彻底损伤蛋白质功能,正如AtGPAT9 (N119H/D230N/A235T/S237N)突变酶中4个氨基酸残基相互间的某种拮抗作用导致该突变酶在酵母ZAFU1中无法发挥功能。230、235和237位氨基酸残基位置相近,且位于酰基转移酶保守结构域Ⅱ中的精氨酸(R215)和Ⅲ中的谷氨酸(E245)之间;鉴于R215和E245这2个氨基酸残基对GPAT的底物结合至关重要[18],推测230、235和237位氨基酸残基与其他位点之间存在的复杂互作效应对酶活性的影响可能与其干扰3-磷酸甘油底物结合区域的三维结构有关[37]。
尽管N237S或A322G单位点突变均能降低BnGPAT9的活性,但无论是S237N和G322A单或双替换均不能增强AtGPAT9的活性,这从一个侧面说明237和322位氨基酸的作用均极大地受到其他氨基酸的理化性质影响。但需要指出的是,两者的作用方式可能不一。如前所述,237位氨基酸的磷酸化状态可能对酶活性产生某种调节作用,而322位的丙氨酸被甘氨酸取代可能会影响蛋白构象的稳定性,这是因为甘氨酸侧链小,仅有1个氢原子,这不利于α-螺旋结构的稳定。与此一致,三维空间结构预测显示:BnGPAT9中的A322与AtGPAT9中的G322相比,前者在空间上更靠近114、119、230、235、237位氨基酸残基(它们可能与酶活性中心形成有关),这可能对GPAT9活性产生正面效应。鉴于237与322位氨基酸的重要作用,将来有必要进一步探究在这2个氨基酸位点的何种替换有利于增强酰基转移酶的活性。
-
本研究首次报道了6个位于酰基转移酶保守区域外的GPAT9酶活性调控位点及其复杂的互作效应,从而深化了对酰基转移酶结构与功能内在关系的认知,为酰基转移酶的分子改造与结构优化提供了理论基础。另外,构建的GPAT9变异基因可用于探索植物中TAG的生物合成机制,特别是磷酸化-非磷酸化调控机制对GPAT酶活性的调节作用。
Identification of key amino acid residues controlling the activities of glycerol-3-phosphate acyltransferases in Arabidopsis thaliana and Brassica napus
-
摘要:
目的 3-磷酸甘油酰基转移酶(GPAT)催化三酰甘油(TAG)生物合成途径中的第1步酰化反应,TAG合成能力是油料作物的关键性状,但亦与人类肥胖症密切相关,了解GPAT结构与功能的内在关系对于这一性状的遗传或化学遗传调控至关重要。本研究旨在鉴定调控植物GPAT活性的关键氨基酸位点。 方法 运用定点突变技术构建了58个GPAT9突变基因,结合GPAT特异的酵母遗传互补法,剖析单一和多个氨基酸位点改变对GPAT9酶活性的影响。 结果 通过对拟南芥Arabidopsis thaliana AtGPAT9和油菜Brassica napus BnGPAT9的19个氨基酸残基进行分析发现:AtGPAT9的N端6个磷酸化位点的单独突变(T10A、S11A、S13A、S28A、S30A和S31A)不能增强AtGPAT9在酵母异源表达时的活性。相反,其他6个位于酰基转移酶保守结构域外的氨基酸残基(85、114、119、230、237、322位)的改变能够显著影响GPAT9酶活性。发现这些氨基酸残基之间存在交互作用,例如,3个位点同时突变(Y85W/N119H/S237N)能使AtGPAT9活性大幅上升,加速酵母的生长并促进TAG的合成,表达这一突变酶的酵母中的TAG含量比表达野生型BnGPAT9的增加了45.7%。更值得注意的是,在114和237位磷酸化氨基酸残基对酰基转移酶活性产生负面效应,暗示植物GPAT9活性可能受磷酸化和非磷酸化机制调节。 结论 本研究获得了6个未经报道的关键GPAT酶活性调控位点,其中W85和H119是GPAT9正常功能所必需的,而L114、D230、N237和A322有利于维持GPAT9活性。图7表2参37 Abstract:Objective Glycerol-3-phosphate acyltransferase (GPAT) catalyzes the first acylation reaction in the triacylglycerol (TAG) biosynthetic pathway. Understanding the structure-function relationship of GPAT is important for genetic or chemogenetic manipulation of the TAG biosynthetic capacity as a key trait in oilseed crops but associated with human obesity disease. This study aimed to identify key amino acid residues controlling the activity of GPAT9 in plants. Method Site-directed mutagenesis was employed to construct 58 GPAT9 mutant genes, and yeast genetic complementation specific for GPAT enzyme was utilized to dissect the effects of alterations of single and multiple amino acid residues on the activities of GPAT9 from Arabidopsis thaliana and Brassica napus. Result By analyzing 19 amino acid residues of AtGPAT9 and BnGPAT9, it was found that single mutations (T10A, S11A, S13A, S28A, S30A, S31A) at the N-terminus of AtGPAT9 could not enhance its activity when the respective variants were expressed heterologously. In contrast, the alteration of the other six amino acid residues at positions 85, 114, 119, 230, 237, and 322, respectively, which are located outside the conserved domains of acyltransferases significantly affected GPAT9 enzymatic activity. Additionally, mutual interactions were evident between these amino acids. For instance, the simultaneous mutation of the three residues (Y85W, N119H, S237N) greatly increased the activity of AtGPAT9, as exemplified by the findings that the expression of the corresponding mutant enzyme could accelerate the growth of yeast cells and enhanced the synthesis of TAG by 45.7%, compared with that of yeast cells expressing BnGPAT9. Noticeably, the presence of potentially phosphorylated amino acids at positions 114 and 237 was detrimental to acyltransferase activity, implying that plant GPAT9 activity may be regulated through protein phosphorylation and non-phosphorylation. Conclusion This study describes six previously unreported amino acid residues key to the regulation of GPAT enzymatic activity. Among them, W85 and H119 are prerequisite for the proper functioning of GPAT9, and L114, D230, N237, and A322 are beneficial to maintaining the activity of GPAT9. [Ch, 7 fig. 2 tab. 37 ref.] -
表 1 拟南芥AtGPAT9和油菜BnGPAT9定点突变所用的引物序列
Table 1. Sequences of the primers used for site-directed mutagenesis of A. thaliana AtGPAT9 and B. napus BnGPAT9
突变位点 引物序列 (5′→3′) 突变位点 引物序列 (5′→3′) BnGPAT9(R40S) AGCCTCGTGGCAAGCTCAGCCTGCGTGATTTGCTAGACAT AtGPAT9(N119H) TTTCATTGTTTATCCCTGTACACGCGTTGCTGAAAGGTCAAG BnGPAT9(W85Y) TCTACTTGTTTCCTTTATACTGCTGTGGTGTTGTTGTTAG AtGPAT9(D230N) TTGTAGCAAAAAAGTTAAGGAACCATGTCCAAGGAGCTGAC BnGPAT9(C87F) TTGTTTCCTTTATGGTGCTTTGGTGTTGTTGTTAGATACT AtGPAT9(A235T) TAAGGGACCATGTCCAAGGAACTGACAGTAATCCTCTTCTC BnGPAT9(I102F) TTCTCTTTCCCTTGAGGTGCTTCACTTTAGCTTTTGGATG AtGPAT9(S237N) ACCATGTCCAAGGAGCTGACAATAATCCTCTTCTCATATTTCC BnGPAT9(F109I) CATCACTTTAGCTTTTGGATGGATTATTTTCCTTTCAACG AtGPAT9(D230N/A235T) TTGTAGCAAAAAAGTTAAGGAACCATGTCCAAGGAACTGAC BnGPAT9(T114L) TGGTTTATTTTCCTTTCATTGTTTATCCCTGTACACTCTC AtGPAT9(A235T/ S237N) TAAGGGACCATGTCCAAGGAACTGACAATAATCCTCTTCTCATATTTC BnGPAT9(H119N) TTCAACGTTTATCCCTGTAAATTCTCTCCTGAAAGGTCAG AtGPAT9(D230N/A235T/
S237N)TTGTAGCAAAAAAGTTAAGGAACCATGTCCAAGGAACTGAC BnGPAT9(N230D) GTAGCAAGAAAGTTAAGGGACCATGTTCAAGGAACTGACA AtGPAT9(G332A) CATAAGGCCCGGTGAAACAGCAATTGAATTTGCAGAGAGGG BnGPAT9(T235A) TAAGGAACCATGTTCAAGGAGCTGACAATAACCCTCTTCT AtGPAT9(L335H) GGTCAGAGACATGATATCTCATCGGGCGGGTCTCAAAAAGG BnGPAT9(N237S) CATGTTCAAGGAACTGACAGTAACCCTCTTCTTATATTTC AtGPAT9(P355S) TGAAGTATTCGAGACCAAGCTCCAAGCATAGTGAACGCAAG BnGPAT9(A322G) AAGGCCTGGTGAAACAGGAATTGAGTTTGCAGAGAGGGTC AtGPAT9(T10A) GTACGGCAGGGAGGCTCGTGGCTTCAAAATCCGAGCTTGAC AtGPAT9(S40R) ATGAACCTCGCGGCAAGCTCCGCCTGCGTGATTTGCTAGA AtGPAT9(S11A) CGGCAGGGAGGCTCGTGACTGCAAAATCCGAGCTTGACCTC AtGPAT9(Y85W) ATTTACTTATTCCCACTATGGTGCTTTGGGGTTGTTGTTAG AtGPAT9(S13A) GGAGGCTCGTGACTTCAAAAGCCGAGCTTGACCTCGATCAC AtGPAT9(F87C) CTTATTCCCACTATACTGCTGTGGGGTTGTTGTTAGATACT AtGPAT9(S28A) AACATCGAAGATTACCTTCCTGCTGGTTCTTCCATCAATGAAC AtGPAT9(F102I) TCCTCTTTCCCTTGAGGTGCATCACTTTAGCTTTTGGGTGG AtGPAT9(S30A) GAAGATTACCTTCCTTCTGGTGCTTCCATCAATGAACCTCGCG AtGPAT9(I109F) TCACTTTAGCTTTTGGGTGGTTTATTTTCCTTTCATTGTTT AtGPAT9(S31A) GATTACCTTCCTTCTGGTTCTGCCATCAATGAACCTCGCGGCA AtGPAT9(L114T) GGGTGGATTATTTTCCTTTCAACGTTTATCCCTGTAAATGCG 表 2 AtGPAT9和BnGPAT9中单和多位点氨基酸残基的定点突变
Table 2. Site-directed mutagenesis of amino acid residues at single and multiple sites in AtGPAT9 and BnGPAT9
单个氨基酸残基突变 AtGPAT9氨基酸残基突变组合 BnGPAT9 AtGPAT9 BnR40S** AtT10A AtF102I/S237N AtY85W/D230N BnW85Y AtS11A AtI109F/S237N AtY85W/A235T BnC87F** AtS13A AtD230N/A235T AtY85W/S237N** BnI102F** AtS28A AtD230N/S237N AtY85W/D230N/A235T BnF109I** AtS30A AtA235T/S237N AtY85W/D230N/S237N BnT114L*** AtS31A AtD230N/A235T/S237N AtY85W/A235T/S237N BnH119N AtS40R AtS237N/G322A AtY85W/D230N/A235T/S237N BnN230D*** AtY85W AtY85W/N119H*** AtS40R/Y85W/S237N** BnT235A** AtF87C AtY85W/L114T/N119H* AtN119H/D230N BnN237S* AtF102I AtY85W/N119H/S237N*** AtN119H/A235T BnA322G* AtI109F AtY85W/L114T/N119H/S237N*** AtN119H/S237N*** AtL114T AtY85W/N119H/D230N** AtN119H/D230N/A235T AtN119H* AtY85W/N119H/A235T** AtN119H/D230N/S237N** AtD230N AtN119H/A235T/S237N*** AtA235T AtN119H/D230N/A235T/S237N AtS237N AtG322A AtL335H AtP355S 说明:每种突变以物种的首字母缩写和突变前后的氨基酸残基缩写表示,如AtS40R/S237N代表AtGPAT9的40位由丝氨酸(S)变为精氨酸(R),237位由丝氨酸(S)变为天冬酰胺(N)。*代表基因的异源表达能够恢复酵母双突变体ZAFU1的生长缺陷;*数目代表恢复能力的大小,数目越多,能力越强。 -
[1] ATHENSTAEDT K, DAUM G. Phosphatidic acid, a key intermediate in lipid metabolism [J]. European Journal of Biochemistry, 1999, 266(1): 1 − 16. [2] MURATA N, TASAKA Y. Glycerol-3-phosphate acyltransferase in plants [J]. Biochimica et Biophysica Acta, 1997, 1348(1/2): 10 − 16. [3] GIMENO R E, CAO Jingsong. Thematic review series: glycerolipids. mammalian glycerol-3-phosphate acyltransferases: new genes for an old activity [J]. Journal of Lipid Research, 2008, 49(10): 2079 − 2088. [4] WENDEL A A, LEWIN T M, COLEMAN R A. Glycerol-3-phosphate acyltransferases: rate limiting enzymes of triacylglycerol biosynthesis [J]. Biochimica et Biophysica Acta, 2009, 1791(6): 501 − 506. [5] ZHENG Zhifu, ZOU Jitao. The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae [J]. The Journal of Biological Chemistry, 2001, 276(45): 41710 − 41716. [6] ZHENG Zhifu, XIA Qun, DAUK M, et al. Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility [J]. The Plant Cell, 2003, 15(8): 1872 − 1887. [7] ZOU Jitao, WEI Yangdou, JAKO C, et al. The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene [J]. The Plant Journal, 1999, 19(6): 645 − 653. [8] DAHLQVIST A, STAHL U, LENMAN M, et al. Phospholipid: diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants [J]. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(12): 6487 − 6492. [9] STÅHL U, CARLSSON A S, LENMAN M, et al. Cloning and functional characterization of a phospholipid: diacylglycerol acyltransferase from Arabidopsis [J]. Plant Physiology, 2004, 135(3): 1324 − 1335. [10] ZHANG Meng, FAN Jilian, TAYLOR D C, et al. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development [J]. The Plant Cell, 2009, 21(12): 3885 − 3901. [11] WYDYSH E A, MEDGHALCHI S M, VADLAMUDI A, et al. Design and synthesis of small molecule glycerol 3-phosphate acyltransferase inhibitors [J]. Journal of Medicinal Chemistry, 2009, 52(10): 3317 − 3327. [12] WYDYSH E A, VADLAMUDI A, MEDGHALCHI S M, et al. Design, synthesis, and biological evaluation of conformationally constrained glycerol 3-phosphate acyltransferase inhibitors [J]. Bioorganic &Medicinal Chemistry, 2010, 18(17): 6470 − 6479. [13] ELLIS J M, PAUL D S, DEPETRILLO M A, et al. Mice deficient in glycerol-3-phosphate acyltransferase-1 have a reduced susceptibility to liver cancer [J]. Toxicologic Pathology, 2012, 40(3): 513 − 521. [14] CAO Jingsong, PEREZ S, GOODWIN B, et al. Mice deleted for GPAT3 have reduced GPAT activity in white adipose tissue and altered energy and cholesterol homeostasis in diet-induced obesity [J/OL]. American Journal of Physiology. Endocrinology and Metabolism, 2014, 306(10): E1176-1187[2022-12-12]. doi: 10.1152/ajpendo.00666.2013. [15] OUTLAW V K, WYDYSH E A, VADLAMUDI A, et al. Design, synthesis, and evaluation of 4- and 5-substituted o-(octanesulfonamido)benzoic acids as inhibitors of glycerol-3-phosphate acyltransferase [J]. Medchemcomm, 2014, 5(6): 826 − 830. [16] PELLON-MAISON M, MONTANARO M A, LACUNZA E, et al. Glycerol-3-phosphate acyltranferase-2 behaves as a cancer testis gene and promotes growth and tumorigenicity of the breast cancer MDA-MB-231 cell line [J/OL]. PLoS One, 2014, 9(6): e100896[2022-12-12]. doi: 10.1371/journal.pone.0100896. [17] TURNBULL A P, RAFFERTY J B, SEDELNIKOVA S E, et al. Analysis of the structure, substrate specificity, and mechanism of squash glycerol-3-phosphate (1)-acyltransferase [J]. Structure, 2001, 9(5): 347 − 353. [18] LEWIN T M, WANG Ping, COLEMAN R A. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction [J]. Biochemistry, 1999, 38(18): 5764 − 5771. [19] YANG S U, KIM J, KIM H, et al. Functional characterization of Physcomitrellapatens glycerol-3-phosphate acyltransferase 9 and an increase in seed oil content in arabidopsis by its ectopic expression [J/OL]. Plants, 2019, 8(8): 284[2022-12-12]. doi: 10.3390/plants8080284. [20] GIDDA S K, SHOCKEY J M, ROTHSTEIN S J, et al. Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functional divergence of the dilysine ER retrieval motif in plant cells [J]. Plant Physiology and Biochemistry, 2009, 47(10): 867 − 879. [21] SHOCKEY J, REGMI A, COTTON K, et al. Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis [J]. Plant Physiology, 2016, 170(1): 163 − 179. [22] SINGER S D, CHEN Guanqun, MIETKIEWSKA E, et al. Arabidopsis GPAT9 contributes to synthesis of intracellular glycerolipids but not surface lipids [J]. Journal of Experimental Botany, 2016, 67(15): 4627 − 4638. [23] 陈丹丹. 拟南芥3-磷酸甘油酰基转移酶2/3/9的结构与功能分析[D]. 杭州: 浙江农林大学, 2019. CHEN Dandan. Structure and Functional Analysis of Arabidopsis thaliana Glycerol-3-phosphate Acyltransferase 2/3/9 [D]. Hangzhou: Zhejiang A&F University, 2019. [24] 段芊芊, 林怡馨, 丁硕, 等. 13个甘蓝型油菜GPATs编码基因的酵母遗传互补功能鉴定[J]. 农业生物技术学报, 2020, 28(7): 1156 − 1164. DUAN Qianqian, LIN Yixin, DING Shuo, et al. Functional identification of 13 Brassica napus GPATs encoding genes by genetic complementation in yeast (Saccharomyces cerevisiae) [J]. Journal of Agricultural Biotechnology, 2020, 28(7): 1156 − 1164. [25] LEI Jie, MIAO Yingchun, LAN Yu, et al. A novel complementation assay for quick and specific screen of genes encoding glycerol-3-phosphate acyltransferases [J/OL]. Frontiers in Plant Science, 2018, 9: 353[2022-12-12]. doi: 10.3389/fpls.2018.00353. [26] WASCHBURGER E, KULCHESKI F R, VETO N M, et al. Genome-wide analysis of the glycerol-3-phosphate acyltransferase (GPAT) gene family reveals the evolution and diversification of plant GPATs [J]. Genetics and Molecular Biology, 2018, 41(suppl 1): 355−370. [27] 邢蔓, 周雪晴, 何婷, 等. 甘蓝型油菜BnGPAT9基因表达模式及其苗期非生物胁迫表达分析[J]. 中国油料作物学报, 2017, 39(4): 454 − 461. XING Man, ZHOU Xueqing, HE Ting, et al. Expression pattern of BnGPAT9 gene in Brassica napus and its expression under abiotic stresses [J]. Chinese Journal of Oil Crop Sciences, 2017, 39(4): 454 − 461. [28] KROGH A, LARSSON B, von HEIJINE G, et al. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes [J]. Journal of Molecular Biology, 2001, 305(3): 567 − 580. [29] OMASITS U, AHRENS C H, MÜLLER S, et al. Protter: interactive protein feature visualization and integration with experimental proteomic data [J]. Bioinformatics, 2014, 30(6): 884 − 886. [30] YANG Jianxi, YAN Renxiang, ROY A, et al. The I-TASSER Suite: protein structure and function prediction [J]. Nature Methods, 2015, 12(1): 7 − 8. [31] 陈丹丹, 刘宏波. 筛选GPAT基因的酵母遗传互补体系的优化[J]. 江苏农业科学, 2019, 47(13): 64 − 66. CHEN Dandan, LIU Hongbo. Optimization of yeast genetic complementary system for screening GPAT genes [J]. Jiangsu Agricultural Sciences, 2019, 47(13): 64 − 66. [32] SUN Baocheng, GUO Xuejie, FAN Chengming, et al. Newly identified essential amino acids affecting Chlorella ellipsoidea DGAT1 function revealed by site-directed mutagenesis [J/OL]. International Journal of Molecular Sciences, 2018, 19(11): 3462[2022-12-12].doi: 10.3390/ijms19113462. [33] HEATH R J, ROCK C O. A conserved histidine is essential for glycerolipid acyltransferase catalysis [J]. Journal of Bacteriology, 1998, 180(6): 1425 − 1430. [34] BRONNIKOV G E, ABOULAICH N, VENER A V, et al. Acute effects of insulin on the activity of mitochondrial GPAT1 in primary adipocytes [J]. Biochemical and Biophysical Research Communications, 2008, 367(1): 201 − 207. [35] BRATSCHI M W, BURROWES D P, KULAGA A, et al. Glycerol-3-phosphate acyltransferases gat1p and gat2p are microsomal phosphoproteins with differential contributions to polarized cell growth [J]. Eukaryotic Cell, 2009, 8(8): 1184 − 1196. [36] MITON C M, BUDA K, TOKURIKI N. Epistasis and intramolecular networks in protein evolution [J]. Current Opinion in Structural Biology, 2021, 69: 160 − 168. [37] HEATH R J, ROCK C O. A missense mutation accounts for the defect in the glycerol-3-phosphate acyltransferase expressed in the plsB26 mutant [J]. Journal of Bacteriology, 1999, 181(6): 1944 − 1946. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.20220764






 下载:
下载: