-
密码子承担着生物体内遗传信息传递的重要功能,是DNA转录与翻译、蛋白质合成与表达过程中的关键单元。在生物体共用的一套密码子中,终止密码子不编码氨基酸,甲硫氨酸(Met)和色氨酸(Trp)分别由1种密码子编码。其余59个密码子具有简并性,即1种氨基酸可由2~6个密码子对应编码,编码相同氨基酸的密码子即为同义密码子[1]。基因并非完全随机地使用同义密码子,而是存在一定的偏好性。特定的密码子偏好性是生物体长期适应性进化的结果,能够反映生物对环境的分子适应机制[2]。分析密码子偏好性及其影响因素,对生物遗传育种、进化基因组学以及系统发育学研究具有深远的意义。1,5-二磷酸核酮糖羧化/加氧酶(Ribulose-1,5-bisphosphate carboxylase/oxygenase, Rubisco酶)是植物叶绿体基质中参与光合作用的关键酶,约占可溶性蛋白质总量的50%[3]。Rubisco酶具有催化1,5-二磷酸核酮糖(Ribulose-1,5-disphosphate, RuBP)与二氧化碳(CO2)羧化反应和光呼吸中RuBP与氧气(O2)加氧反应的双重活性,对净光合率有决定性影响[4]。Rubisco酶由8个大亚基(催化亚基)和8个小亚基(调节亚基)组成,前者是固定CO2的活性位点和催化位点,由叶绿体基因组大单拷贝区的rbcL基因编码[5-6]。环境的变化会导致rbcL基因产生适应性进化,从而影响植物光合效率[7]。因此,研究rbcL基因的密码子使用模式有利于理解高等植物对环境的适应机制。千屈菜科Lythraceae包括许多重要的园林植物,具有重要的观赏价值和经济价值[8]。目前,rbcL基因在千屈菜科中的研究应用仅局限于系统发育[9-10],对于该科密码子使用偏好性的相关研究尚未见报道。本研究选取了千屈菜科具有代表性的10属20种植物,分析rbcL基因的碱基组成、密码子使用偏好性及其影响因素,并与模式物种进行比较,为该科物种rbcL基因异源高效表达提供理论基础。
HTML
-
20条rbcL基因全长编码区序列(CDS)数据来源于美国国家生物技术信息中心(NCBI)的GenBank数据库(https://www.ncbi.nlm.nih.gov/),详见表1。
物种 GenBank登录号 CDS位置 物种 GenBank登录号 CDS位置 萼距花 Cuphea hyssopifolia MN833211 58955~60382 南洋紫薇 Lagerstroemia siamica MK881628 55129~56556 八宝树 Duabanga grandiflora MK881638 56823~58250 绒毛紫薇 Lagerstroemia tomentosa MK881632 54873~56300 黄薇 Heimia myrtifolia MG921615 58612~60039 西双紫薇 Lagerstroemia venusta MK881630 55159~56586 副萼紫薇 Lagerstroemia calyculata MK881636 54873~56300 散沫花 Lawsonia inermis MK881631 58836~60263 川黔紫薇 Lagerstroemia excelsa MK881635 54910~56337 千屈菜 Lythrum salicaria MK881629 59099~60526 屋久岛紫薇 Lagerstroemia fauriei NC_029808 54810~56237 石榴 Punica granatum NC_035240 59017~60444 多花紫薇 Lagerstroemia floribunda NC_031825 54776~56203 圆叶节节菜 Rotala rotundifolia MK881626 58835~60262 桂林紫薇 Lagerstroemia guilinensis NC_029885 54697~56124 细果野菱 Trapa maximowiczii NC_037023 58322~59770 云南紫薇 Lagerstroemia intermedia NC_034662 54948~56375 欧菱 Trapa natans MK881634 58387~59814 福建紫薇 Lagerstroemia limii MK881627 54830~56257 虾子花 Woodfordia fruticosa MK881637 59444~60871 Table 1. Information of rbcL genes from 20 Lythraceae species
-
通过CodonW 1.4.4软件和在线工具EMBOSS explorer中的CUSP和CHIPS程序,统计rbcL基因密码子末端各类型碱基含量(A3s、T3s、C3s和T3s)、GC总含量(GC)、密码子各位点GC含量(GC1s、GC2s和GC3s)、有效密码子数(ENC)和密码子适应指数(CAI)。利用SPSS 22.0软件,选用皮尔森相关系数评估碱基组成和密码子偏好性相关显著水平[11]。
-
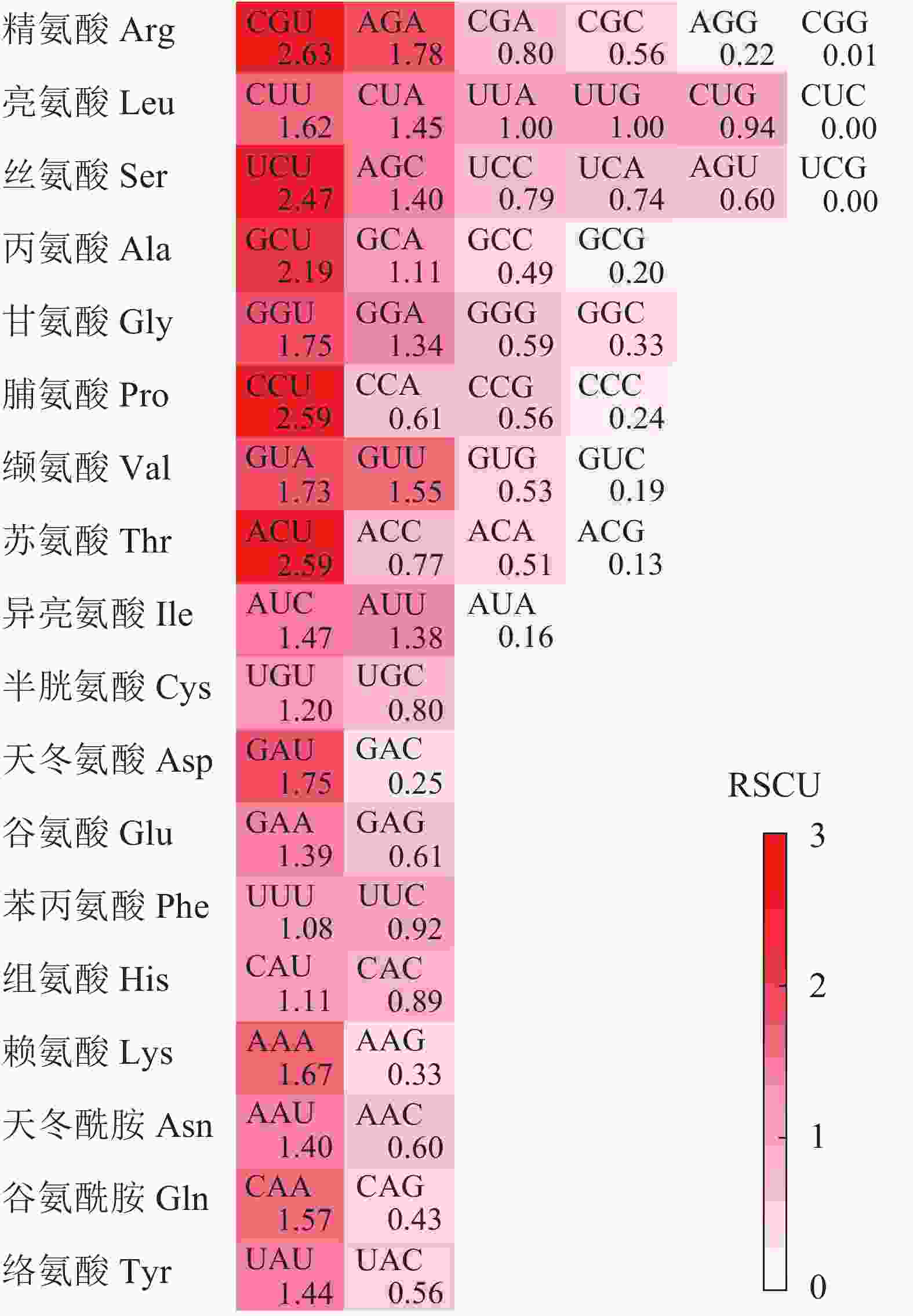
同义密码子相对使用度(RSCU)是同义密码子的实际使用频次与无使用偏好性时期望频次的比率,去除了碱基成分对密码子使用产生的影响。RSCU>1,表示该密码子在同义密码子中使用相对较多;RSCU=1,表示该密码子在同义密码子中使用无偏好性;RSCU<1表示该密码子在同义密码子中使用相对较少[12]。通过CodonW 1.4.4软件计算千屈菜科植物的RSCU,并利用TBtools 0.6软件绘图。
-
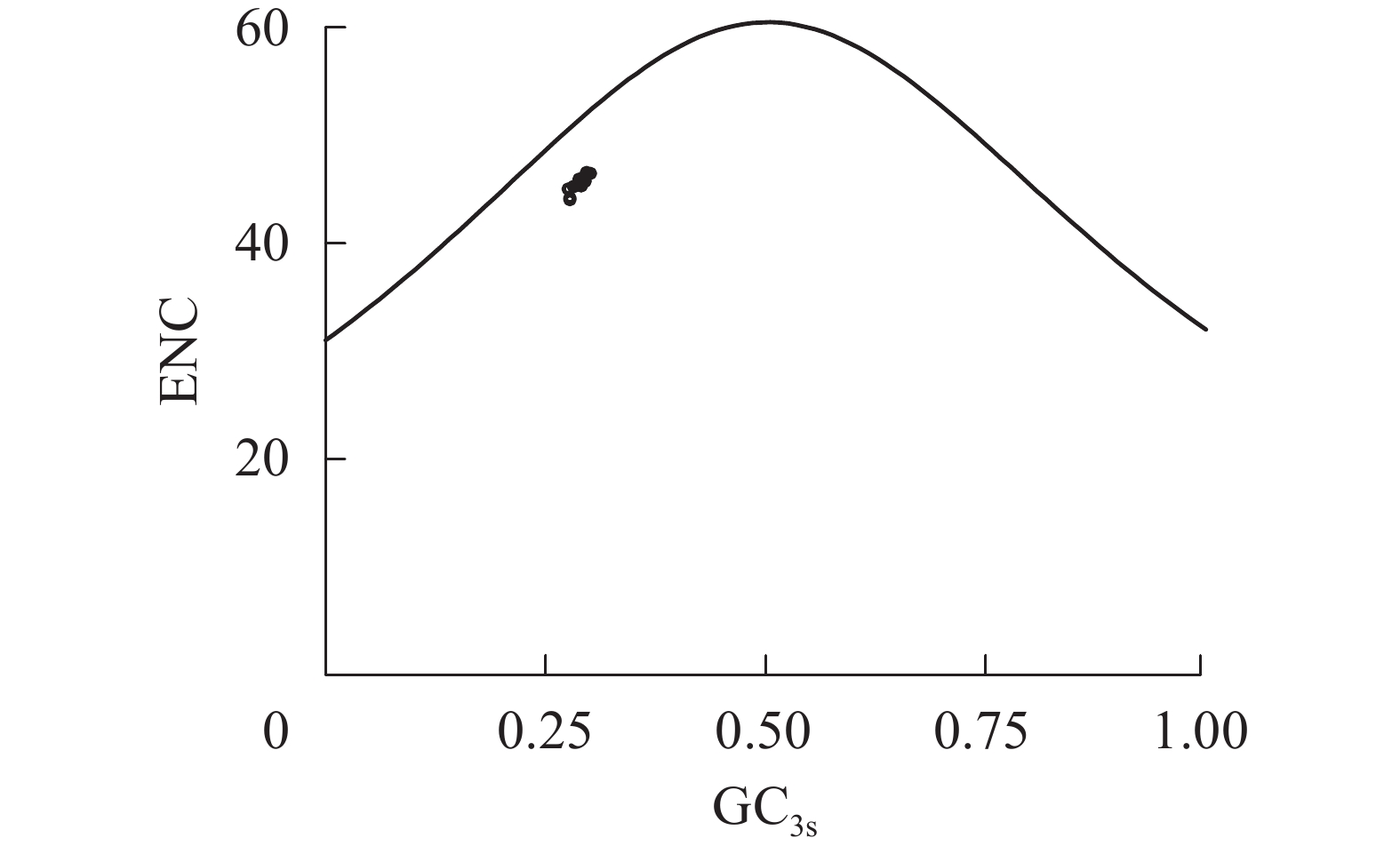
以GC3s和ENC为横、纵坐标,通过Origin 9.1绘制ENC-GC3s散点图。标准曲线为ENC期望值,即NENC=2+MGC3s+29/[MGC3s2+(1−MGC3s)2],其中NENC表示有效密码子数,MGC3s表示密码子第3位碱基平均GC含量,该公式的成立表示密码子的偏好性仅受突变压力约束[13],此条件下,散点应位于标准曲线上部或紧贴标准曲线下部;当散点分布于曲线下方较远距离的区域时,表明除突变压力作用外,选择压力对偏好性产生主要影响。
-
以GC3s为横坐标,密码子第1、2位点GC含量平均值(GC12)为纵坐标,利用Origin 9.1绘制散点图并做线性回归分析,分析密码子不同位点碱基组成差异性[14]。当回归曲线斜率趋近1时,密码子各位点碱基成分差异不大,偏好性主要受到突变的影响;当斜率趋近0时,密码子第3位点和第1、2位点碱基变异模式差异较大,偏好性主要受到选择压力影响。
-
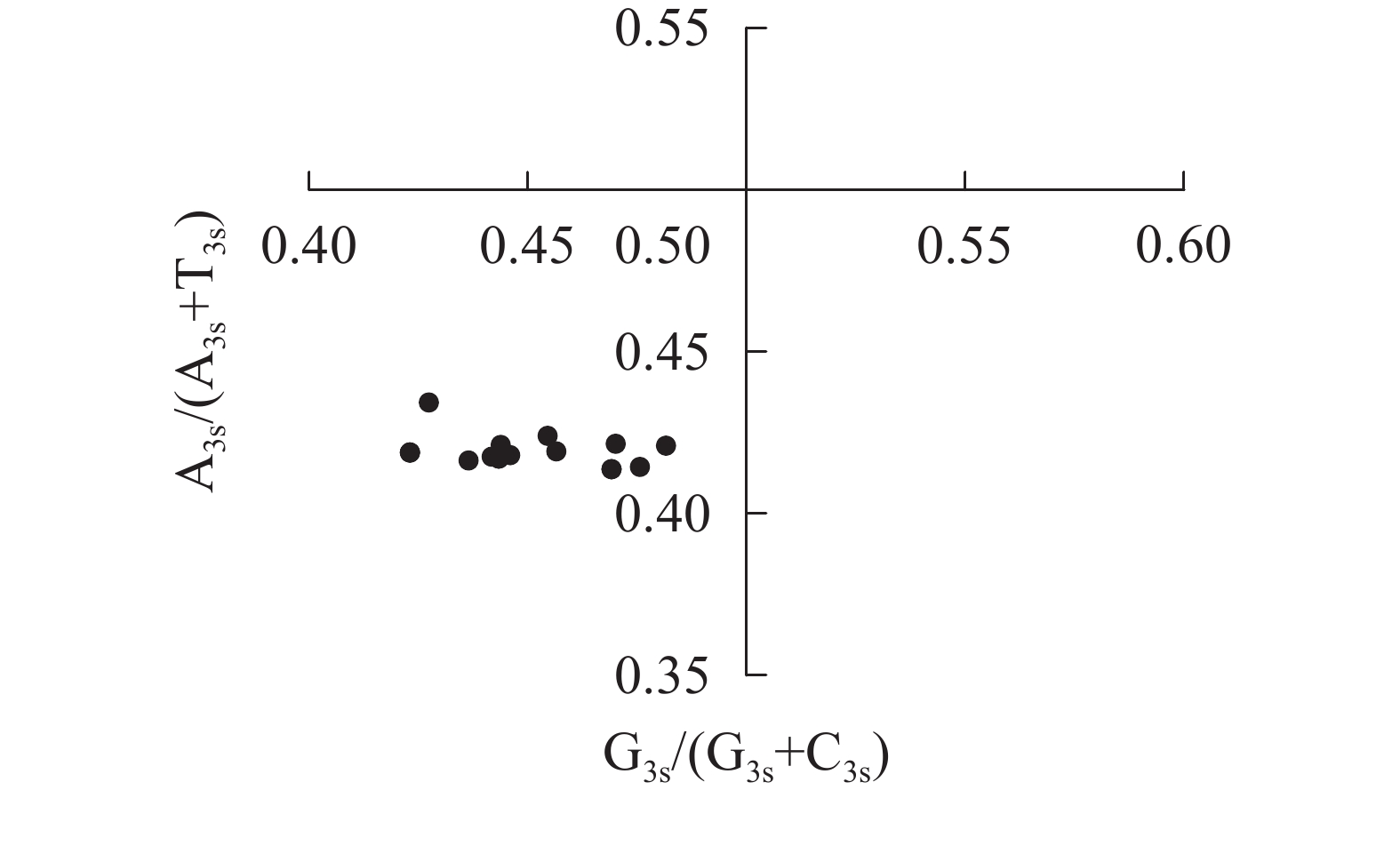
奇偶偏差分析可评估密码子第3位点嘌呤和嘧啶组成偏差对密码子使用偏好性的影响[15]。以G3s/(G3s+C3s)和A3s/(A3s+T3s)为横、纵坐标,利用Origin 9.1绘制奇偶偏差图,交点(0.50, 0.50)表示无碱基突变和选择压力下,A=T且G=C。
-
参照巫伟峰等[16]方法,以59个密码子(去除AUG、UGG和3个终止密码子UAA、UAG、UGA)的RSCU为变量,20条CDS为个体,通过SPSS进行系统聚类,类间距离为组内联接法,基因间距离为平方欧式距离。分别利用DAMBE 5.2.73和MEGA-X软件对CDS进行碱基替换饱和度检测和总体平均距离(d)计算,同时满足替换饱和度指数(Iss)小于饱和度标准指数(Iss.c),即Iss<Iss.c,表明碱基替换未饱和,且P=0.000和0<d<1后,通过MEGA-X软件邻接法(NJ)构建系统发生树,重复1 000次。
-
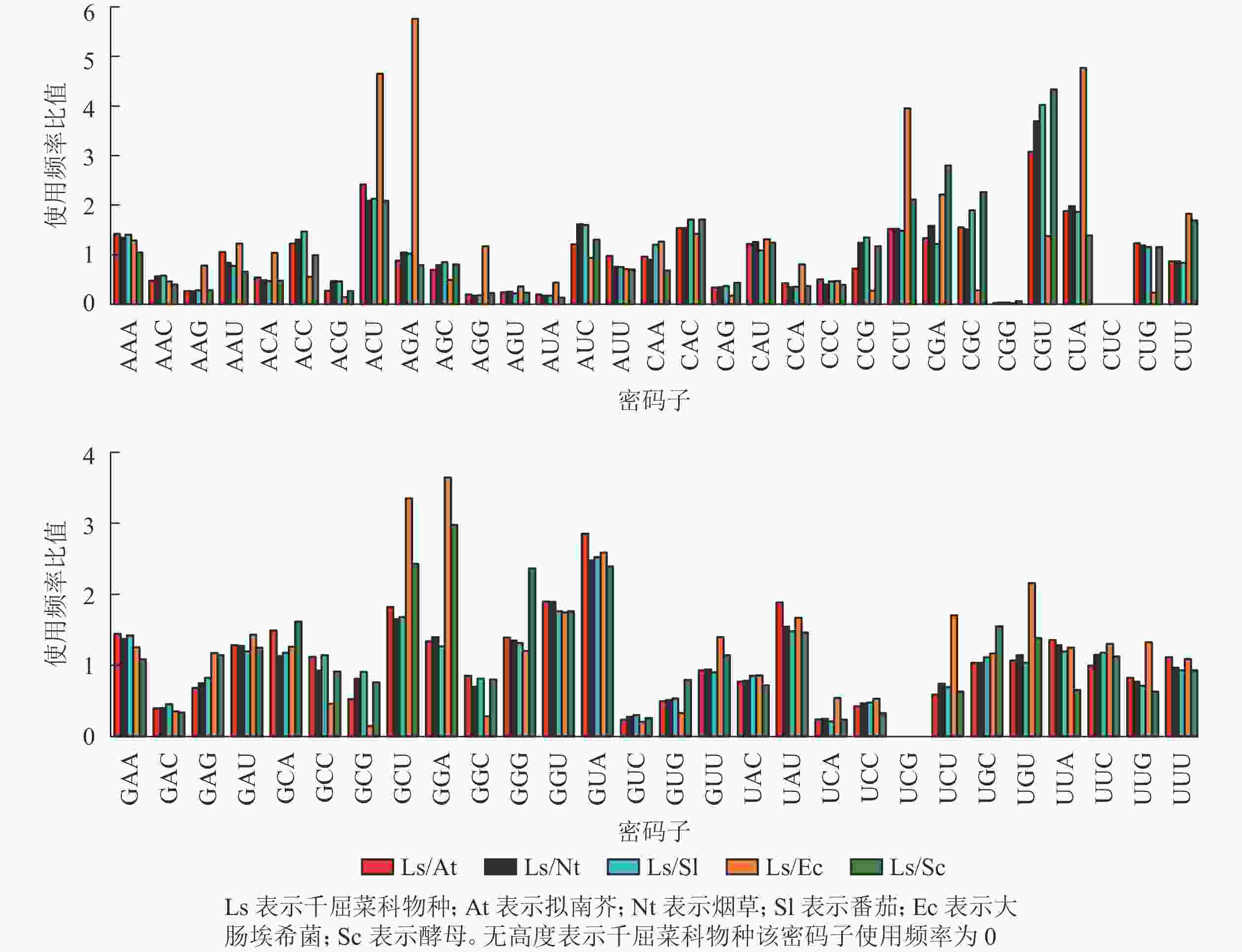
密码子相对使用频率比值是评估不同生物密码子使用偏好性差异程度的重要参数。当比值为0.5~2.0时,认为物种密码子偏好性差异较小[17]。拟南芥Arabidopsis thaliana、烟草Nicotiana tabacum、番茄Solanum lycopersicum、大肠埃希菌Escherichia coli和酵母Saccharomyces cerevisiae的基因组密码子使用频率来源于密码子使用数据库(http://www.kazusa.or.jp/codon/)。千屈菜科物种整体密码子平均使用频率通过EMBOSS explorer中CUSP计算获得[18]。利用Origin 9.1进行绘图。
1.1. 基因序列和密码子使用频率数据获取
1.2. CDS碱基组成和密码子使用偏好性参数统计
1.3. 同义密码子相对使用度统计与分析
1.4. ENC绘图分析
1.5. 中性绘图分析
1.6. 奇偶偏差(PR2)分析
1.7. 基于RSCU和CDS的聚类分析
1.8. 密码子使用频率比较分析
-
从表2可见:GC含量为0.425~0.437,平均为0.431。结合密码子各位点GC含量(GC1s为0.567~0.582,平均0.573;GC2s为0.429~0.437,平均0.432;GC3s为0.275~0.300,平均0.288),表明rbcL基因CDS在组成上更倾向于使用A/T碱基。第3位点各类型碱基含量从大到小依次为T3s、A3s、C3s、G3s,表明rbcL基因更偏向于使用A/T碱基结尾的密码子。
物种 A3s T3s G3s C3s GC GC1s GC2s GC3s CAI ENC 萼距花 0.376 0.531 0.157 0.173 0.435 0.582 0.437 0.286 0.276 45.392 八宝树 0.380 0.526 0.152 0.180 0.431 0.571 0.433 0.288 0.278 45.942 黄薇 0.390 0.508 0.145 0.194 0.434 0.571 0.433 0.296 0.283 46.540 副萼紫薇 0.377 0.525 0.148 0.186 0.432 0.576 0.429 0.292 0.277 45.635 川黔紫薇 0.376 0.526 0.149 0.187 0.432 0.571 0.431 0.294 0.275 45.743 屋久岛紫薇 0.379 0.529 0.146 0.184 0.431 0.571 0.431 0.290 0.272 45.659 多花紫薇 0.378 0.526 0.148 0.184 0.432 0.576 0.429 0.292 0.276 45.625 桂林紫薇 0.376 0.526 0.149 0.187 0.432 0.571 0.431 0.294 0.275 45.743 云南紫薇 0.379 0.526 0.140 0.191 0.431 0.571 0.431 0.290 0.275 45.340 福建紫薇 0.379 0.531 0.142 0.184 0.430 0.571 0.431 0.288 0.274 45.564 南洋紫薇 0.379 0.526 0.140 0.191 0.431 0.571 0.431 0.290 0.275 45.340 绒毛紫薇 0.377 0.525 0.148 0.186 0.432 0.576 0.429 0.292 0.277 45.635 西双紫薇 0.379 0.526 0.140 0.191 0.431 0.571 0.431 0.290 0.275 45.340 散沫花 0.379 0.536 0.151 0.171 0.429 0.569 0.435 0.282 0.276 45.264 千屈菜 0.389 0.535 0.138 0.173 0.428 0.576 0.433 0.275 0.285 45.007 石榴 0.381 0.518 0.153 0.184 0.436 0.578 0.437 0.294 0.275 46.153 圆叶节节菜 0.379 0.536 0.151 0.171 0.429 0.569 0.435 0.282 0.276 45.264 细果野菱 0.387 0.532 0.154 0.165 0.425 0.567 0.431 0.277 0.274 44.181 欧菱 0.387 0.532 0.154 0.165 0.426 0.569 0.431 0.277 0.274 44.029 虾子花 0.376 0.516 0.163 0.184 0.437 0.576 0.435 0.300 0.270 46.458 Table 2. Base composition and codon usage characteristics of rbcL genes from 20 Lythraceae species
ENC和CAI是衡量密码子使用偏好性程度的主要指标。ENC从20(氨基酸只由1种同义密码子编码)至61(同义密码子的使用没有偏好性),越接近20偏好性越强。一般认为,ENC<35表示密码子的使用偏好性较强[19]。20种千屈菜科植物ENC为44.029~46.540,平均45.493,分布范围较小且均远大于35,表明rbcL基因整体偏好性不强。CAI取值0~1,越接近1密码子偏好性越强[20]。20种植物CAI为0.270~0.285,平均0.276,同样说明偏好性强度不大。一般情况下,基因的密码子使用偏好性越强,在生物体内的表达水平越高[21],可推测rbcL基因在千屈菜科植物中表达水平较低。
-
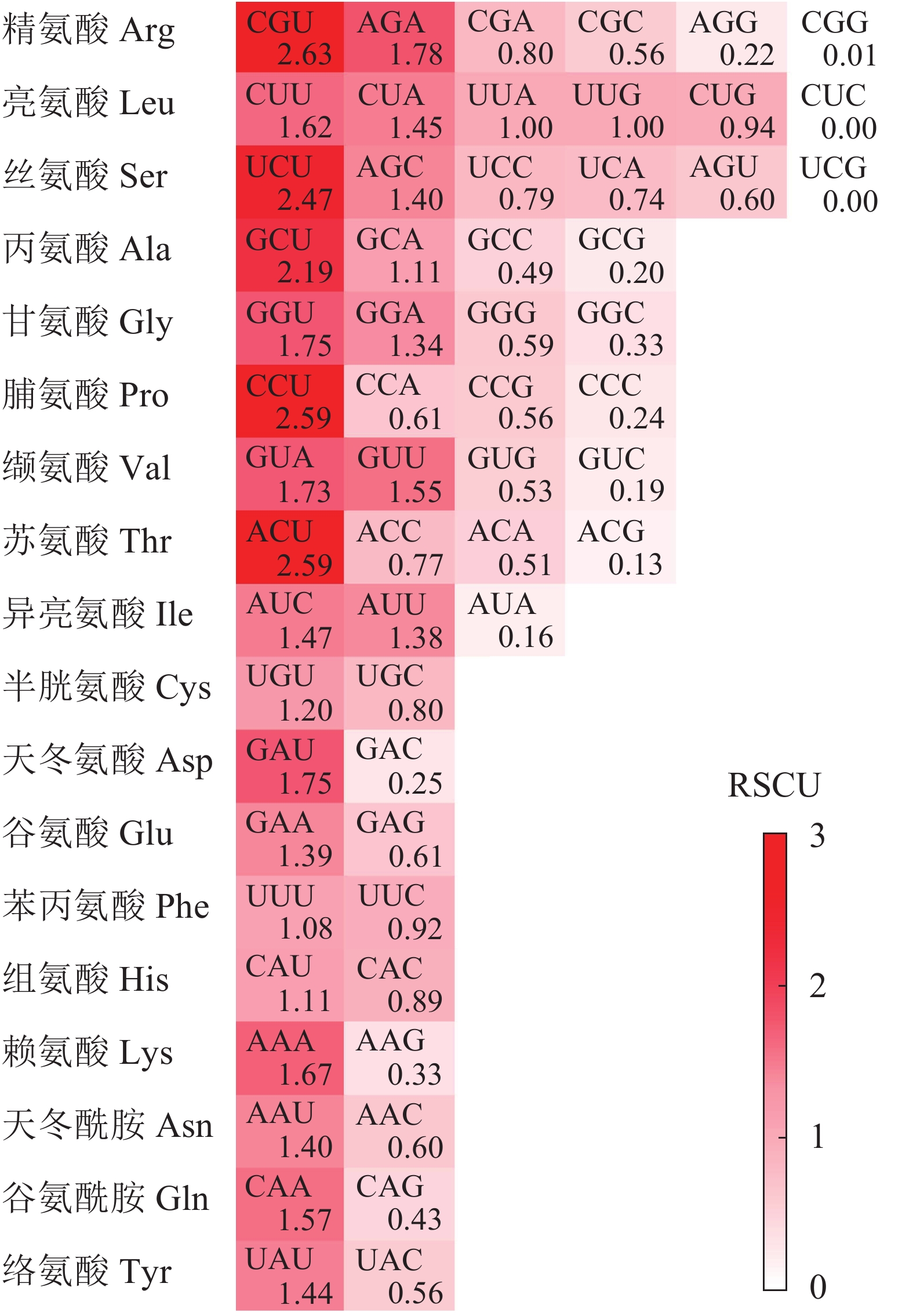
图1显示:在25个高频密码子(RSCU>1)中,23个以A/U结尾,仅2个由C(AUC和AGC)结尾。其中RSCU最高的5个密码子(RSCU>2)末尾均为U碱基,表明rbcL基因CDS对于末端A/U(T)密码子具有的使用偏好性。
-
相关分析(表3)表明:ENC和GC、GC3s在0.01水平上显著相关(Pearson相关系数分别为0.855和0.856),表明碱基组成,尤其是密码子第3位点碱基类型对千屈菜科rbcL基因的密码子偏好性有明显影响。GC3s和GC12相关不显著,说明不同位点组成上关联不大,碱基变异模式存在差异,rbcL基因较保守,突变偏性较小。
参数 CAI ENC GC GC1s GC2s GC3s ENC 0.062 GC − 0.136 0.855** GC1s 0.138 0.403 0.712** GC2s 0.029 0.229 0.348 0.314 GC3s − 0.264 0.856** 0.846** 0.324 − 0.074 GC12 0.112 0.403 0.684** 0.869** 0.743** 0.190 说明:**表示在0.01水平上显著相关(双尾) Table 3. Correlation between base composition and codon usage bias
-
图2显示了rbcL基因ENC和GC3s的关系。所有散点分布在标准曲线下方一定距离处,表明千屈菜科植物rbcL基因的密码子偏好性除了受到碱基突变压力外,更主要受自然选择压力的约束;散点集中分布在较小范围内说明自然选择压力强度相近。
-
中性分析结果(图3)显示:所有散点均落在直线y=x(GC12)上方。GC3s与GC12的回归曲线(斜率为0.069 4,R2=0.036 1)近似平行于X轴,表明千屈菜科植物rbcL基因密码子第1、2位点与第3位点碱基类型相差较大。结合表3,GC3s与GC12相关性较低(Pearson相关系数为0.190),说明碱基突变对于密码子第3位点的作用比第1、2位点弱,密码子偏好性主要受自然选择压力的作用,受突变压力的影响则较小。
-
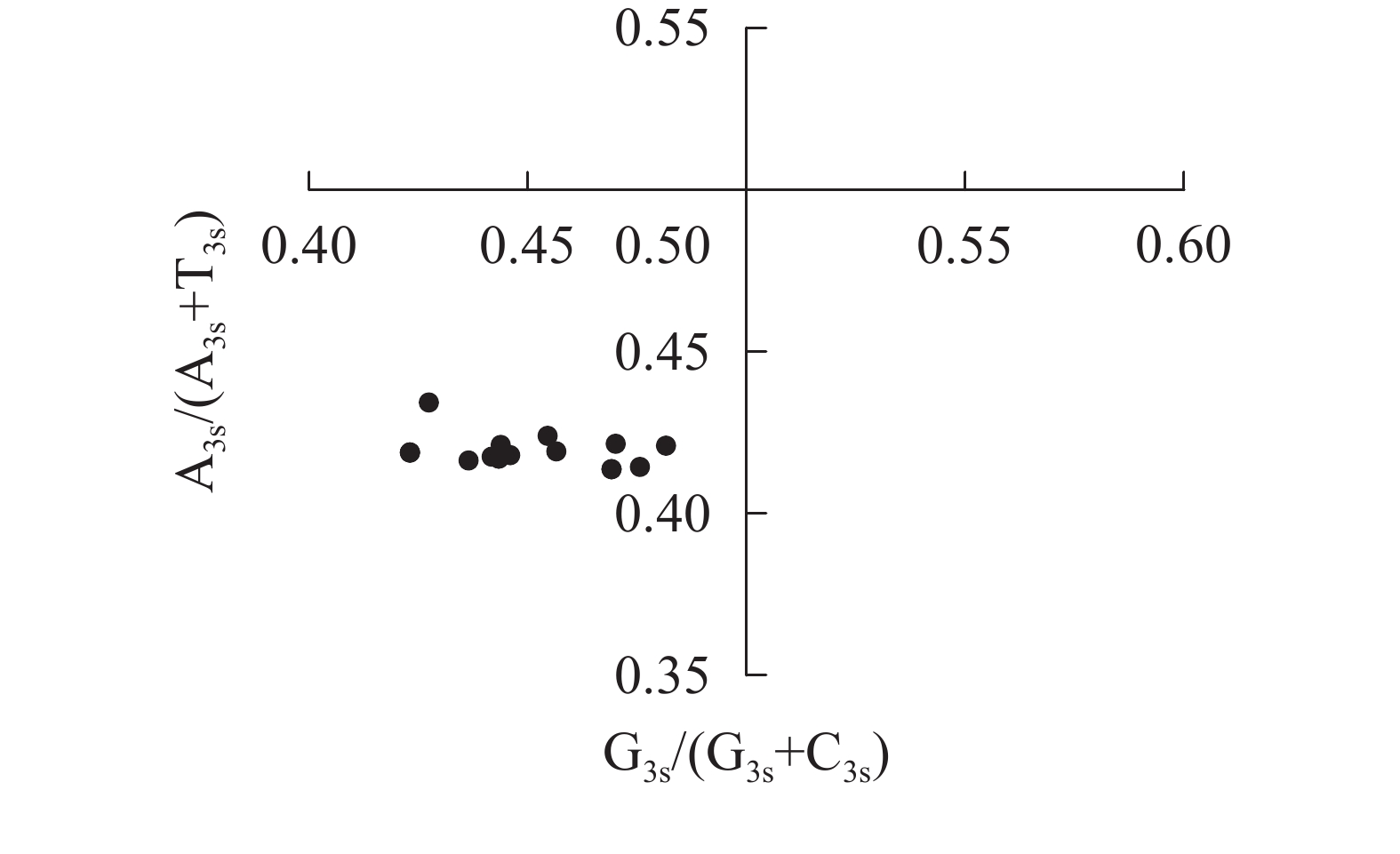
图4显示:当密码子偏好性只受碱基突变影响时,密码子第3位点上嘌呤和嘧啶含量应相同,即A3s=T3s或C3s=G3s[22]。所有散点均明显偏离交点(0.50, 0.50),且都分布在左下象限[G3s/(G3s+C3s)<0.5,A3s/(A3s+T3s)<0.5],密码子第3位点上嘧啶含量高于嘌呤[(A3s+G3s)<(T3s+C3s)]。4种碱基在密码子第3位点上分布不均匀,说明相较于碱基突变压力,自然选择压力对rbcL密码子偏好性有更强的影响。
-
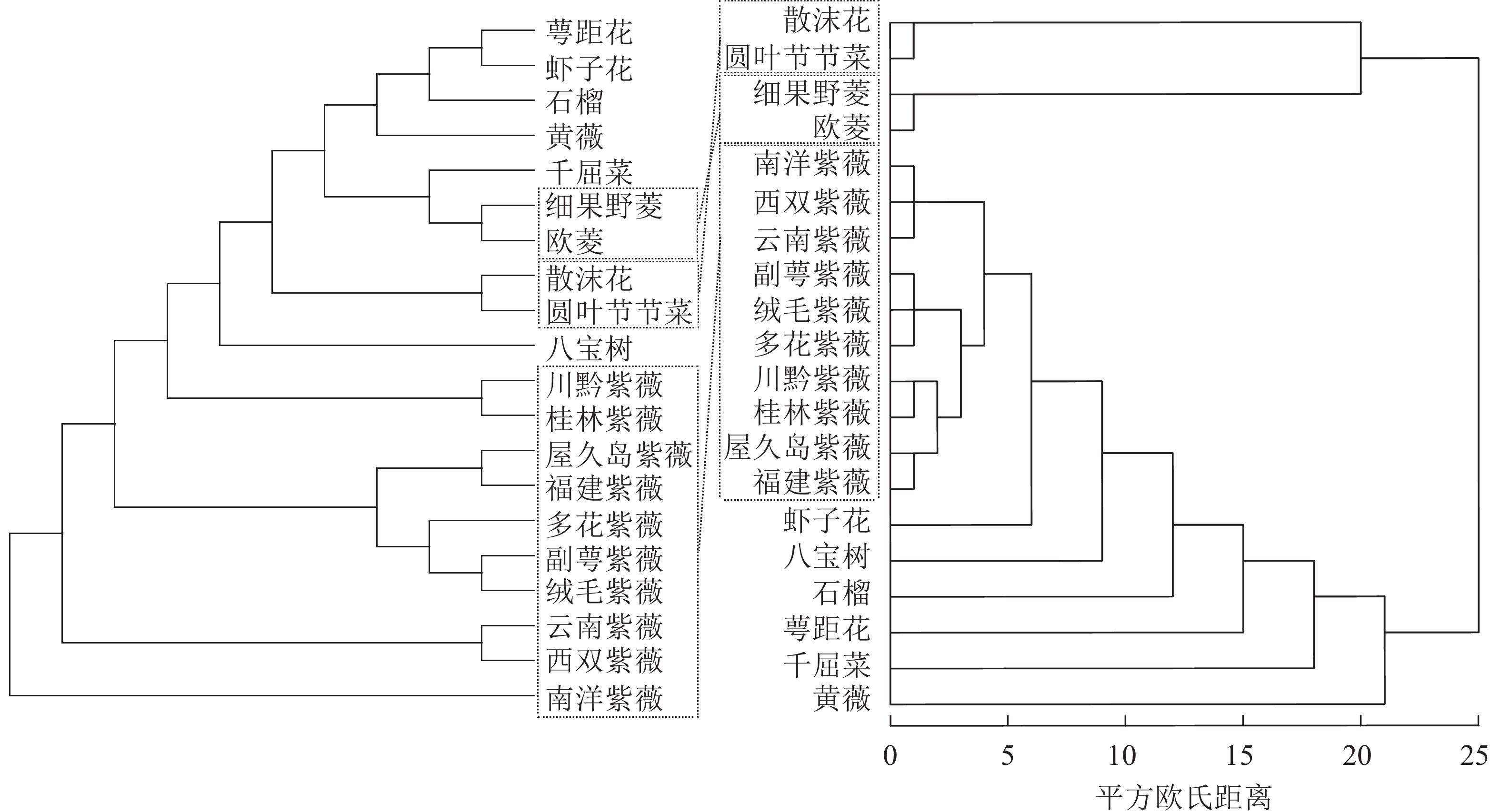
20条CDS碱基替换未饱和(Iss=0.025 3,Iss.c=0.785 2,P=0.000),总体平均遗传距离为0.2。系统聚类树状图和邻接树均将20种千屈菜科植物聚成了4~5个支系(图5),说明不同支系的植物密码子使用特性存在一定区别。虽然两者在部分支系的内部结构上存在较大矛盾,但在支系水平(属)上,两者对10个紫薇属Lagerstroemia植物、散沫花和圆叶节节菜以及2个菱属Trapa植物之间的聚类结果相对一致,说明基于密码子RSCU的系统聚类能在某种程度上反映千屈菜科植物属间水平的亲缘关系,即不同植物密码子的使用偏好性与亲缘关系存在局部对应。
-
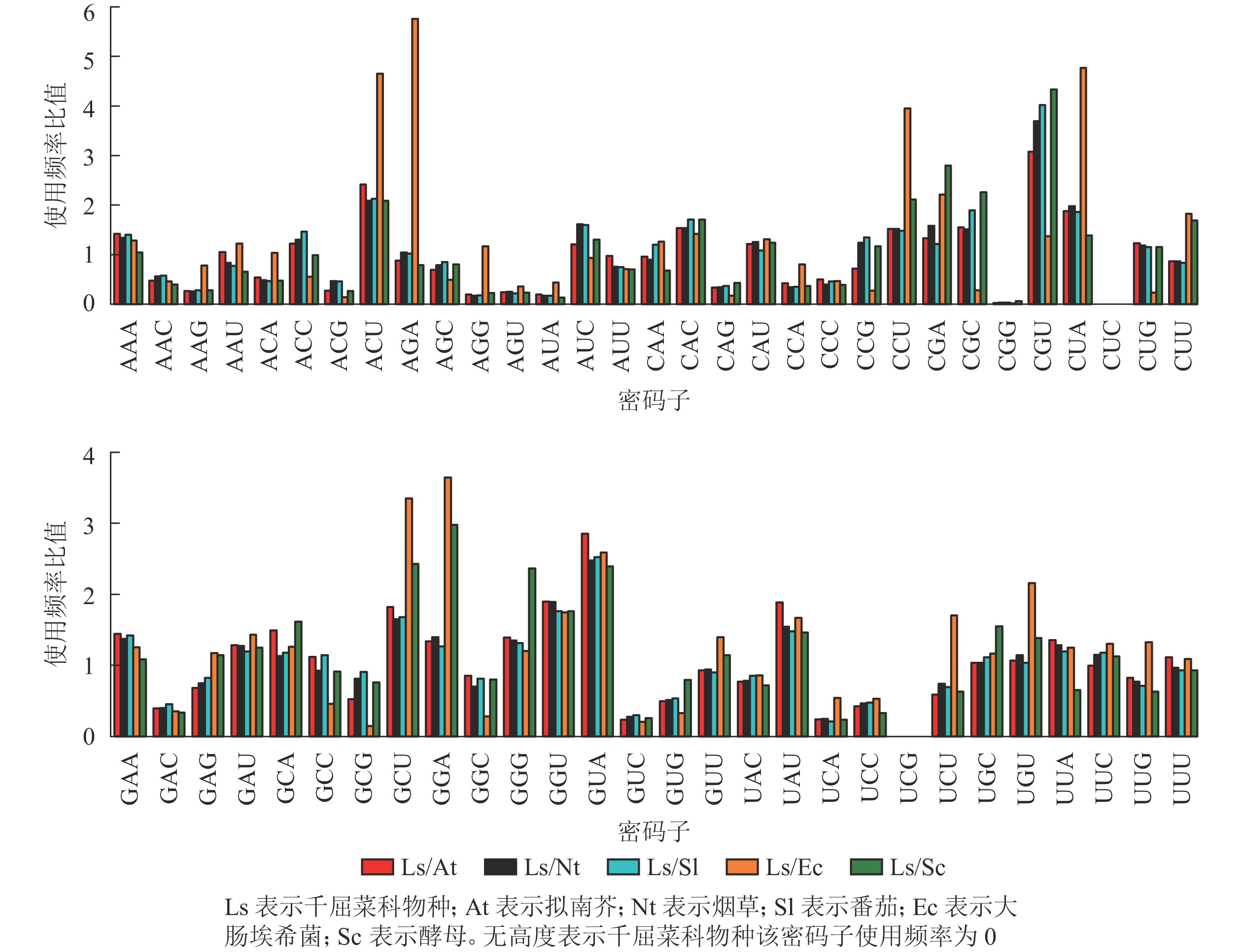
从图6可以看出:与千屈菜科植物rbcL基因密码子平均使用频率相比,大肠埃希菌有28个密码子相差较大,最大值5.76(AGA);酵母有26个密码子相差较大,最大值4.33(CGU),说明酵母更适合作为千屈菜科植物rbcL基因异源表达的受体。拟南芥、烟草和番茄分别存在20、19和17个使用频率相差较大的密码子,且最大值均出现在CGU,初步说明相较于拟南芥和烟草,番茄更适合作为千屈菜科植物rbcL基因遗传转化的受体。
2.1. rbcL基因碱基组成和密码子使用偏好性
2.2. rbcL基因同义密码子相对使用度分析
2.3. 密码子碱基组成和使用偏好相关分析
2.4. ENC绘图分析
2.5. 中性绘图分析
2.6. 奇偶偏差(PR2)分析
2.7. 基于RSCU和CDS的聚类分析
2.8. 千屈菜科植物与模式物种密码子使用频率比较分析
-
特定的密码子使用偏好性是生物对环境变化适应性的体现,不同物种、不同功能基因的密码子偏好性存在明显差异。大部分双子叶植物密码子偏好A/T碱基结尾,单子叶植物则偏好G/C结尾[23],与本研究中千屈菜科植物rbcL基因密码子A3s+T3s远远大于G3s+C3s的偏好性结果一致。李国灵等[13]对红藻门Rhodophyta植物rbcL基因密码子偏好性研究也得到了类似结果,虽然红藻科和千屈菜科植物生活型、生理特性等相差较大,但千屈菜科也包括许多水生或湿生植物。两者研究结果显示:植物从水生向陆生过渡过程中,rbcL基因密码子使用偏好性的变化可能较为稳定,这也许是rbcL基因受到强烈自然选择作用的结果。生物体内高表达的基因,其密码子偏好性也相对较强,反之亦然[24]。千屈菜科植物rbcL基因ENC较高,CAI较低,说明千屈菜科植物rbcL基因整体的密码子使用偏好性不强,在植物体内表达水平也不高。但仍存在CGU、CCU、ACU等13个偏好性相对较强的密码子(RSCU>1.5),其在氨基酸中残基含量也相对丰富。
密码子使用偏好性的影响因素包括碱基组成、突变、自然选择、漂变、基因长度、tRNA丰度以及基因表达水平的高低等,但最主要的压力来自于突变和自然选择[25]。本研究中,千屈菜科植物rbcL基因GC3s和GC、ENC的相关性显著,表明密码子偏好性在一定程度上受到了碱基组成的影响,之前的研究也证明GC3s和GC含量之间存在明显的线性关系[26]。但GC3s与GC12相关程度较低,且GC3s集中分布在0.275~0.300内,KAWABE等[23]研究表明:密码子使用偏好性主要受自然选择的影响,而碱基突变的影响则较小,ENC分析、中性分析、奇偶偏差分析也得出相同的结论。这可能是由于rbcL基因本身为叶绿体基因,分子进化速率相较于核基因更慢,且编码的二磷酸核酮糖羧化酶是参与光合作用的关键蛋白,相对比较保守,所以突变压力对其密码子使用偏好性的作用相对较弱;而正选择、协同进化等作用在陆生植物的rbcL基因中被证明广泛存在,也表明rbcL基因密码子使用偏好性可能广泛受到选择约束[27-28]。
与RSCU聚类分析结果相比,基于CDS的邻接树在理论上更接近真实的物种系统发育关系。两者相对一致的部分说明千屈菜科植物rbcL基因密码子使用特性与属间亲缘关系存在一定程度的对应;两者之间较为矛盾的分支可能是系统聚类仅选取单一RSCU数据分析导致的,结合密码子偏好性的其他参数,或许能获得更加一致的结果。由于单基因建树也可能会受到旁系同源基因干扰、水平基因转移等多种因素影响产生误差[29],因此基于密码子偏好性的聚类分析也可对系统发生的研究内容进行一定补充。
转基因过程中,选择密码子使用偏好性相近的物种作为异源表达受体,有利于外源基因的高效表达[30]。千屈菜科植物多数都是木本植物,遗传转化体系尚未成熟,由于受限于同源物种生活史长、生长速度慢等因素,其基因功能研究十分依赖模式物种。通过与模式物种密码子使用频率的初步比较,酵母更适合作为千屈菜科植物rbcL基因的异源表达受体;与拟南芥、烟草相比,番茄的密码子使用频率与千屈菜科植物rbcL基因差异性最小,更适合作为rbcL基因功能验证的理想受体材料。但相对于番茄,拟南芥和烟草遗传转化体系建立相对较早,发展较为完善,已实现了多种木本植物叶绿体基因的遗传转化,积累的技术经验较多,遗传转化的难度也相对较小[31]。在观赏植物研究中,番茄更多作为植物呈色相关基因的遗传转化受体,验证其在色素积累与代谢中的调控作用[32]。因此,密码子使用频率的比较结果仅能为千屈菜科植物rbcL基因异源表达受体选择提供初步的预测,受限于该科木本植物当前采样难度较大,且遗传转化体系尚未成熟建立等因素,最适的异源表达受体仍须在进一步的实验中进行深入研究和严格筛选。











 DownLoad:
DownLoad: