-
实时荧光定量PCR(qRT-PCR)是在普通PCR技术基础上发展起来的核酸定量技术,具有灵敏度高、定量准确、特异性强、重复性好及高通量等优点,已被广泛应用于基因表达分析的相关研究中[1-2]。但在实验过程中,目的基因的表达结果经常受RNA的提取质量、反转录合成效率等的影响,使实验结果出现偏差[3],因此需要引入内参基因对基因表达结果进行校正和标准化以消除背景误差的影响[4]。理想的内参基因是指在不同的组织,不同时期,不同环境条件下甚至是不同植物下皆可相对稳定表达的基因。它们是构成细胞的基本转录组成分,参与维持细胞的基本功能,且与要分析的目标基因具有相似的表达水平[5]。但大量实验研究表明,许多内参基因的转录水平会因为植物的发育阶段或实验条件的不同而出现差异,这种差异会影响试验结果的准确性,甚至得出错误的结论[6-7]。因此,在进行qRT-PCR试验前,选择合适的内参基因变得尤为重要。景宁木兰Magnolia sinostellata是木兰科Magnoliaceae木兰属Magnolia新种,落叶灌木,先花后叶,花色艳丽,花瓣数量9~18瓣,观赏价值极高,分布于浙江南部地区海拔1 000 m以上的区域,是浙江省特有种,属于极度濒危物种[8]。景宁木兰性喜温凉湿润、水分充足的环境,但随着全球气候变暖,城市热岛效应的加剧,高温环境严重影响着植物的生长。目前对景宁木兰的研究集中在花色和花芽分化的转录组分析上[9],抗逆性研究鲜有报道。通常情况下,采用遮阳等措施来降低高温对景宁木兰的伤害,但植物自身的抗热胁迫的遗传特性才是起决定作用的因素。因此,研究景宁木兰对高温胁迫的抗逆机制,解析其响应高温的基因表达调控网络,有针对性地发掘相关抗性基因,对景宁木兰的保护、分子育种有积极的意义。目前,尚未见到基于qRT-PCR技术筛选景宁木兰内参基因的相关报道。因此,本研究以热胁迫下景宁木兰1年生实生苗的根、茎、叶组织为材料,选取13个内参基因,利用实时荧光定量PCR(qRT-PCR)技术和geNorm,NormFinder和BestKeeper等3个表达分析软件筛选热胁迫下表达稳定的内参基因,以期为景宁木兰热胁迫下相关目的基因的表达提供依据。
HTML
-
采用的植物材料为种植于浙江农林大学苗圃内景宁木兰1年生实生苗。选取长势一致,无病虫害的植株16盆,分别置于2个人工气候箱内(POX-1000A-12H),气候箱光合光子照度为800 μmol·m-2·s-1,湿度控制在75%~80%,温度分别为25 ℃(对照)和40 ℃(高温处理),光周期为12 h/12 h。分别在处理24和48 h时采取根、叶片和茎段,在液氮中速冻后于-80 ℃中保存,用于RNA的提取。
-
将冻存的样品在液氮中研磨成粉后,用RNAperp Pure Plant Kit(天根,北京)试剂提取材料的总RNA,通过质量浓度为1.2%琼脂糖凝胶电泳检测RNA的完整性,紫外分光光度计来检测RNA的纯度[D(260)/D(280)=1.8~2.1,D(260)/D(230)>1.8]及质量浓度[终质量浓度(mg·L-1)=D(260)×n×40。其中,D(λ)为波长λ处的吸光度,n为稀释倍数]。全长cDNA第1条链的合成按照PrimeScriptTM RT Master Mix(Takara,大连)试剂说明书操作,将得到的cDNA在-20 ℃保存。
-
从前期构建的景宁木兰转录组数据库中,选取13个功能基因作为候选内参基因的荧光定量PCR引物。引物信息见表 1。
基因 基因描述 正向引物 反向引物 ACTIN-7 肌动蛋白基因 ACGAATCCGGTCCATCCATT CCGTTCCACCAGGCAATATG CYP 亲环蛋白基因 ATCTTTAAGTGTGGCCCGCC CAACCGACCCAGATCAGTGC EF-1α 延伸因子1α蛋白基因 GATGATTCCAACCAAGCCCA CACCCACTGCAACAGTCTGG GADPH 甘油醛-3-磷酸脱氢酶基因 TTGAAGACGCCGATCTGGAC CAGCAACGGATTCCATCACC GTB GTP结合蛋白 TTCTGCCTAGCTTCGTCGGA GCGAGAACGCCATAGATCCA NAC NAC域蛋白基因 TTCCATCCAACCGACGAAGA CGGATTTGTACGCATCGAGC NADP 异柠檬酸脱氢酶基因 GAGATGAAATGACCCGCGTT ATCACGATGAGGAAGGCCAA TEF 翻译延伸因子 TCTGAGGTCACAGCCGCTCT GGCCTGATCCTTTCTCCCAG UBC 泛素化酶基因 CGAACTCCCCTGCGAATTCT TCAGTCTGCCGTCCAGCTCT UBQ 多聚泛素酶基因 TCCATGCCCTTAAGCCAAAA AATGACAGAGCGGTCGTGCT α-TUB α微管蛋白基因 TGCCCCGGTTATATCTGCTG TGTACTTCCCATGCCTCGGA β-TUB β微管蛋白基因 AGCAGTTCACCGCCATGTTC GTCTGCCGTTGCATCCTGAT 18S 18S核糖体RNA AGCCTGAGAAACGGCTACCAC ATACGCTATTGGAGCTGGAA CSLD3 纤维素合成酶类似蛋白D-3基因 GGACGGCCACTTCCATTACC GTCTGGCTCCGCATCAACAC KOR 1, 4-β-d-葡聚糖酶基因 CCGGACTTCACCAGCTTCAA GAAGCAAGGAAAGCCGCATT Table 1. Primer sequences of the 13 candidate reference genes and target genes
-
将反转录产物cDNA稀释10倍,使用SYRB Premix Ex Tap Ⅱ(Takara,大连)实时定量PCR试剂盒,在Light Cycler 480 Ⅱ(Roche)实时定量PCR仪进行定量分析。反应体系为SYRB Premix Ex Tap Ⅱ10.0 μL,cDNA模板2.0 μL,上下游引物各0.8 μL(10 μm·L-1),双蒸水(ddH2O)6.4 μL,共20.0 μL体系。每个样品3次重复,扩增反应程序为:95 ℃预变性30 s,95 ℃变性5 s,60 ℃预变性30 s,共40个循环,然后60~95 ℃持续15 s作为溶解曲线分析程序。
-
将12个样品的cDNA等量混合作为模板,按照5倍梯度稀释产物,共5个梯度进行荧光定量PCR,制作标准曲线。利用公式E=(10-1/K-1)×100%计算得出内参引物的扩增效率。其中E为扩增效率,K为标准曲线斜率。R2=(总平方和-残差平方和)/总平方和,总平方和为标准曲线中响应浓度Ct的实际值与平方值的平方差之和,残差平方和为相应浓度下Ct的估计值与实际值的平方差之和[10]。
-
利用geNorm,NormFinder和BestKeeper 3个软件分析13个候选内参基因在不同处理下不同组织中的表达稳定性进行分析,从而得到热胁迫下景宁木兰表达相对稳定的内参基因。其中,BestKeeper软件直接通过计算Ct对每个候选内参基因进行分析;geNorm和NormFinder将Ct根据公式Q=ECt min-Ct sample进行转化,式中E为扩增效率,当其接近100%时,默认为2进行计算[11];Ct min为该基因在所有组织中的最小Ct;Ct sample为该基因在各个组织中的Ct。
1.1. 试验材料
1.2. 方法
1.2.1. 总RNA的提取和反转录cDNA的合成
1.2.2. 候选内参基因及定量引物设计
1.2.3. 内参基因荧光定量PCR扩增程序
1.2.4. 引物扩增效率的计算
1.2.5. 数据分析和处理
-
提取的RNA条带清晰,没有弥散片状或条带消失的现象,说明RNA完整性良好,且没有降解。景宁木兰RNA样品的D(260)/D(230)皆约2.0,>1.8,说明RNA纯度较高,符合后续实验的要求。
-
将12个样品的cDNA等量混合,按照10倍梯度稀释产物作为模板,进行普通PCR和定量PCR扩增。琼脂糖凝胶电泳结果显示:13个引物的产物只有单一条带,无可见引物二聚体,且13个引物的溶解曲线都只有明显的单一峰,说明引物的特异性良好。计算扩增效率结果显示:扩增效率为97.36%~110.91%,UBC最高,GADPH最低,相关系数R2为0.990~0.999(表 2)。符合qRT-PCR试验的要求。
基因 扩增效率/% 线性相关系数R2 曲线斜率 ACTIN-7 110.07 0.994 -3.102 CYP 103.23 0.990 -3.247 EF-1α 106.98 0.998 -3.165 GADPH 97.36 0.994 -3.387 GTB 109.43 0.998 -3.115 NAC 99.27 0.999 -3.340 NADP 100.42 0.998 -3.312 TEF 105.84 0.996 -3.190 UBC 110.91 0.996 -3.086 UBQ 109.28 0.997 -3.118 α-TUB 105.02 0.996 -3.207 β-TUB 110.22 0.999 -3.099 18S 104.54 0.996 -3.218 Table 2. Candidate reference genes amplification specificity
-
利用比较Ct的方法评估13个候选内参基因在不同处理下平均表达水平(图 1)。18S的Ct最小,为4.601,平均表达水平最高;余下12个候选内参基因的Ct为15.869~23.072,表达水平相差不大。
-
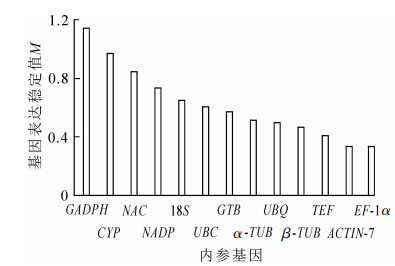
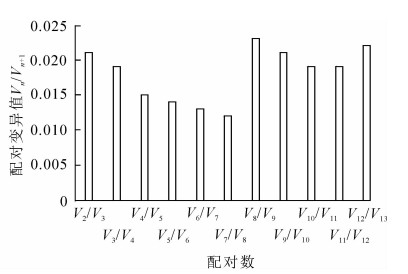
geNorm软件根据计算得到的平均表达稳定指数M来进行稳定性排序,M越大表明基因越不稳定,以M=1.5为标准,M<1.5认为是理想内参基因的稳定表达标准。同时,该软件还通过内参基因标准化的配对差异分析(Vn/Vn+1)来得到内参基因的合适数量,软件默认以Vn/Vn+1=0.15为标准,当Vn/Vn+1<0.15时,表明n个内参基因已经可以稳定作为内参基因,没必要引入第n+1个基因[12]。通过图 2可知:13个基因的M为0.050 0~0.200 0,<1.5,说明13个候选内参基因都达到了作为内参基因的标准,其中ACTIN-7和EF-1α最为稳定,M=0.334 49。GADPH的M最大,为1.141 66,是所有候选内参基因中表达最不稳定的。图 3表明:V2/V3为0.021<0.15,说明2个基因可稳定作为内参基因,结果可靠,无需引入第3个基因。进一步分析表明:ACTIN-7和EF-1α为最佳内参组合。
BestKeeper软件通过计算内参基因Ct的标准偏差(SD)对内参基因的的稳定性进行排序,标准偏差越小表明内参基因的稳定性越好,如果标准偏差>1,则认为该基因不稳定[8]。表 3显示:在不同组织中,NADP和GADPH的标准偏差大于1,说明这2个基因表达不稳定。余下的11个内参基因的标准偏差皆小于1,其中UBC,EF-1α,ACTIN-7和GTB稳定性最好,排名最高。
基因名 NormFinder BestKeeper 稳定值 排名 标准偏差 排名 UBQ 0.107 1 0.643 11 8 EF-1α 0.112 2 0.407 26 2 TEF 0.127 3 0.508 73 5 GTB 0.134 4 0.488 21 4 UBC 0.134 5 0.338 45 1 18S 0.142 6 0.576 29 7 NADP 0.146 7 1.082 30 12 ACTIN-7 0.147 8 0.418 78 3 NAC 0.149 9 0.964 34 11 β-TUB 0.159 10 0.518 46 6 GADPH 0.161 11 1.629 78 13 α-TUB 0.107 12 0.739 43 9 CYP 0.112 13 0.959 90 10 Table 3. Reference genes expression stability and ranking by BestKeeper and NormFinder
NormFinder软件通过计算组内和组间的方差得到表达稳定值,并对候选内参基因进行排序,稳定值越小,表明内参基因越稳定。表 3显示:UBQ,EF-1α和TEF分别排在前3位,表达稳定值较为接近,表达最为稳定;CYP的稳定值最大,为0.237,表达稳定性最差。
-
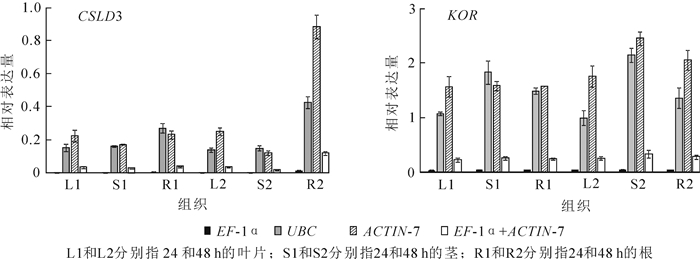
综合以上3个软件分析的结果,本研究选择了稳定性较好的3个基因(UBC,EF-1α和ACTIN-7)和1个基因组合(EF-1α和ACTIN-7)。通过分析CSLD3和KOR基因在景宁木兰不同组织的表达模式来进一步验证筛选的候选内参基因的稳定性。CSLD3是植物合成根毛细胞壁的重要纤维素合成酶基因,可以引起细胞壁形成的缺陷[13]。研究表明此基因在根部中表达最高[14]。KOR基因是植物合成细胞壁中发挥重要作用的纤维素合成酶基因,在茎中和木质部中表达量最高[6, 15]。对3个内参基因进行相对表达量量化(图 4),CSLD3在根中表达最高,KOR在茎中表达最高。以EF-1α和ACTIN-7内参基因组合标准化,目的基因也显示出一致的表达水平,进一步表明了这3个内参基因的可靠性。
2.1. 景宁木兰RNA的质量检测
2.2. 景宁木兰候选内参基因引物特异性及扩增效率检测
2.3. 景宁木兰候选内参基因表达水平分析
2.4. 景宁木兰候选内参基因表达稳定性分析
2.5. 景宁木兰候选内参基因表达稳定性验证
-
SPIEGELAERE等[16]研究认为软件间的算法和判断标准不同导致结果的差异,因此在评估内参基因时应先综合分析各软件的结果,再进行比较排序,最终筛选出最适宜的内参基因。理想的内参基因应在各种类型的组织、细胞和各种实验因素条件下均恒定表达[17-18]。但大量实验表明:内参基因只是在一定的范围内可以稳定表达,在特定的实验条件下,应该筛选特定的内参基因来进行实验[19]。因此,本研究选择了景宁木兰在热胁迫下的根、茎、叶组织进行内参基因的筛选研究。得到UBC,EF-1α和ACTIN-7为热胁迫下不同组织的最佳内参基因,这3个内参基因均是传统内参基因,其中UBC基因是泛素蛋白翻译后修饰的的基因,参与细胞多种调控过程,如细胞周期,细胞的增值与分化,信号转导等[20],在桂花Osmanthus fragrans花芽的不同发展阶段[21]、桉树Eucalyptus robusta不同组织[22]中均表达稳定;ACTIN是细胞的骨架成分,几乎在所有的真核细胞中均有表达[23],在杨树Populus tomentosa不同光周期和低温条件下的茎尖、幼叶、成熟叶和树皮组织中、油菜Brassica campestris的营养组织中以及番茄Lycopersicon esculentum的叶子和根组织中也表现出最佳的稳定性[24-26]。EF-1α是一种重要的多功能蛋白,参与细胞转导、翻译控制、骨架组成等多种重要的细胞学过程[27]。ACTIN和EF-1α在毛果杨Populus trichocarpa不同组织为材料[11]和梧桐Firmiana platanifolia种子的不同发育阶段[28]中均表现出较高的稳定性。SILVEIRA等[29]在一种重要牧草珊状臂形草Brachiaria brizantha的内参基因筛选中同样得到EF-1α稳定性最佳的结果。
理想的内参基因的表达水平应是适中的,不宜过高,也不宜过低,Ct应为15~30[30]。本研究中18S rRNA基因的Ct远低于15,表达丰度过高,影响其稳定性,不适宜作为内参基因。在光皮桦Betula luminifera的根、茎、叶和种子等不同组织中也出现了18S的Ct过低的情况[10]。在牡丹Paeonia suffruticosa不同组织不同花期[31]以及水稻Oryza sativa受到稻纵卷叶螟Chaphalocrocis medinalis虫害诱导后内参基因研究[32]中也得到了相同结果。而周良云等[33]对黄花嵩Artemisia annua进行不同浓度的镉处理,最终选取了ACTIN-7,18S和PAL为内参基因来校正目的基因的表达量;苏晓娟等[11]也发现毛果杨在锌胁迫下18S rRNA稳定性较好,可用作内参基因。进一步表明了不同植物在不同条件下,内参基因稳定性差异较大。
研究中选择2个或2个以上的基因可以有效增加结果的可靠度及精确度,以此来减弱单一内参基因可能造成的结果偏差[17]。刘文哲等[10]在光皮桦中通过目的基因的表达分析验证内参基因的可靠性,结果显示了3个内参基因及其组合对目的基因定量标准化差异较小。PARK等[34]在不同品种甘薯Dioscorea esculenta的非生物胁迫条件下,使用单个稳定的内参基因ARF和最佳内参基因组合(ARF/COX)对2个目的基因IbLEA14和swpa2进行表达验证,结果也显示了单个内参基因和组合验证对目的基因表达水平的校准结果基本一致。本研究以EF-1α和ACTIN-7为组合内参基因进行表达验证,发现单个内参EF-1α与其有微小差异,2个基因组合可使基因定量结果更加准确。但是与单个基因做内参相比,2个或2个以上的基因做内参需要增加更多的样本量和耗费大量成本,且实验结果并无明显差异。因此,在实验过程中可综合考虑后再进行选择。














 DownLoad:
DownLoad: