-
高等植物成花受到自身遗传因素和外界环境因素的共同影响[1-2]。当植物完成成花诱导,花器官原基开始分化,逐渐形成花萼、花瓣、雄蕊和心皮等组织。在这期间,植物的茎尖分生组织形态发生了很大的变化,同时成花素FT(FLOWERING LOCUS T)的表达量显著增加并参与调控花芽分化[3]。在模式植物拟南芥Arabidopsis thaliana以及许多物种中已经证实,FT可以通过激活花芽分化组织特异性基因AP1(APETALA1)的表达促使植物完成开花进程[4]。过表达AP1基因,不论在长、短日照下拟南芥都会出现早花表型[5],其突变体ap1则出现明显的晚花表型[6]。在茎尖分生组织,AP1基因不仅在植物成花诱导的调控网络中起核心作用,而且也决定花器官的形成。AP1基因是植物ABCDE分化模型中的A类基因,控制第一轮花萼和第二轮花被片组织的形成[7],同时AP1也调控SEP3(SEPALLATA3)与LFY(LEAFY)基因的表达,促进花萼和花瓣的分化[8-9]。另外在拟南芥ap1突变体研究中发现,AP1突变会引起花器官的异常发育,如花萼发育成叶片、花瓣缺失等[10-11]。人们已从多种观赏植物中分离得到了AP1基因,并且对多个物种的AP1基因进行了功能验证和分析。超表达AP1及其同源基因都能促进开花,在拟南芥中异源表达百合Lilium longiflorum,蝴蝶兰Phalaenopsis aphrodite以及山茶Camellia japonica的AP1同源基因都能引起早花现象[12-14]。同时,在拟南芥AP1突变体中异源表达百合,枇杷Eriobotrya japonica以及麻疯树Jatropha curcas中的AP1基因,都可以回补其突变体花器官缺陷的性状[12, 15-16]。桂花Osmanthus fragrans是中国十大传统名花之一,也是常见的园林绿化树种。按照开花习性不同,可分为秋桂和四季桂,秋桂仅在每年秋季开花,花序常为无总梗的聚伞花序[17];关于其成花过程以及花芽分化和发育报道较少。木本植物桂花成花的机制与草本植物拟南芥等有一定差异。为深入了解桂花的成花机制,有必要分离成花相关基因并展开相关研究。本研究以秋桂品种‘堰虹桂’Osmanthus fragrans ‘Yanhonggui’为材料,克隆桂花AP1基因(OfAP1),并对其序列进行生物信息学分析,同时运用荧光定量对其组织和时空表达进行分析,以明确OfAP1基因的基本特征和表达模式,为今后桂花的开花分子机理研究提供科学依据。
HTML
-
以盆栽‘堰虹桂’为材料,选取相同株龄且生长一致的植株。取桂花不同组织的样品,包括根、茎、叶、叶芽、花芽、盛开期的花朵;同时根据桂花花芽分化进程取叶芽、花序分化期、花萼花瓣分化期、雌雄蕊分化期以及花开放不同阶段圆珠期、铃梗期、初花期、盛花期和盛花末期的样品[18],用于基因克隆、组织特异性和时空表达分析。所有材料使用液氮冷冻并于-80 ℃保存备用。
RNA提取试剂盒(RNAprep pure Plant Kit)购自天根公司(北京)。反转录试剂盒、荧光定量试剂盒、胶回收试剂盒、Premix Taq酶、pMD-18T载体和大肠埃希菌Escherichia coli DH5α均购自Takara公司(大连),运用实时荧光定量PCR(Applied Biosystems,Foster City,美国)分析OfAP1基因的组织特异性及时空表达变化。
-
按照RNAprep pure Plant Kit试剂盒说明书提取各样品的总RNA,并参照Reverse Transcriptase M-MLV反转录酶说明书合成cDNA的第一链后,储存于-20 ℃备用。
-
利用前期转录组测序获得的AP1 Unigene序列,设计特异性引物AP1-F:5′-TAGAGTGAGAAAATGGGGAGA-3′;AP1-R:5′-AATACAATCCCTGGCTACTT-3′。以花芽分化期茎尖RNA反转录的cDNA为模板进行PCR扩增,PCR反应体系为:上下游引物各1 μL,cDNA 1 μL,Premix Taq DNA 10 μL和去离子水7 μL。PCR反应条件:95 ℃预变性5 min;然后进行35个循环,每个循环包括95 ℃变性30 s,60 ℃退火30 s,72 ℃延伸1 min;最后72 ℃保温10 min,4 ℃保存。PCR反应产物经质量分数为1%的琼脂糖凝胶电泳检测后回收、纯化,与pMD18-T载体连接,转化大肠埃希菌DH5α感受态细胞,蓝白斑筛选阳性克隆,经PCR鉴定后送生工生物工程(上海)股份有限公司测序。
-
对获得的基因序列进行生物信息学分析,其中编码区分析采用美国国家生物技术信息中心(NCBI)在线网站ORFFinder(http://www.ncbi.nlm.nih.gov/gorf/gorf.html),编码蛋白质的保守区段分析采用NCBI Conserved Domain Search软件(http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi),编码区蛋白质特征分析采用在线软件ProtParam(http://web.expasy.org/protparam/)和TMHMM Server v2.0(http://www.cbs.dtu.dk/services/TMHMM/)。多序列比对采用软件DNAman,系统进化树构建采用Clustal X+MEGA 7.0软件并选择邻位连接法(neighbor joining method)用Bootstrap法检验1 000次。
-
按照SYBR® Premix Ex Taq TM Ⅱ(Tli RNaseH Plus)试剂盒的说明书,检测桂花OfAP1基因不同组织以及不同时期的表达情况。OfAP1定量引物为qAP1-F:5′-GCAGAAGTGGCTTTGATTTG-3′;qAP1-R:5′-GTTTGCTGGTGACTGAGGTT-3′。同时以OfACT为内参qACT-F:5′-CCCAAGGCAAACAGAGAAAAAAT-3′;qACT-R:5′-ACCCCATCACCAGAATCAAGAA-3′[19]。qRT-PCR反应体系:2×SYBR Premix Ex TaqⅡ(Tli RNaseH Plus)10 μL,上下游定量引物(10 μmol·L-1)各0.8 μL,cDNA 2 μL,50×ROX Reference Dye 0.4 μL,双蒸水补齐至20 μL。qRT-PCR程序如下:95 ℃预变性30 s;95 ℃变性5 s,60 ℃退火30 s,72 ℃延伸1 min,35个循环;72 ℃延伸10 min。3次生物学重复。反应后根据熔解曲线分析qRT-PCR产物的特异性,并采用2-△△CT法计算相对表达量。
1.1. 材料
1.2. 方法
1.2.1. RNA提取及反转录
1.2.2. 桂花OfAP1基因的克隆
1.2.3. 序列生物信息学分析
1.2.4. 实时荧光定量PCR
-
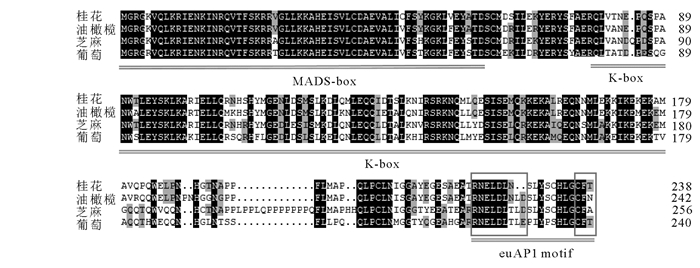
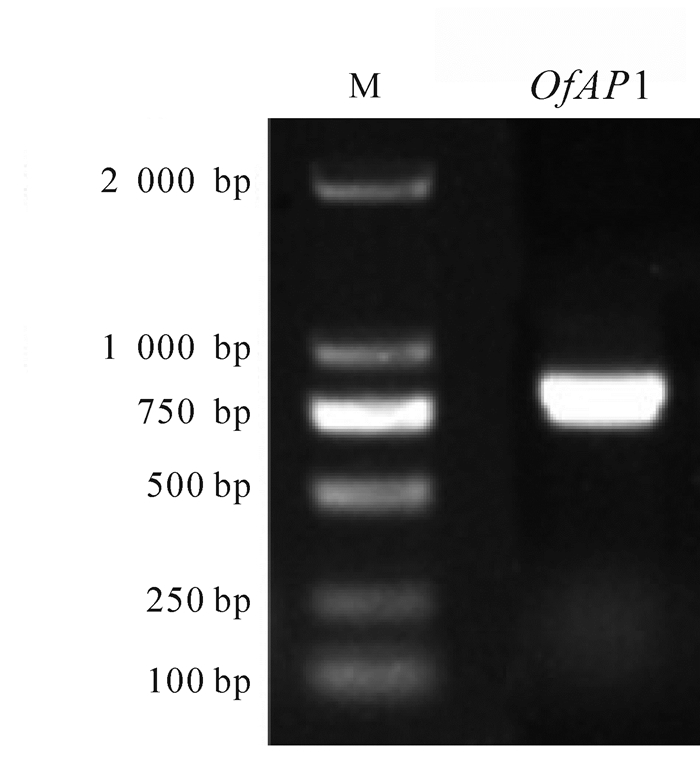
以‘堰虹桂’花芽分化期cDNA为模板,扩增得到约750 bp片段(图 1),其开放阅读框长为720 bp,编码239个氨基酸(图 2)。该基因片段具有MADS-box基因的保守区,在NCBI上进行BLASTX在线分析,发现该序列与芝麻Sesamum indicum(AIS82596.1),葡萄Vitis vinifera(ACZ26528.1)和油橄榄Olea europaea(XP_022878350.1)等中AP1基因的同源性最高,命名为OfAP1,并在GenBank注册,登录号为MH593222。
-
OfAP1蛋白氨基酸序列运用NCBI在线CDD(Conserved Domain Datebase)分析发现,OfAP1具有典型的MEF2_like MADS结构域和K-box结构域,MADS结构域位于N端第2~74位氨基酸,K-box结构域位于第90~168位氨基酸(图 2)。OfAP1蛋白的分子式为C1199H1941N341O368S16,其相对分子质量为27 534.62,理论等电点为8.4。负电荷残基总数(天冬氨酸Asp+谷氨酸Glu)为32,正电荷残基总数(精氨酸Arg+赖氨酸Lys)为35。在组成OfAP1蛋白的20种氨基酸中,亮氨酸(Leu)所占比例最高,为11.7%,其次为谷氨酸(Glu),占10.0%,色氨酸(Trp)所占比例最低,为0.8%。预测亲水性指数(GRAVY)为-0.665,表明OfAP1具有较好的亲水性。
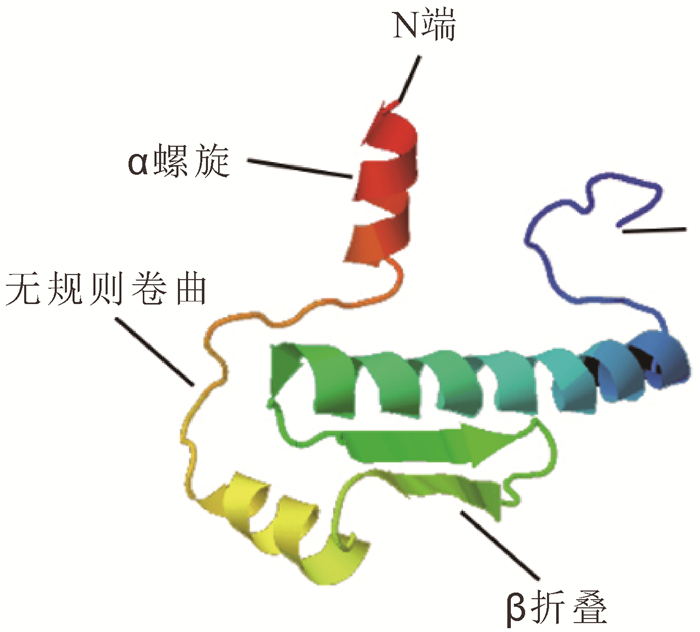
采用GOR4软件对OfAP1蛋白二级结构预测发现,OfAP1蛋白二级结构由α螺旋、伸展链和无规则卷曲组成,其中,α螺旋(Hh)占56.49%,β折叠(Ee)占10.46%,无规则卷曲(Cc)占33.05%。同时采用Phyre 2在线工具对OfAP1蛋白三级结构进行预测,OfAP1有3个典型的α螺旋和2个β折叠(图 3)。NetPhos 2.0 Server预测结果表明:OfAP1蛋白序列中存在11个潜在的磷酸化位点,包括6个Ser位点(22Ser,61Ser,74Ser,87Ser,110Ser,121Ser,135Ser,150Ser,216Ser)和1个Thr位点(220Thr)。NetNGlyc 1.0 Server预测结果表明:OfAP1蛋白存在1个潜在的N-糖基化位点,90Asn。
-
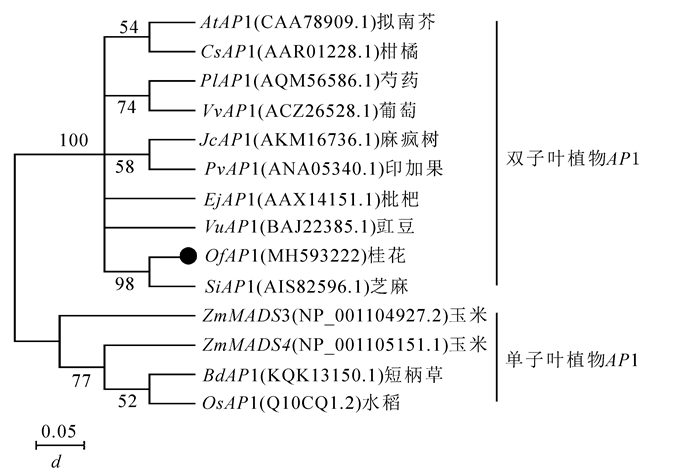
为了分析OfAP1基因与其他物种中AP1基因的系统进化关系,采用MEGA 7.0软件构建系统进化树(图 4)。单子叶植物(如短柄草Brachypodium distachyon,水稻Oryza sativa,玉米Zea mays)和双子叶植物可以明显地分为2个大类。桂花OfAP1与芝麻,豇豆Vigna unguiculata,枇杷,印加果Plukenetia volubilis,麻疯树,葡萄,芍药Paeonia lactiflora,柑橘Citrus sinensis以及拟南芥中的AP1基因聚在一起,说明这些基因亲缘关系较近且功能具有一定的相似性。
-
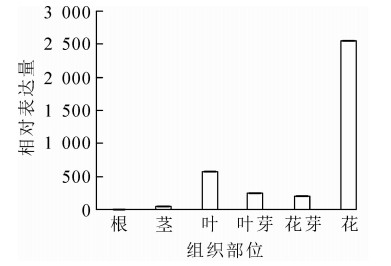
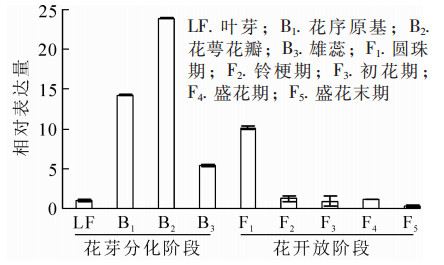
为明确OfAP1基因在桂花不同组织以及花芽分化不同时期的表达情况,对不同组织和器官(根、茎、叶、叶芽、花芽以及花)及不同发育时期的样品进行荧光定量RT-PCR分析。结果表明:OfAP1基因在花芽中的表达量最高,其次为叶,在茎中的表达比较弱,在根中几乎不表达(图 5)。同时,与叶芽时期相比,OfAP1基因在分化初期(即花原基的形成以及花萼花瓣分化期)表达量最高,随后呈下降趋势(图 6),到花开放阶段表达量均较低。
2.1. 桂花OfAP1基因的克隆
2.2. 桂花OfAP1基因的氨基酸序列分析
2.3. 桂花OfAP1系统进化关系分析
2.4. 桂花OfAP1基因的组织特异性与时空表达分析
-
AP1及其同源基因已在多个物种中克隆得到,但桂花中的AP1至今没有相关报道。本研究克隆得到OfAP1基因,发现其与芝麻、葡萄以及油橄榄等AP1基因高度相似,OfAP1氨基酸序列含有高度保守的MEF2_like MADS结构域和次级保守的K-box结构域以及可变C,MADS结构域具有结合DNA、蛋白质二聚化以及与其他因子结合的功能;K-box结构域的蛋白二级结构为3个α螺旋,具有介导蛋白-蛋白之间的相互作用[20]。AP1基因C末端的变化较大,但是不同类的MADS-box基因常含有一些保守的基序(motif),这些基序在蛋白复合体的形成和转录激活中起重要作用[21]。
开花是植物重要的发育阶段,是多条基因网络共同调控的复杂过程[2]。AP1基因是植物特有的MIKCC-Type MADS-box基因,既参与花分生组织的形成,又是花器官形成的重要基因,在植物成花中起关键调控作用[3]。根据在模式植物拟南芥中的研究,AP1是花分生组织形成的标志物,成花整合基因FT和SOC1能够激活其表达并使其参与对成花抑制基因TFL1基因的拮抗,促进花分生组织的形成。在花器官发育中,AP1一方面通过促进AP3和PI的表达诱导花瓣和雄蕊的发育,另一方面受到AG基因的抑制作用控制萼片和花瓣的发育[22]。在百合中AP1的同源基因LMADS5/6转基因拟南芥后发现其能够造成早花,并且花器官中萼片变成心皮状,花瓣变成雄蕊状结构[12];同样地,在洋桔梗Eustoma grandiflorum中超表达AP1发现:转基因植株花期提前,部分花瓣出现雄蕊类似的结构,进一步突变体互补实验表明:AP1基因对花瓣形成,尤其是第2轮花被片的发育有重要作用[23]。在建兰Cymbidium ensifolium,芍药以及菊花Chrysanthemum morifolium等中,AP1基因在花芽中的表达量显著高于其他器官组织,尤其在花芽分化的初期阶段[24-26]。在桂花中,OfAP1在花芽中有较高的表达,其在花芽分化初期表达量显著上调,尤其是成花转变以及花瓣、花萼分化时期,而当完成花萼花瓣分化后,表达量则显著下调。这充分说明桂花OfAP1基因参与了桂花成花转变和花瓣、花萼等器官分化,但其具体作用机制还有待进一步通过转化实验加以验证。















 DownLoad:
DownLoad: