-
植物生长激素能够调节或者影响植物不同的生理过程,比如顶端优势、侧根萌发、导管分化、胚胎发育和芽的伸长,也可以促进细胞的分裂、延伸和分化[1]。在分子水平上,生长素能够特异性地调节基因的表达[2]。生长素信号转导相关的3类主要蛋白是Aux/IAAs,ARFs和SCF复合体[3-4]。作为生长素信号途径一个重要的成员,ARFs基因能够通过结合生长素启动子上的AuxRE来调控生长素反应基因的表达[5]。这些ARFs基因调控不同的发育过程,比如顶芽形成,导管组织的形成,胚胎、花和果实的形成[6]。一个经典的ARF蛋白,结构相对简单,大部分蛋白的分子量为70~130 kD,含有3个保守的结构域,它们分别是N末端DNA结合结构域(DBD),中间区域(MR)和C末端二聚体结构域(CTD)。不同的结构域也决定ARF不同的功能,DBD在信号转导中起着鉴别作用,CTD可以使ARF之间形成二聚体,而MR则起着转录激活或抑制的功能[7]。ULMASOV等[8]鉴定出第1个ARF基因,即AtARF1,先后在拟南芥Arabidopsis thaliana和水稻Oryza sativa中鉴定出23和25个ARF基因[9-10]。ARFs大部分在植物中发现,比如在双子叶、单子叶、裸子和蕨类植物中都有发现,但是在动物和微生物中至今没有发现。因此,ARFs是植物中特有的一类转录因子[11]。ARF的生物学功能主要来自拟南芥ARF基因功能缺失的表型,到目前为止,已经发现很多ARF基因及其功能,比如ARF3和ARF7[12-14]。研究表明:拟南芥arf3的突变体会出现花蕊基部和顶端的发育不良,说明ARF3在调节花器官发育上起作用;缺失ARF7,导致上胚轴的向光性和下胚轴的向地性功能的消失。这2个ARF基因有着不同的功能,很少在功能上出现冗余现象。毛竹Phyllostachys edulis与水稻、玉米Zea mays等同属于禾本科Gramineae单子叶植物,但是,毛竹营养生长周期长,开花时期不确定,开花后死亡,导致竹林面积减少,对经济发展和生态环境造成重大损失和破坏,竹子开花的调控机制一直是竹类植物研究中的难点和热点。目前,ARF家族基因在模式植物拟南芥、水稻、玉米中已有研究,而毛竹ARF家族基因在花器官和幼胚发育上鲜有报道。本研究通过生物信息学的方法,根据毛竹的基因组,鉴定ARF基因家族,进行进化树分析、基序分析、基因差异表达模式分析,为研究ARF基因在毛竹花和种子发育过程中的功能奠定基础。
-
毛竹开花实验地位于广西壮族自治区桂林市南岭山系的西南部。该毛竹林属于自然生长状态,基本无人为干扰。以毛竹的花器官为材料,进行解剖,分离出花芽、苞片、颖片、稃片、雄蕊、雌蕊和幼胚以及未开花的成熟叶片,建立8个样本进行转录组高通量测序。
-
从拟南芥基因组数据库()和水稻基因组数据库()中分别检索拟南芥和水稻中ARF蛋白序列;毛竹ARF相关蛋白序列从毛竹基因组数据库()中获得。毛竹PheARF家族蛋白的序列号,开放阅读框的长度、氨基酸数目、分子质量和等电点在表 1中提供。毛竹ARF分子量及等电点数据通过ExPASY(http://web.expasy.org/compute_pi/)获得。
表 1 毛竹中ARF成员数量及其属性
Table 1. Properties and numbers of ARF identified from Phyllostachys edulis
ARF蛋白名称 毛竹ARF序列号 氨基酸长度 开放阅读框/bp 等电点 分子量/kD PheARFl PH0l0l3505G00l0 278 837 7.61 31.67 PheARF2 PH0l008675G00l0 260 780 6.70 28.56 PheARF3 PH0l0076l2G00l0 672 2 0l9 5.72 74.77 PheARF4 PH0l005322G00l0 792 2 379 6.09 87.49 PheARF5 PH0l00AEAlG0lA0 832 2 499 5.84 92.70 PheARF6 PH0l00A096G0200 92l 2 766 5.3l l02.29 PheARF7 PH0l002857G0060 708 2 l27 5.99 78.96 PheARF8 PH0l002850G0020 476 l 43l 9.23 54.32 PheARF9 PH0l002806G0200 840 2 523 6.97 93.0l PheARFl0 PH0l002685G0l20 605 l 8l8 6.29 65.89 PheARFll PH0l002498G0280 658 l 977 6.46 72.04 PheARF12 PH0l002l60G0l80 757 2 274 6.l6 84.64 PheARFlA PH0l00l899G0250 728 2 l87 5.68 80.88 PheARF14 PH0l00l690G0Al0 70l 2 l06 6.50 76.99 PheARFl5 PH0l00l555G0390 878 2 637 6.l6 97.8l PheARFlE PH0l00l285G0430 603 l 8l2 7.02 65.37 PheARFl7 PH0l00l2l2G0l90 364 l 095 7.25 39.9l PheARFl8 PH0l00l026G0A00 744 2 235 7.55 8l.07 PheARFl9 PH0l000667G0020 637 l 9l4 5.22 7l.47 PheARF20 PH0l000626G0040 693 2 082 5.34 77.55 PheARF2l PH0l00062AG0440 l l86 3 56l 8.87 l32.0l PheARF22 PH0l000548G0300 985 2 958 6.05 ll0.ll PheARF23 PH01000483G0220 866 2 60l 5.74 96.l5 PheARF24 PH0l000384G0l70 763 2 292 5.36 84.80 PheARF25 PH0l000305G0690 635 l 908 7.25 69.46 PheARF26 PH0l000277G0820 59l l 776 6.90 64.69 PheARF27 PH0l000259Gll20 904 2 7l5 6.60 l00.7l PheARF28 PH0l000227G0020 908 2 727 5.87 l00.72 PheARF29 PH0l000222G0l80 750 2 253 6.ll 83.00 PheARF30 PH0l000l83G0570 897 2 694 6.03 99.ll PheARF3l PH0l000l76G0540 l l2l 3 366 6.2l l24.73 PheARF32 PH0l000ll6G09l0 8l7 2 454 7.6l 90.ll PheARF33 PH0l000ll4G0050 l 257 3 774 6.23 l39.07 PheARF34 PH0l000093G0670 750 2 253 6.82 83.28 PheARF35 PH0l000087Gl340 738 2 2l7 5.85 82.09 PheARF36 PH0l000057Gl420 928 2 787 5.54 l02.92 PheARF37 PH0l000046G0220 734 2 205 6.70 80.5l PheARF38 PH0l000044G0540 439 l 320 5.87 47.45 PheARF39 PH0l0000l8G0940 667 2 004 6.96 74.36 PheARF40 PH0l0000l4G06l0 427 l 284 9.00 47.32 PheARF4l PH0l0000llG0660 554 l 665 5.66 6l.35 PheARF42 PH0l000002G3ll0 828 2 487 6.4l 92.55 PheARF43 PH0l000093G0690 l92 579 5.44 2l.02 PheARF44 PH0l000237G0420 5l4 l 545 8.52 56.44 -
用ClustalX 1.83()[15]软件对蛋白全长的多序列比对进行分析,进化树分析前去掉比对序列的差异和不明确序列。用no-rooted neighbor-joining方法通过MEGA 6.0()[16]构建系统进化树。
-
MEME version 4.11.2(http://meme-suite.org/)[17]在线工具鉴定候选蛋白序列的保守区域,公式为any,maximum number of motifs = 20,minimum width≥ 6和maximum width ≤ 200。
-
将花芽、苞片、颖片、稃片、雄蕊、雌蕊和幼胚以及未开花的成熟叶片的FPKM值输入到Cluster 3.0,用Java TreeView生成热点图[18]。
-
通过23个拟南芥和25个水稻ARF蛋白序列检索毛竹基因组数据库,共得到44个ARF蛋白(表 1)。从表 1可以看出,ARF家族的蛋白的等电点为5.22~9.23,蛋白序列的长度为192~1 257个氨基酸,分子量的大小为31.70 ~139.07 kD。
-
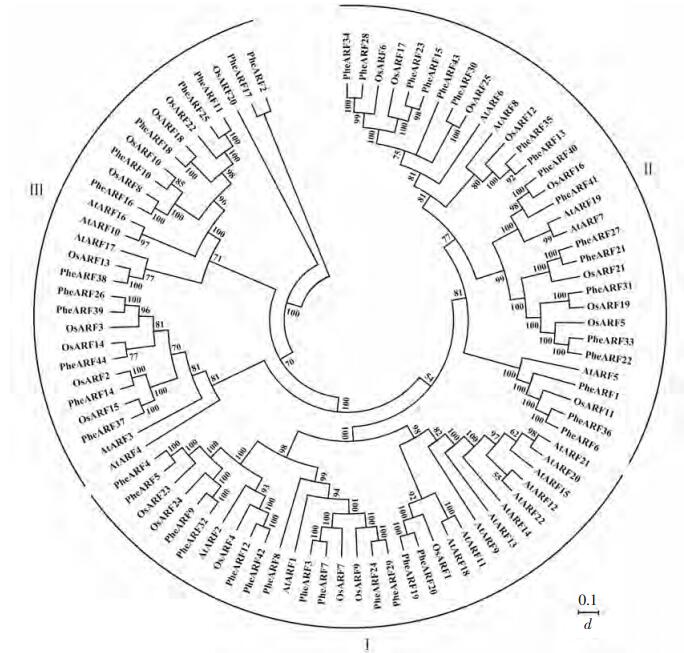
系统进化结果显示:44个基因之间相似性比较高(图 1)。将44个毛竹的ARF蛋白列同23个拟南芥和25个水稻的蛋白序列同时比对,发现可以将这3个物种的ARF分为三大类,分别为Ⅰ,Ⅱ和Ⅲ。其中有13个基因分布在Ⅰ类,17个基因分布在Ⅱ类,还有14个基因属于Ⅲ类。大部分的PheARFs包含3个经典的结构域:DBD,结构域Ⅱ和AUX/IAA家族结构域。有11个与水稻的同源关系较近的基因对:osAR25/PheARF30(Ⅱ),OsARF16/PheARF40(Ⅱ),OsARF19/PheARF31(Ⅱ),OsARF15/PheARF37(Ⅲ),OsARF2/PheARF14(Ⅲ),OsARF14/PheARF39(Ⅲ),OsARF3/PheARF26(Ⅲ),OsARF13/PheARF38(Ⅲ),OsARF8/PheARF16(Ⅲ),OsARF10/PheARF10(Ⅲ)和OsARF18/PheARF18(Ⅲ)。这些基因大部分出现在Ⅱ类和Ⅲ类中,说明这2类的基因相对比较保守。
-
为了进一步了解毛竹ARF保守区域的结构,通过MEME在线工具构建基序分析图,结果如图 2所示。毛竹ARF成员含有的基序结构不一,大概有20个不同的基序组成。其中每个基因都包含不同种类的基序,大部分的基序为4~17个。大部分的PheARF基因都包含Motif1~8,Motif10,Motif11,Motif14,Motif18和Motif20等,这些基序出现的次数较多,而Motif13,Motif16和Motif17等基序在这些基因中不常见。从进化角度来看,同源关系较近的Ⅰ,Ⅱ,Ⅲ类的基因在基序的长度和种类较为相似,比如Ⅰ类的PheARF4,PheARF9和PheARF32等,Ⅱ类的PheARF22,PheARF31和PheARF33等,Ⅲ类的PheARF17,PheARF18和PheARF37等。以上结果说明大部分的毛竹ARF家族基因是相当保守的。
-
为了研究毛竹PheARF基因在花器官发育中的作用和调控机制,进一步分析PheARF基因在花芽、苞片、颖片、稃片、雌蕊、雄蕊、幼胚和未开花的叶中表达分析。结果表明:PheARF12,PheARF13,PheARF14,PheARF15,PheARF24,PheARF35,PheARF43,PheARF23和PheARF32在花芽中高量表达,说明这些ARF基因可能在开花初期起着重要的调控作用(图 3)。在雄蕊中,PheARF9,PheARF29,PheARF31,PheARF10,PheARF4,PheARF22,PheARF11,PheARF25,PheARF40,PheARF8,PheARF39,PheARF1,PheARF44,PheARF45,PheARF42和PheARF41高量表达,但是这些基因在幼胚中表达量极低。PheARF2,PheARF30,PheARF14,PheARF13,PheARF35,PheARF7,PheARF37和PheARF38在雌蕊中表达量较高,同时在幼胚中也高量表达,说明这些基因同时调控毛竹雌蕊形成和幼胚的发育。PheARF13,PheARF14和PheARF35同时在花芽、雌蕊和幼胚中高表达,这3个基因可能既调控开花又能控制花的发育。此外,PheARF5,PheARF18,PheARF27,PheARF10和PheARF4在颖片和稃片中都高量表达,可能调节稃片和颖片的发育。
-
生长素是植物器官发育和模式形态形成和发育非常重要的信号分子。生长素转导途径中最重要的2类家族分别为ARFs和Aux/IAAs[19]。在发育过程中,ARFs能直接地调控下游靶基因的表达[6, 20],但大部分的ARF和Aux/IAA都是以家族的形式存在,所以它们在植物体内的调控机制是相当复杂的[21-23]。ARFs基因也参与生殖过程[24]。木瓜Carica papaya和番茄Solanum lycopersicum的ARFs基因家族的分析和鉴定揭示了该家族基因在花和果实的发育过程中具有调控作用[25-26]。在本研究中,通过生物信息学工具,检索44个毛竹的ARF基因。在拟南芥和水稻中的ARF基因分别为23和25个。ARF基因家族在不同植物中成员数量不同,这可能与植物体内的基因组复制有关[27],而基因数量多少由该基因复制事件的频率决定。毛竹基因组为2 G左右[28],大于水稻和拟南芥的基因组。以上分析表明:毛竹ARF基因复制事件的频繁发生有可能导致毛竹ARF家族成员数量增多。通过系统进化分析,毛竹ARF家族基因大致分为3个亚组(图 1),亚组内的基因相似性比较高,但是亚组之间相似性不高,或许在基因功能上也有差异。毛竹与拟南芥和水稻的ARF家族的系统进化关系表明,毛竹与水稻之间有11对同源基因对,这些基因对都属于Ⅱ类和Ⅲ类,一方面说明毛竹的ARF家族与水稻有很高的同源性(图 1),另一方面说明毛竹ARF家族Ⅱ类和Ⅲ类的基因保守性很高。
研究人员对水稻和拟南芥的ARF基因进行了广泛地研究[29-31]。ARF基因在花和种子发育中起着非常重要的作用,为研究毛竹的开花及种子发育提供了许多有用的信息。据报道,拟南芥的AtARF3和AtARF4参与花的发育,在花中高量表达[32]。与AtARF3和AtARF4同源的毛竹PheARF14和PheARF37在幼胚和雌蕊中都高量表达,说明2个毛竹ARF基因可能与AtARF3和AtARF4基因功能相似,推测它们在毛竹花和种子发育过程中起着关键的作用。
Bioinformatic analysis and differential expression of auxin response factor (ARF) gene in Phyllostachys edulis
-
摘要: 生长素反应因子(ARF)基因家族在植物的生长发育中起至关重要的作用。关于毛竹Phyllostachys edulis ARF基因家族在花器官中生物信息学分析未见报道。以毛竹花器官为材料,采用生物信息学的方法,对毛竹中ARF基因进行筛选,并对其系统进化关系、保守基序及在花器官中的差异表达模式进行了初步的分析。结果表明:毛竹全基因组中含有44个ARF基因,分为Ⅰ,Ⅱ,Ⅲ类。与拟南芥Arabidopsis thaliana和水稻Oryza sativa分别比较,发现毛竹与水稻存在11个姐妹同源基因对。此外,PheARF13,PheARF14和PheARF35在花芽、雌蕊和幼胚中高量表达,同时PheARF2,PheARF30,PheARF14,PheARF13,PheARF35,PheARF7,PheARF37,PheARF38在雌蕊和幼胚中高量表达,推测这些基因可能在花发育和种子发育中发挥重要的作用。Abstract: The auxin response factor (ARF) gene family plays a key role in plant growth and developmental processes, such as root and shoot development as well as flower and fruit development. To provide a theoretical basis for flower and seed development of Phyllostachys edulis, a genome-wide analysis of the previously undocumented ARF gene family for Ph. edulis was conducted. In this study a whole-genome survey of Ph. edulis was performed and a detailed analysis of the gene motif and phylogenetic classification was provided. Results showed 44 ARF genes which were classified into three groups. A comparative analysis of the ARF genes among Ph. edulis, rice, and Arabidopsis suggested a total of 11 sister pairs (OsARF-PheARF) providing insights into various orthologous relationships between OsARFs and PheARFs. For ARF expression patterns of diverse floral organs, PheARF13, PheARF14, and PheARF35 showed the highest expression in the flower bud, pistil, and young embryo; whereas, PheARF2, PheARF30, PheARF14, PheARF13, PheARF35, PheARF7, PheARF37, and PheARF38 were highly expressed in the pistil and the young embryo. This study suggested that ARF genes may play a very critical role during flower and fruit development of Ph. edulis.
-
Key words:
- forest tree breeding /
- Phyllostachys edulis /
- auxin response factors /
- floral development
-
表 1 毛竹中ARF成员数量及其属性
Table 1. Properties and numbers of ARF identified from Phyllostachys edulis
ARF蛋白名称 毛竹ARF序列号 氨基酸长度 开放阅读框/bp 等电点 分子量/kD PheARFl PH0l0l3505G00l0 278 837 7.61 31.67 PheARF2 PH0l008675G00l0 260 780 6.70 28.56 PheARF3 PH0l0076l2G00l0 672 2 0l9 5.72 74.77 PheARF4 PH0l005322G00l0 792 2 379 6.09 87.49 PheARF5 PH0l00AEAlG0lA0 832 2 499 5.84 92.70 PheARF6 PH0l00A096G0200 92l 2 766 5.3l l02.29 PheARF7 PH0l002857G0060 708 2 l27 5.99 78.96 PheARF8 PH0l002850G0020 476 l 43l 9.23 54.32 PheARF9 PH0l002806G0200 840 2 523 6.97 93.0l PheARFl0 PH0l002685G0l20 605 l 8l8 6.29 65.89 PheARFll PH0l002498G0280 658 l 977 6.46 72.04 PheARF12 PH0l002l60G0l80 757 2 274 6.l6 84.64 PheARFlA PH0l00l899G0250 728 2 l87 5.68 80.88 PheARF14 PH0l00l690G0Al0 70l 2 l06 6.50 76.99 PheARFl5 PH0l00l555G0390 878 2 637 6.l6 97.8l PheARFlE PH0l00l285G0430 603 l 8l2 7.02 65.37 PheARFl7 PH0l00l2l2G0l90 364 l 095 7.25 39.9l PheARFl8 PH0l00l026G0A00 744 2 235 7.55 8l.07 PheARFl9 PH0l000667G0020 637 l 9l4 5.22 7l.47 PheARF20 PH0l000626G0040 693 2 082 5.34 77.55 PheARF2l PH0l00062AG0440 l l86 3 56l 8.87 l32.0l PheARF22 PH0l000548G0300 985 2 958 6.05 ll0.ll PheARF23 PH01000483G0220 866 2 60l 5.74 96.l5 PheARF24 PH0l000384G0l70 763 2 292 5.36 84.80 PheARF25 PH0l000305G0690 635 l 908 7.25 69.46 PheARF26 PH0l000277G0820 59l l 776 6.90 64.69 PheARF27 PH0l000259Gll20 904 2 7l5 6.60 l00.7l PheARF28 PH0l000227G0020 908 2 727 5.87 l00.72 PheARF29 PH0l000222G0l80 750 2 253 6.ll 83.00 PheARF30 PH0l000l83G0570 897 2 694 6.03 99.ll PheARF3l PH0l000l76G0540 l l2l 3 366 6.2l l24.73 PheARF32 PH0l000ll6G09l0 8l7 2 454 7.6l 90.ll PheARF33 PH0l000ll4G0050 l 257 3 774 6.23 l39.07 PheARF34 PH0l000093G0670 750 2 253 6.82 83.28 PheARF35 PH0l000087Gl340 738 2 2l7 5.85 82.09 PheARF36 PH0l000057Gl420 928 2 787 5.54 l02.92 PheARF37 PH0l000046G0220 734 2 205 6.70 80.5l PheARF38 PH0l000044G0540 439 l 320 5.87 47.45 PheARF39 PH0l0000l8G0940 667 2 004 6.96 74.36 PheARF40 PH0l0000l4G06l0 427 l 284 9.00 47.32 PheARF4l PH0l0000llG0660 554 l 665 5.66 6l.35 PheARF42 PH0l000002G3ll0 828 2 487 6.4l 92.55 PheARF43 PH0l000093G0690 l92 579 5.44 2l.02 PheARF44 PH0l000237G0420 5l4 l 545 8.52 56.44 -
[1] DAVIES P J. Plant Hormones: Physiology, Biochemistry and Molecular Biology[M]. Dordrecht: Kluwer Academic Publishers, 2013. [2] THEOLOGIS A. Rapid gene regulation by auxin[J]. Ann Rev Plant Physiol, 1986, 37(1): 407-438. [3] GUILFOYLE T J, ULMASOV T, HAGEN G. The ARF family of transcription factors and their role in plant hormone-responsive transcription[J]. Cell Mol Life Sci, 1998, 54(7): 619-627. [4] SHEN Chenjia, YUE Runqing, SUN Tao, et al. Genome-wide identification and expression analysis of auxin response factor gene family in Medicago truncatula[J]. Front Plant Sci, 2015, 6(11): 3932-3935. [5] TIWARI S B, HAGEN G, GUILFOYLE T. The roles of auxin response factor domains in auxin-responsive transcription[J]. Plant Cell, 2003, 15(2): 533-543. [6] GUILFOYLE T J, HAGEN G. Auxin response factors[J]. Curr Opin Plant Biol, 2007, 10(5): 453-460. [7] RAMOS J A, ZENSER N, LEYSER O, et al. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain Ⅱ and is proteasome dependent[J]. Plant Cell, 2001, 13(10): 2349-2360. [8] ULMASOV T, HAGEN G, GUILFOYLE T J. ARF1, a transcription factor that binds to auxin response elements[J]. Science, 1997, 276(5320): 1865-1868. [9] HAGEN G, GUILFOYLE T. Auxin-responsive gene expression: genes, promoters and regulatory factors[J]. Plant Mol Biol, 2002, 49(3/4): 373-385. [10] REMINGTON D L, VISION T J, GUILFOYLE T J, et al. Contrasting modes of diversification in the Aux/IAA and ARF gene families[J]. Plant Physiol, 2004, 135(3): 1738-1752. [11] LISCUM E, REED J W. Genetics of Aux/IAA and ARF action in plant growth and development[J]. Plant Mol Biol, 2002, 49(3/4): 387-400. [12] SESSIONS R. Arabidopsis (Brassicaceae) flower development and gynoecium patterning in wild type and ettin mutants[J]. Am J Bot, 1997, 84(9): 1179-1179. [13] HARDTKE C S, BERLETH T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development[J]. EMBO J, 1998, 17(5): 1405-1411. [14] HARPER R M, STOWE-EVANS E L, LUESSE D R, et al. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue[J]. Plant Cell, 2000, 12(5): 757-770. [15] THOMPSON J D, GIBSON T J, PLEWNIAK F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools[J]. Nucl Acids Res, 1997, 25(24): 4876-4882. [16] TAMURA K, STECHER G, PETERSON D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0[J]. Mol Biol Evol, 2013, 30(12): 2725-2729. [17] BAILEY T L, ELKAN C. The value of prior knowledge in discovering motifs with MEME[C]. Proc Third Int Conf Intel Syst Mol Biol, California: AAAI Press, 1995: 21-29. [18] SALDANHA A J. Java Treeview-extensible visualization of microarray data[J]. Bioinformatics, 2004, 20(17): 3246-3248. [19] FARCOT E, LAVEDRINE C, VERNOUX T. A modular analysis of the auxin signalling network[J]. PloS One, 2015, 10(3): 864-865. [20] ULMASOV T, MURFETT J, HAGEN G, et al. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements[J]. Plant Cell, 1997, 9(11): 1963-1971. [21] 王垒, 陈劲枫, 娄丽娜, 等.黄瓜果实中ARF和Aux/IAA基因对外源激素的应答[J].西北植物学报, 2011, 31(6): 1127-1131. WANG Lei, CHEN Jinfeng, LOU Lina, et al. Expression analysis about some ARF and Aux/IAA family members in fruits of cucumber with exogenous hormones[J]. Acta Bot Boreal-Occident Sin, 2011, 31(6): 1127-1131. [22] 史梦雅, 李阳, 张巍, 等.生长素反应因子作用机制研究进展[J].生物技术通报, 2012(8): 24-28. SHI Mengya, LI Yang, ZHANG Wei, et al. Progress in mechanism of auxin response factors[J]. Biotechnol Bull, 2012(8): 24-28. [23] AUDRAN-DELALANDE C, BASSA C, MILA I, et al. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato[J]. Plant Cell Physiol, 2012, 53(4): 659-672. [24] CHO H, RYU H, RHO S, et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development[J]. Nat Cell Biol, 2014, 16(1): 66-76. [25] PAULL R E, IRIKURA B, WU Pingfang, et al. Fruit development, ripening and quality related genes in the papaya genome[J]. Trop Plant Biol, 2008, 1(3/4): 246-277. [26] KUMAR R, TYAGI A K, SHARMA A K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development[J]. Mol Gen Genomics Mgg, 2011, 285(3): 245-260. [27] OHNO S. Evolution by Gene Duplication[M]. Heidelberg: Springer Verlag, 2013. [28] PENG Zhenhua, LU Ying, LI Lubin, et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla)[J]. Nat Gen, 2013, 45(4): 456-461. [29] QI Yanhua, WANG Suikang, SHEN Chenjia, et al. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa)[J]. New Phytol, 2012, 193(1): 109-120. [30] COLE M, CHANDLER J, WEIJERS D, et al. DORNRÖSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo[J]. Development, 2009, 136(10): 1643-1651. [31] LI Jisheng, DAI Xinhua, ZHAO Yunde. A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis[J]. Plant Physiol, 2006, 140(3): 899-908. [32] HUNTER C, WILLMANN M R, WU Gang, et al. Trans-acting siRNA-mediated repression of Ettin and ARF4 regulates heteroblasty in Arabidopsis[J]. Development, 2006, 133(15): 2973-2981. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.2017.04.002







 下载:
下载: