-
LBD转录因子是一类在其蛋白质N端具有侧生器官边界(LOB)蛋白质结构域的植物特有转录因子[1]。SHUAI等[2]首次发现了LBD基因,发现它在植物侧生器官的边界细胞中表达并且参与侧生器官的发育形成。根据侧生器官边界(LOB)域的结构,拟南芥Arabidopsis thaliana中的LBD转录因子家族通常可被分为2类。类1(Group 1)具有1个完整的高度保守的CX2CX6CX3C锌指结构,通常能够结合特定DNA序列。同时类1的LBD基因具有高度保守的甘氨酸GAS结构和亮氨酸拉链(zipper-like)结构(LX6LX3LX6L),它们之间会形成卷曲-螺旋结构的蛋白质并相互作用。而类2(Group 2)只含有1个保守的CX2CX6CX3C锌指结构[1-4]。大多数的LBD转录因子基因从属于类1。类2的LBD蛋白质往往保存有1个可能不具有功能的残缺的亮氨酸拉链结构,它会形成1个盘绕-线圈结构[4]。最近,CHEN等[5]通过研究小麦Triticum aestivum LBD蛋白质中LOB结构域的蛋白质结晶学,揭示了之前提到的CX2CX6CX3C锌指结构实际是C4型锌指结构。C4锌指结构常常与GAS结构以及α4和α5之间的垂直构象共同作用,从而精确识别DNA,并能对结合位点的空间构型进行定位。最初的一些LBD基因功能研究表明:LBD基因通常在侧生器官底部的边界新生细胞中表达,在器官分化和侧生器官发育中具有潜在功能[1]。同时,LBD基因还能够参与植物花青素、氮元素代谢和器官再生等关键过程[6]。类1的LBD基因大多数参与植物发育过程[4, 7]和由生长素信号转导级联反应介导的侧根形成过程。相反,类2基因往往作为花青素合成的阻遏物和有效性信号参与了新陈代谢过程[8]。在拟南芥LBD转录因子的表达模式研究中,病原体几乎诱导了类2的所有LBD基因的表达,这表明LBD基因能够在植物病原防御反应中发挥作用[9]。LBD基因参与到拟南芥许多组织发育过程中,比如叶片发育[10]、根部侧生器官发育[11-13]、细胞分裂素信号转导[14]和赤霉素途径[15]。值得一提的是,拟南芥中的AthLBD16基因会促进侧根的起始发育[16],AthLBD29则能够抑制拟南芥茎纤维壁增厚的生长素信号转导过程[17]。在尖孢镰刀菌Fusarium oxysporum中AtLBD20作为易感基因似乎能够调节茉莉酸(JA)信号传导和激活尖孢镰刀菌中JA信号转导过程依赖的反应[18]。除拟南芥之外LBD基因功能在许多物种中同样被研究过,如OsIG1基因会参与配子发生过程并影响水稻Oryza sativa的花器官数量[19];MdLBD13蛋白质则可以抑制苹果Malus × domestica中的花色苷合成和氮吸收[20]。与此同时,在许多物种中也开展了利用基因组数据资源对LBD基因家族鉴定和分析的研究,如茶树Camellia sinensis[21]、马铃薯Solanum tuberosum[22]、桉树Eucalyptus robusta[23]、芸薹Brassica campestris[24]、葡萄Vitis vinifera[25]、大豆Glycine max[26]。但是目前薄壳山核桃Carya illinoensis中的LBD基因研究却鲜有报道。在薄壳山核桃全基因组测序组装完成后LBD基因家族仍然没有进行系统的研究[27]。本研究通过生物信息学手段鉴定了薄壳山核桃全基因组内的LBD基因,分析其基因结构、基序(motif)分布、转录组表达模式并构建系统进化树,预测LBD基因家族在薄壳山核桃中可能的功能,并为进一步研究提供理论基础。
HTML
-
薄壳山核桃、山核桃Carya cathayensis、核桃Juglans reiga的全基因组蛋白质序列来源于胡桃科Juglandaceae数据库[28]。银杏Ginkgo biloba、无油樟Amborella trichopoda、蓝星睡莲Nymphaea coloratar、大豆、葡萄、水稻的全基因组蛋白质序列来源于美国国立生物技术信息中心(NCBI)。拟南芥的全基因组蛋白质序列来源于拟南芥信息资源库(TAIR,https://www.arabidopsis.org/)。
-
以LBD为关键词,搜索获取LBD在拟南芥中的10条核酸编码序列(CDS),并下载这10条基因的FASTA格式的核酸序列。将获得的10条拟南芥编码序列通过本地NCBI-BLAST 2.9.0软件的BLASTX程序与1.1中获得的全基因组蛋白质序列进行同源比对,在Pfam数据库[29]中下载LOB结构域的种子文件,得到的候选LBD蛋白质序列用HMMER软件[30]使用隐马尔科夫模型算法筛选出LBD基因,利用ExPASy蛋白质在线分析网站对蛋白质序列进行理化性质的鉴定和分析。
-
通过MCSCAN软件[31],将薄壳山核桃全基因组核酸编码序列CDS文件和基因组注释GFF文件输入执行Pairwise Synteny Search程序,得到的Anchor Pairwise结果以30个基因为阈值过滤小片段block。在最后的gene block中寻找1.2中鉴定到的LBD基因。
-
利用ENSEMBL的CLUSTALW软件[32]对获取的52条薄壳山核桃LBD基因进行多重序列比对,将比对后的ALN文件输入至ENDscript网站进行多序列比对的可视化。
-
利用MUSCLE v3.8.31软件[33]分别对获取的52条薄壳山核桃和547条多物种LBD基因的蛋白质序列进行多序列比对。获得的多序列比对结果文件使用FastTree V2.1.11软件[34]的JTT+CAT算法[35]构建最大似然树,获得的树文件通过Evolview在线网站[36]可视化。在胡桃科数据库下载薄壳山核桃基因组结构注释GFF文件,并利用Gene Structure Display Server 2.0在线网站[37]可视化LBD基因结构图。在MEME网站上传52条薄壳山核桃蛋白质序列分析基序。
-
转录组原始测序数据从NCBI (https://www.ncbi.nlm.nih.gov/) PRJNA435846下的SRA数据库获取:SRR6793964、SRR6793963、SRR6793962、SRR6793961、SRR6793960、SRR6793959、SRR6793958、SRR6793957、SRR6793956、SRR6793955。从GIGADB (http://gigadb.org/)获取薄壳山核桃全基因组序列和基因组注释GFF文件,利用fastp软件进行质控和过滤,STAR软件进行序列比对,RSEM软件对基因进行定量分析,计算TPM值并生成基因表达数据框。得到的表达数据通过R软件将表达值进行对数转换后利用pheatmap包绘制表达热图。
1.1. 多物种全基因组蛋白质序列获取
1.2. LBD转录因子基因的鉴定与蛋白质理化性质分析
1.3. LBD基因组共线性区块分析
1.4. LBD基因多重序列比对
1.5. LBD基因系统发育、基因结构和基序分析
1.6. LBD基因表达模式分析
-
通过BLASTX初筛出LBD的同源基因,经HMM-SEARCH程序[29]分析和筛选,供试的10个物种中共获得551条LBD基因(表1)。其中大豆中LBD基因的数量是无油樟的4.46倍、核桃的1.61倍,推测这可能和大豆是由古四倍体演化而来的二倍体有关[38]。根据薄壳山核桃基因组GFF注释文件,发现有pecanLBD50、pecanLBD51和pecanLBD52等3个基因是位于scaffold123681的短串联重复基因。
鉴定程序 基因数量/个 银杏 无油樟 蓝星睡莲 水稻 拟南芥 葡萄 大豆 核桃 山核桃 薄壳山核桃 BLASTX 90 45 51 61 91 98 153 87 89 80 HMMER 50 26 35 29 58 57 116 72 56 52 Table 1. Identify result of LBD genes in multi-species
MCSCAN分析显示:薄壳山核桃基因组共有211个共线性的区块。在这些共线性的区块中找到了13对在可能全基因组加倍事件中发生重复的同源LBD基因对:pecanLBD1和pecanLBD25、pecanLBD4和pecanLBD17、pecanLBD5和pecanLBD16、pecanLBD11和pecanLBD33、pecanLBD11和pecanLBD32、pecanLBD12和pecanLBD33、pecanLBD12和pecanLBD32、pecanLBD14和pecanLBD43、pecanLBD14和pecanLBD42、pecanLBD19和pecanLBD26、pecanLBD20和pecanLBD29、pecanLBD21和pecanLBD36、pecanLBD34和pecanLBD41。共线性分析共得到基因23个,占所有LBD基因的44.2%,这说明了全基因组加倍事件使LBD基因家族得到了扩张。
通过理化性质分析(表2):在氨基酸数量、分子量、等电点、不稳定系数和脂肪系数等方面存在差异。氨基酸数量为 92~326个,大部分为 170~300个;等电点为4.48~9.85,>7.5的有24个,多在碱性范围内;不稳定系数<40的有51个,为稳定蛋白质;>40仅1个,为不稳定蛋白质;蛋白质的脂肪系数大多<100,为疏水性蛋白质,仅有pecanLBD50脂肪系数>100,为亲水性蛋白质。
蛋白质 氨基酸残基数/个 分子量/kD 理论等电点 酸性氨基酸/个 碱性氨基酸/个 不稳定系数 脂肪系数 总平均亲水性 pecanLBD1 210 23 309.19 8.24 17 19 47.65 73.52 −0.485 pecanLBD2 208 22 353.77 7.55 20 21 58.32 88.65 0 pecanLBD3 288 31 617.01 8.99 29 35 61.92 76.88 −0.326 pecanLBD4 223 24 069.37 7.69 17 18 73.89 77.89 −0.171 pecanLBD5 188 20 808.56 7.63 17 18 67.24 69.15 −0.490 pecanLBD6 260 28 451.22 8.23 31 34 52.55 69.38 −0.501 pecanLBD7 156 17 540.20 9.03 12 18 55.87 78.91 −0.235 pecanLBD8 172 18 611.24 8.59 14 17 61.24 70.35 −0.238 pecanLBD9 276 30 985.11 8.55 28 32 30.84 76.96 −0.522 pecanLBD10 326 36 029.86 6.91 23 22 64.45 77.58 −0.472 pecanLBD11 204 22 376.57 7.53 18 19 60.78 77.99 −0.233 pecanLBD12 232 25 857.68 5.90 21 14 50.52 65.22 −0.425 pecanLBD13 170 18 808.03 6.42 18 17 65.04 57.47 −0.607 pecanLBD14 202 21 823.68 8.59 14 17 57.41 80.20 −0.175 pecanLBD15 203 23 706.08 8.15 27 29 52.00 77.39 −0.556 pecanLBD16 191 20 998.77 6.08 19 17 70.05 72.04 −0.423 pecanLBD17 230 25 054.58 9.22 17 23 78.94 70.04 −0.284 pecanLBD18 172 18 626.23 8.82 14 18 67.64 73.14 −0.296 pecanLBD19 306 33 113.72 7.95 33 35 46.03 81.54 −0.280 pecanLBD20 162 18 029.72 6.70 14 14 51.76 88.52 −0.133 pecanLBD21 127 14 182.02 4.48 14 7 36.18 92.20 −0.246 pecanLBD22 176 19 971.98 5.17 25 18 44.88 80.34 −0.186 pecanLBD23 130 15 042.74 9.84 10 25 45.43 77.23 −0.541 pecanLBD24 162 18 285.75 8.23 13 15 57.65 78.40 −0.337 pecanLBD25 214 23 771.70 8.25 17 19 49.42 74.44 −0.472 pecanLBD26 310 33 384.03 8.22 34 37 47.44 80.87 −0.302 pecanLBD27 168 18 892.28 6.28 17 13 72.86 63.87 −0.476 pecanLBD28 215 23 539.68 5.33 18 14 75.47 70.88 −0.272 pecanLBD29 167 18 504.18 6.94 15 15 59.00 81.80 −0.217 pecanLBD30 176 19 230.73 6.50 18 17 69.15 73.75 −0.242 pecanLBD31 169 18 881.58 6.49 17 16 51.28 79.59 −0.219 pecanLBD32 236 26 157.91 5.74 22 13 46.73 65.76 −0.405 pecanLBD33 213 23 405.50 7.06 19 19 65.23 67.37 −0.380 pecanLBD34 326 37 266.20 5.47 54 35 49.05 74.29 −0.813 pecanLBD35 251 27 236.49 6.44 24 22 59.55 80.08 −0.322 pecanLBD36 228 24 949.42 6.29 17 16 83.40 71.05 −0.271 pecanLBD37 228 24 709.22 8.06 22 24 59.20 76.14 −0.244 pecanLBD38 216 23 449.53 6.03 17 13 64.11 75.05 −0.227 pecanLBD39 158 17 663.15 6.27 15 13 53.44 80.25 −0.243 pecanLBD40 221 24 171.04 4.97 21 12 58.45 78.60 −0.143 pecanLBD41 265 30 244.09 6.42 33 29 46.86 77.66 −0.594 pecanLBD42 237 24 801.38 8.20 13 15 69.07 79.87 −0.065 pecanLBD43 213 23 452.55 6.74 19 18 56.45 80.61 −0.203 pecanLBD44 224 24 373.77 8.93 17 22 69.41 73.62 −0.276 pecanLBD45 202 21 927.15 8.56 19 23 54.66 83.47 −0.159 pecanLBD46 221 24 114.37 9.01 18 23 73.18 66.29 −0.421 pecanLBD47 289 32 466.21 5.27 29 21 67.19 65.92 −0.600 pecanLBD48 176 19 971.98 5.17 25 18 44.88 80.34 −0.186 pecanLBD49 170 18 770.95 6.19 19 17 60.44 55.18 −0.615 pecanLBD50 92 10 319.08 6.26 11 10 39.17 115.65 0.018 pecanLBD51 313 34 650.03 7.29 20 20 64.34 73.07 −0.490 pecanLBD52 320 35 091.25 6.83 23 21 66.59 67.53 −0.593 Table 2. Physicochemical properties of LBD transcription factor family protein in C. illinoensis
-
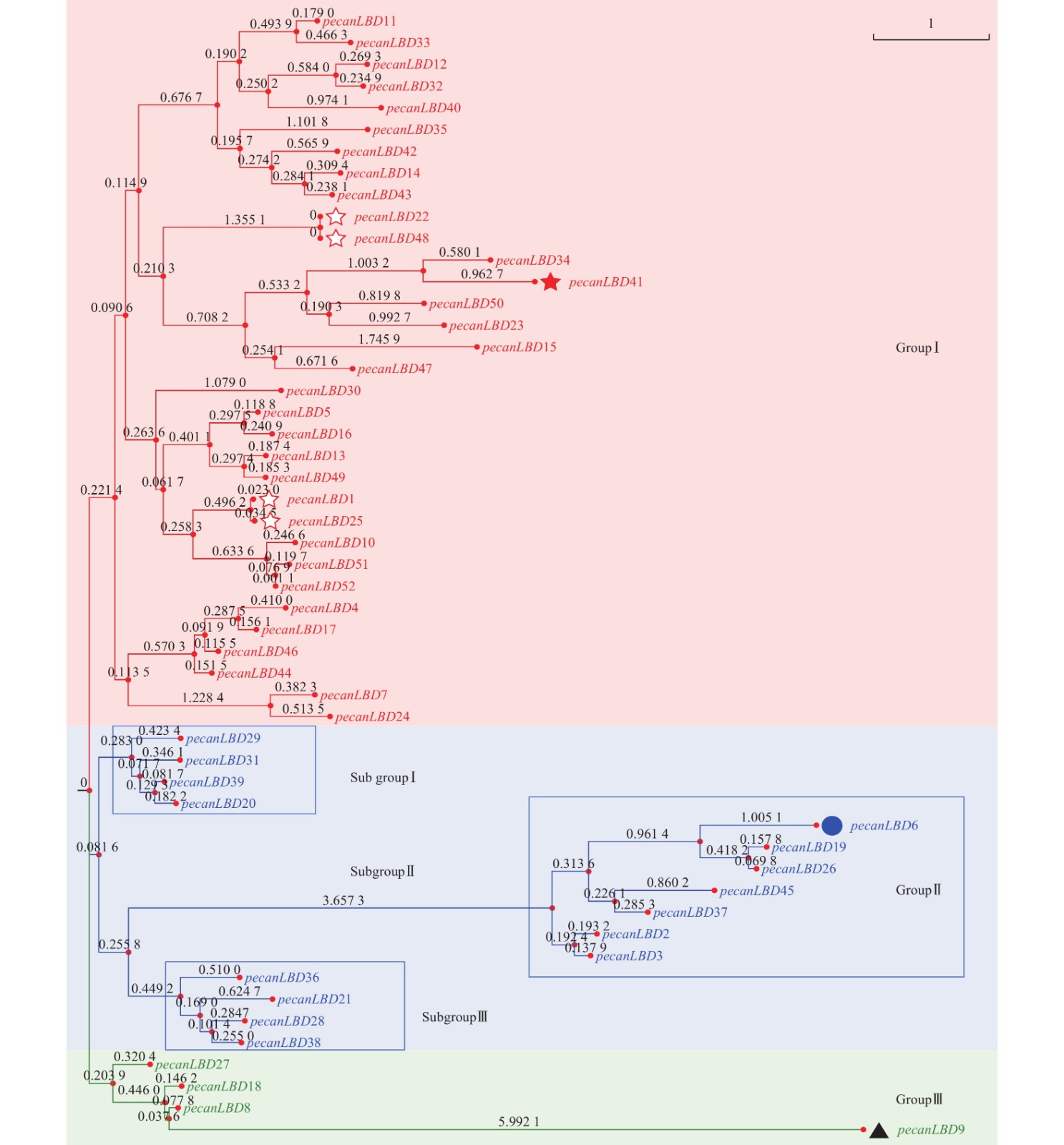
如图1所示:52个薄壳山核桃LBD基因聚成3类:GroupⅠ、GroupⅡ、GroupⅢ。其中,GroupⅠ含有最多的33个LBD基因,它们可能承担了LBD基因促进侧生器官发育的主要功能。GroupⅠ中有1对蛋白质序列完全相同的基因pecanLBD22和pecanLBD48,导致它们在进化树上没有显示变异,而pecanLBD41的进化枝长度达到3.7346,是GroupⅠ中相对于其他LBD基因变异程度最大的。GroupⅡ细分为3支基因簇,其中 Sub groupⅠ和Sub groupⅢ的进化枝相对较短,因此序列和功能变异不大;而Sub groupⅡ有明显的进化变异,其中pecanLBD6的枝长为6.2748,是Sub groupⅡ中变异最大的基因,它可能已经出现了功能上的分化变异。GroupⅢ是3类中最保守的一支,其中的pecanLBD9基因的枝长在所有52个LBD基因中最长,达6.764 2。
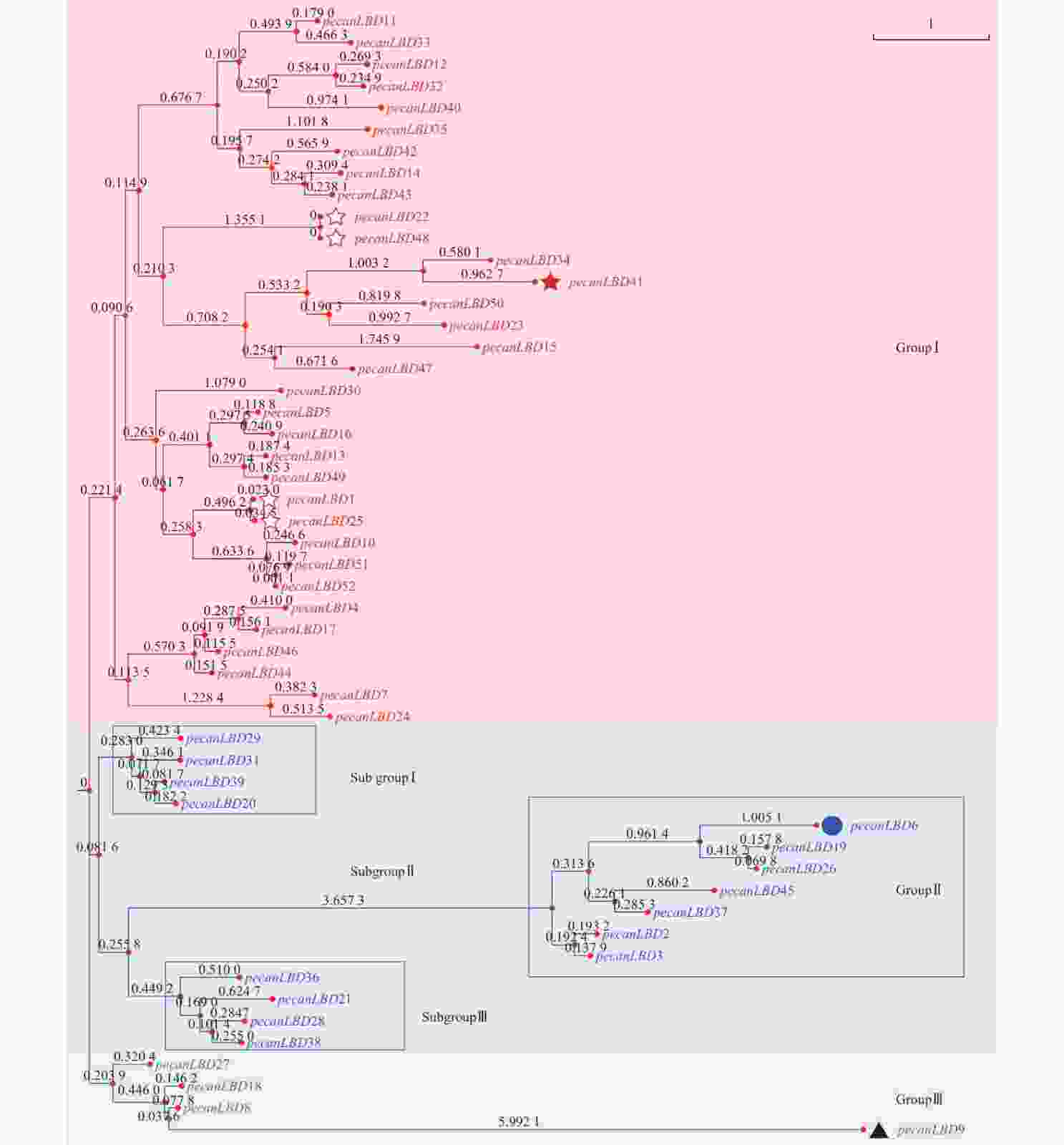
Figure 1. Phylogenetic tree constructed based on the full-length sequences of pecan LBD genes using JTT+CAT algorithm with FastTree software
如图2中52个薄壳山核桃LBD基因基序分析结果所示:共发现了10个基序并命名为Motif1~Motif10。对LBD转录因子家族较重要的有3个基序:Motif1含GAS(Gly-Ala-Ser)甘氨酸保守结构;Motif 2拥有完整的高度保守的CX2CX6CX3C锌指结构,通常具有结合特定DNA序列的能力;Motif 3 为亮氨酸拉链结构(LX6LX3LX6L)。Motif1和Motif3往往会形成卷曲-螺旋结构的蛋白质并相互作用。52个薄壳山核桃LBD基因中的39个具有完整的Motif1、Motif2和Motif3的顺序基序结构;而pecanLBD34、pecanLBD41和pecanLBD9在进化过程中丢失了Motif1的GAS甘氨酸保守结构;pecanLBD6、pecanLBD19、pecanLBD26、pecanLBD45、pecanLBD37、pecanLBD2和pecanLBD3丢失了Motif3亮氨酸zipper-like结构(LX6LX3LX6L),尽管它们仍具有GAS结构但已经丧失了和Motif1结合生成卷曲-螺旋蛋白质结构的功能;pecanLBD50则丢失了所有的基序,可能丧失了LBD基因的基本功能。
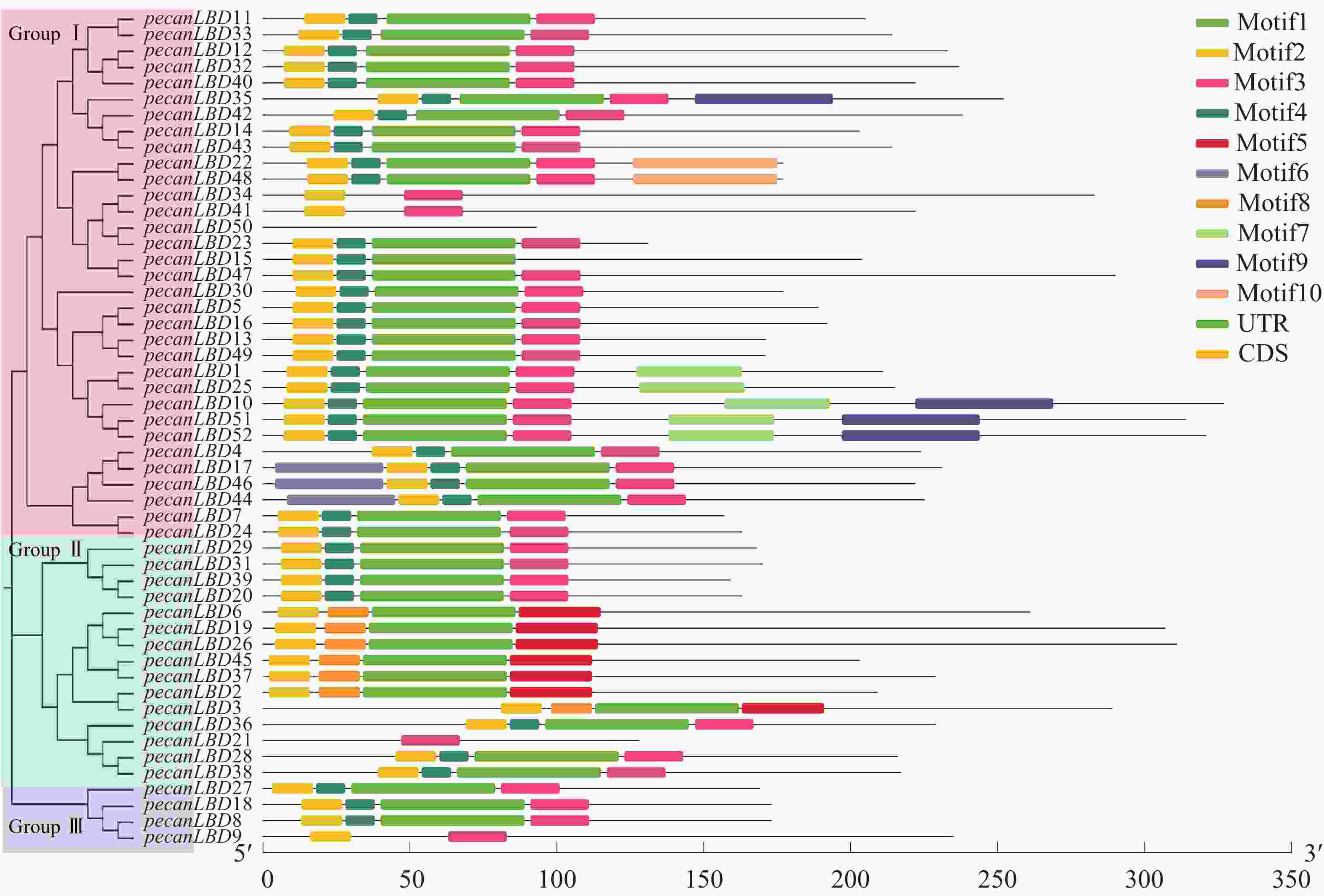
Figure 2. Phylogenetic tree constructed based on the full-length sequences of pecan LBD genes using JTT+CAT algorithm with FastTree software
基因结构分析显示:薄壳山核桃LBD基因一般具有1~3个外显子。其中pecanLBD41~pecanLBD52都只具有1个外显子(除pecanLBD47有2个外显子除外),并且都属于GroupⅠ;具有3个外显子的基因只有pecanLBD3、pecanLBD34和pecanLBD9,在每类都有部分;剩余的LBD基因都为2个外显子,占所有薄壳山核桃LBD基因的67.3%。因此,薄壳山核桃LBD转录因子家族的大多数基因较为保守,有着相似的基序、基因结构,执行并发挥LBD的主要功能,而其发生变异的小部分可能已经丢失了原来的基因功能或者特化出新的功能。
-
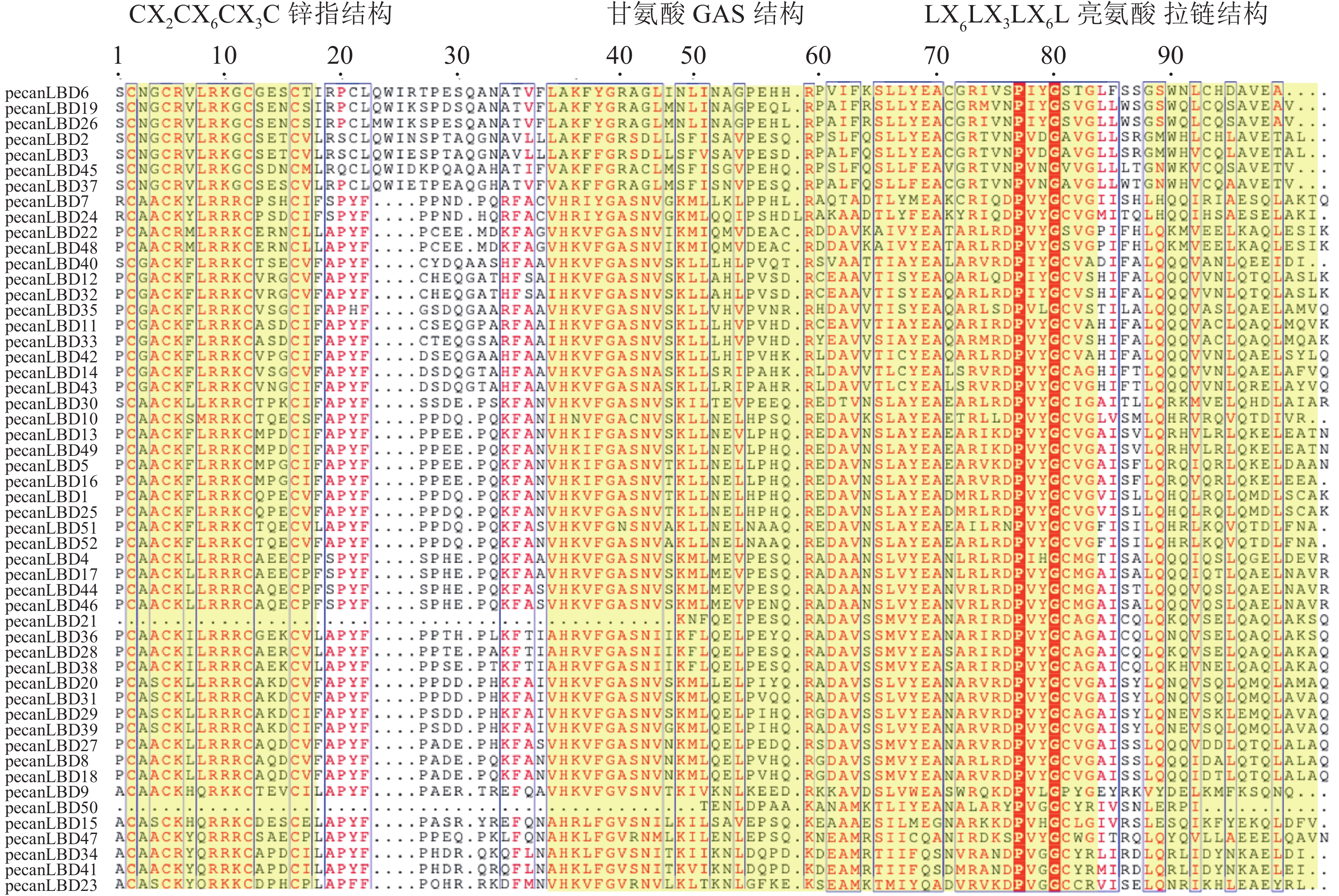
对52个薄壳山核桃LBD转录因子基因的LOB蛋白质结构域多序列比对(图3)发现:LOB蛋白质结构域主要由3个保守序列模式组成:长度为16个氨基酸的CX2CX6CX3C锌指结构、长度为50个氨基酸的甘氨酸GAS结构和LX6LX3LX6L亮氨酸拉链结构。其中pecanLBD50和pecanLBD21完全缺失了锌指结构和部分的甘氨酸GAS结构,而其他的LBD的锌指结构则全部保存下来没有发生缺失或者突变,较为保守。甘氨酸GAS结构相对锌指结构变异程度较大,GroupⅠ中pecanLBD23~pecanLBD47、pecanLBD10、pecanLBD51发生了1~2个氨基酸的变异,GroupⅡ中pecanLBD6、pecanLBD19、pecanLBD26、pecanLBD45、pecanLBD37、pecanLBD2、pecanLBD3等7个转录因子GAS中的丙氨酸突变为精氨酸、丝氨酸突变为丙氨酸,52个LBD转录因子中有12个发生了突变,2个缺失了GAS结构。LX6LX3LX6L亮氨酸拉链结构中,上述7个转录因子的第1个亮氨酸位置都突变为甘氨酸,第3个亮氨酸位置突变为甘氨酸或苏氨酸,有5个转录因子的第2个亮氨酸位置发生了突变,由此可见上述7个LBD转录因子与进化模式(图1)一致,已经发生了较大程度的变异,可能已经分化出新的功能。pecanLBD15、pecanLBD47、pecanLBD10、pecanLBD51、 pecanLBD52、pecanLBD7和pecanLBD24的第2个亮氨酸位置发生了突变,第3个亮氨酸位置只有pecanLBD40、pecanLBD23和pecanLBD10发生了突变,相对保守。
-
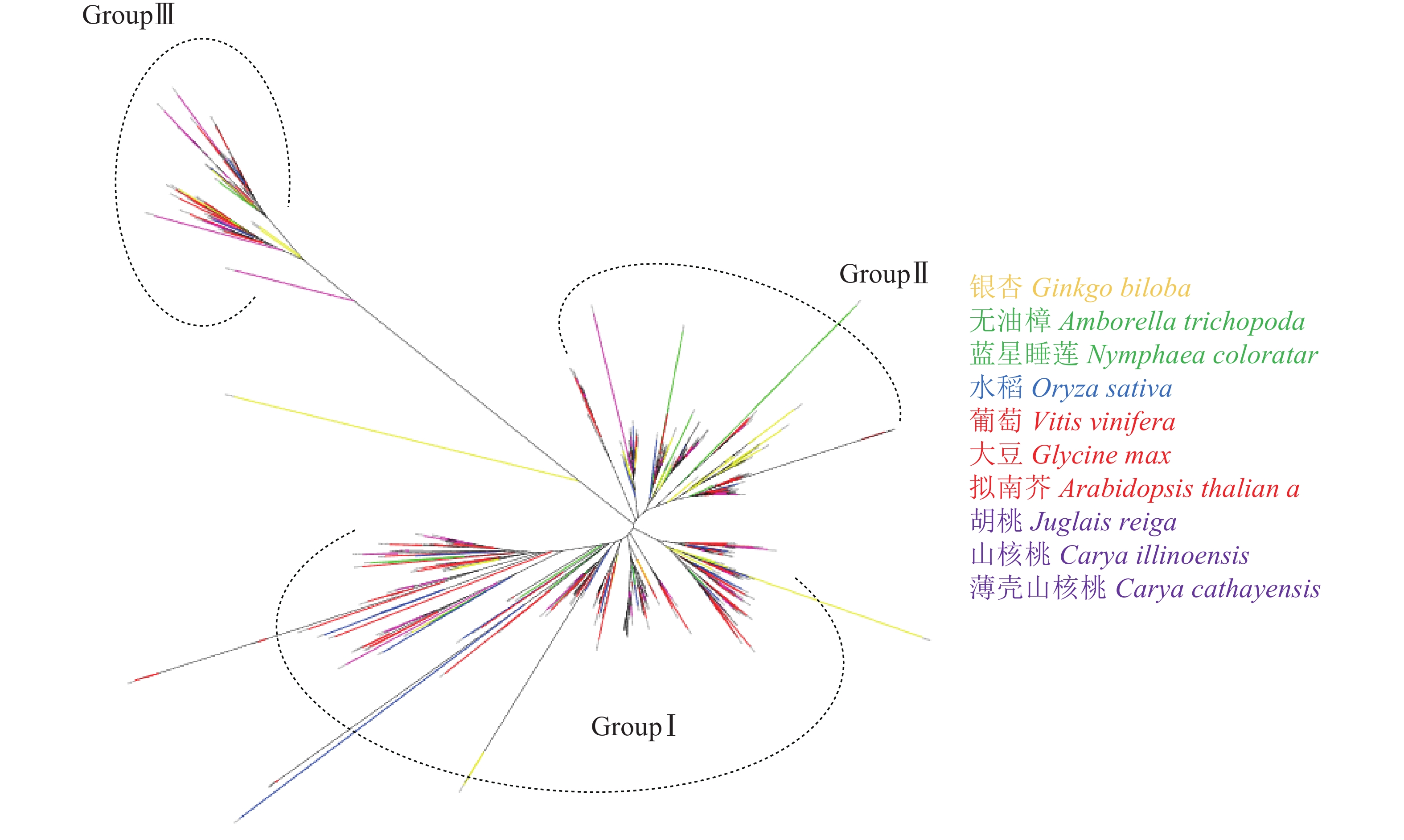
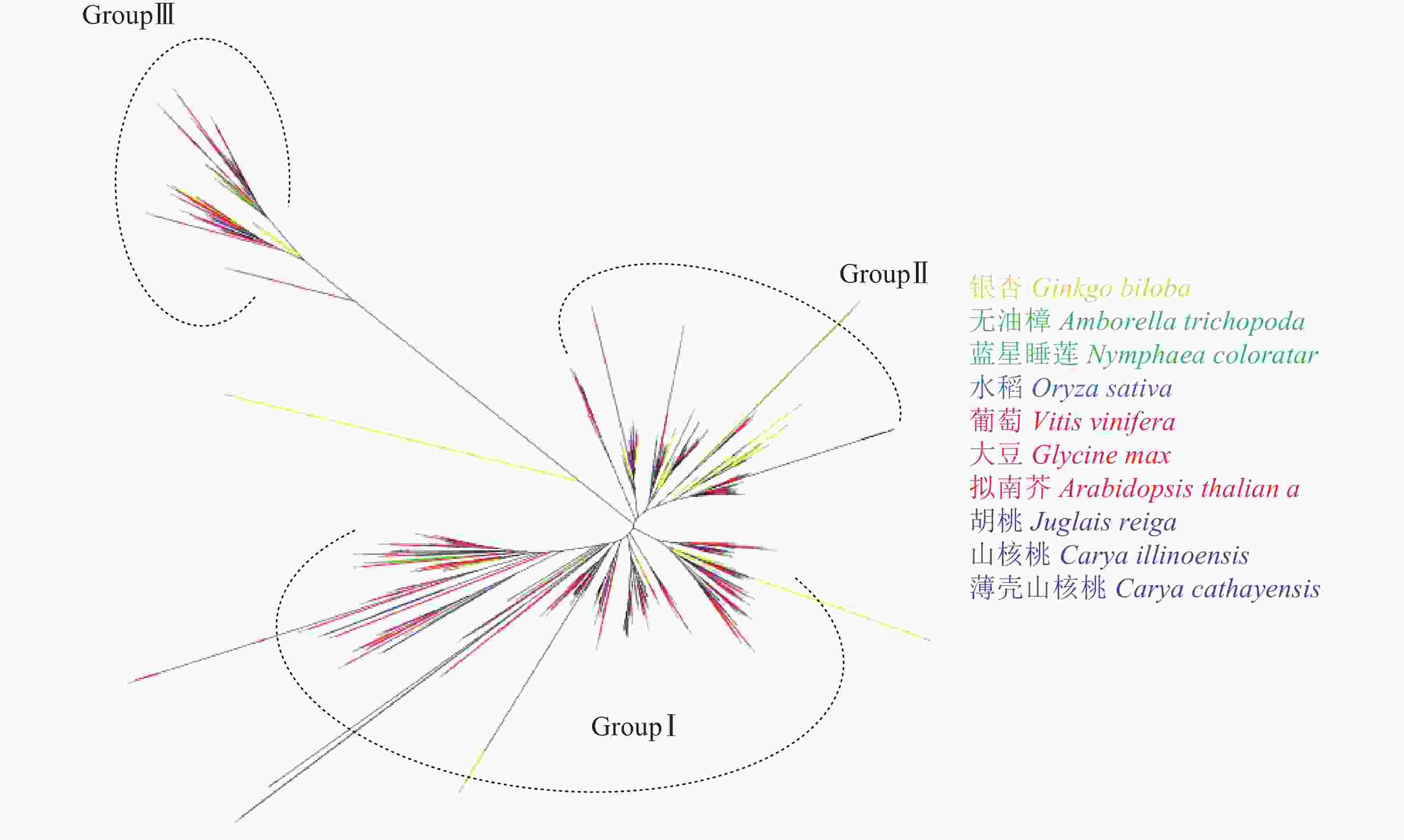
由图4可见:与薄壳山核桃LBD系统发育树相似,图4中各物种LBD转录因子可以聚为3类:GroupⅠ、GroupⅡ和GroupⅢ。其中GroupⅠ有314个LBD基因,为总数的57.0%,占比最多;GroupⅡ有166个LBD基因,占总数的30.1%;而GroupⅢ的基因数量最少,只有71个,占总数的12.9%。从表1鉴定到的LBD转录因子的数量来看,裸子植物银杏的数量是早期被子植物类群的2倍,这与银杏最近一次特异性的WGD事件有关[39];早期被子植物类群和单子叶植物水稻的LBD基因数量接近;而双子叶植物的LBD基因数量明显又发生了1次倍增。从分布来看,GroupⅠ、GroupⅡ和GroupⅢ中每个植物类群都存在LBD基因,因此LBD转录因子家族可能在裸子植物和被子植物未分化前已经分为3类,分化后3类LBD继续各自进化。从变异程度看GroupⅠ和GroupⅡ相对较为保守,而GroupⅢ内的所有LBD基因共享一支较长的分支,说明它们已发生了较大的变异,可能已经分化出新的功能。
-
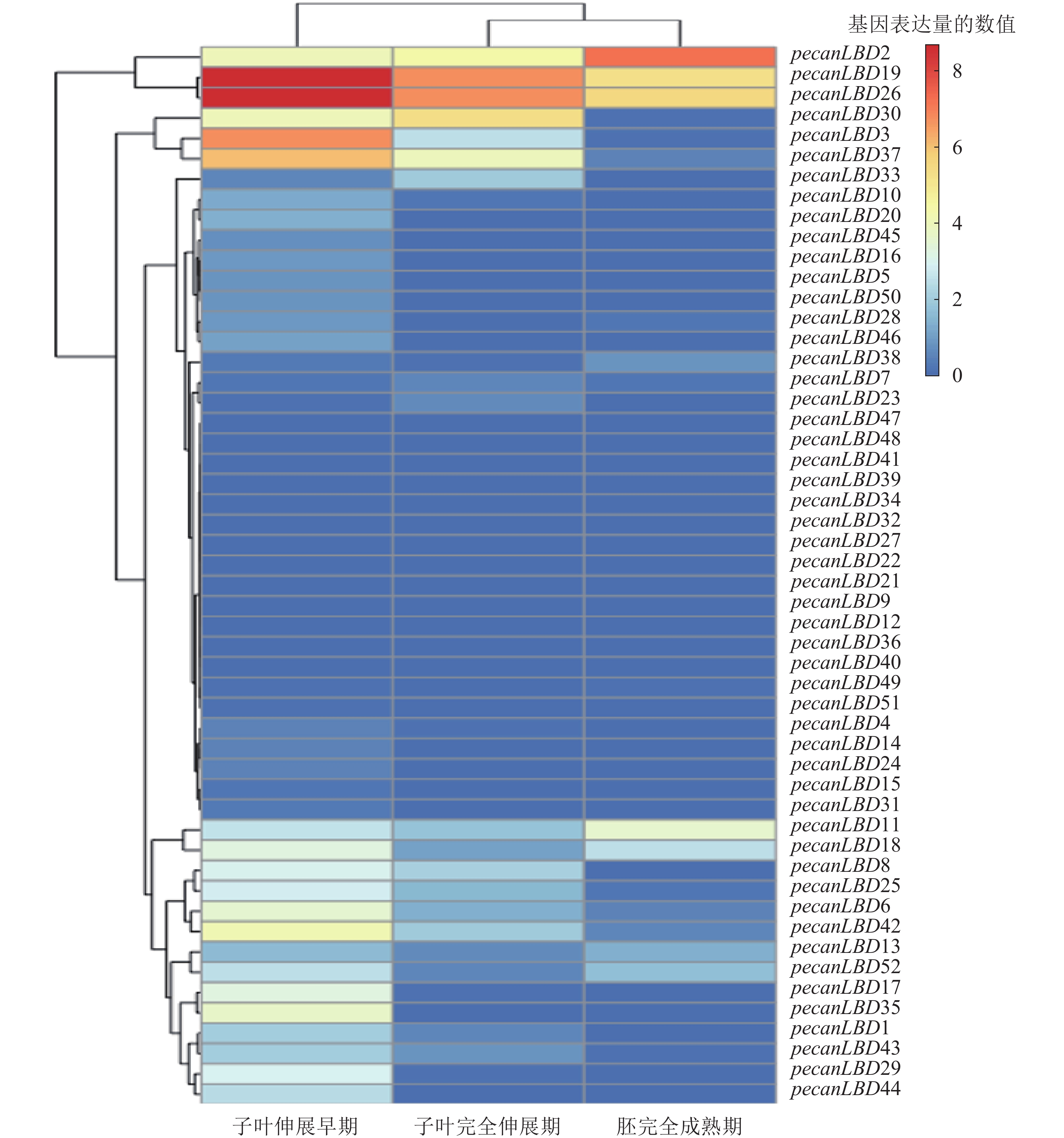
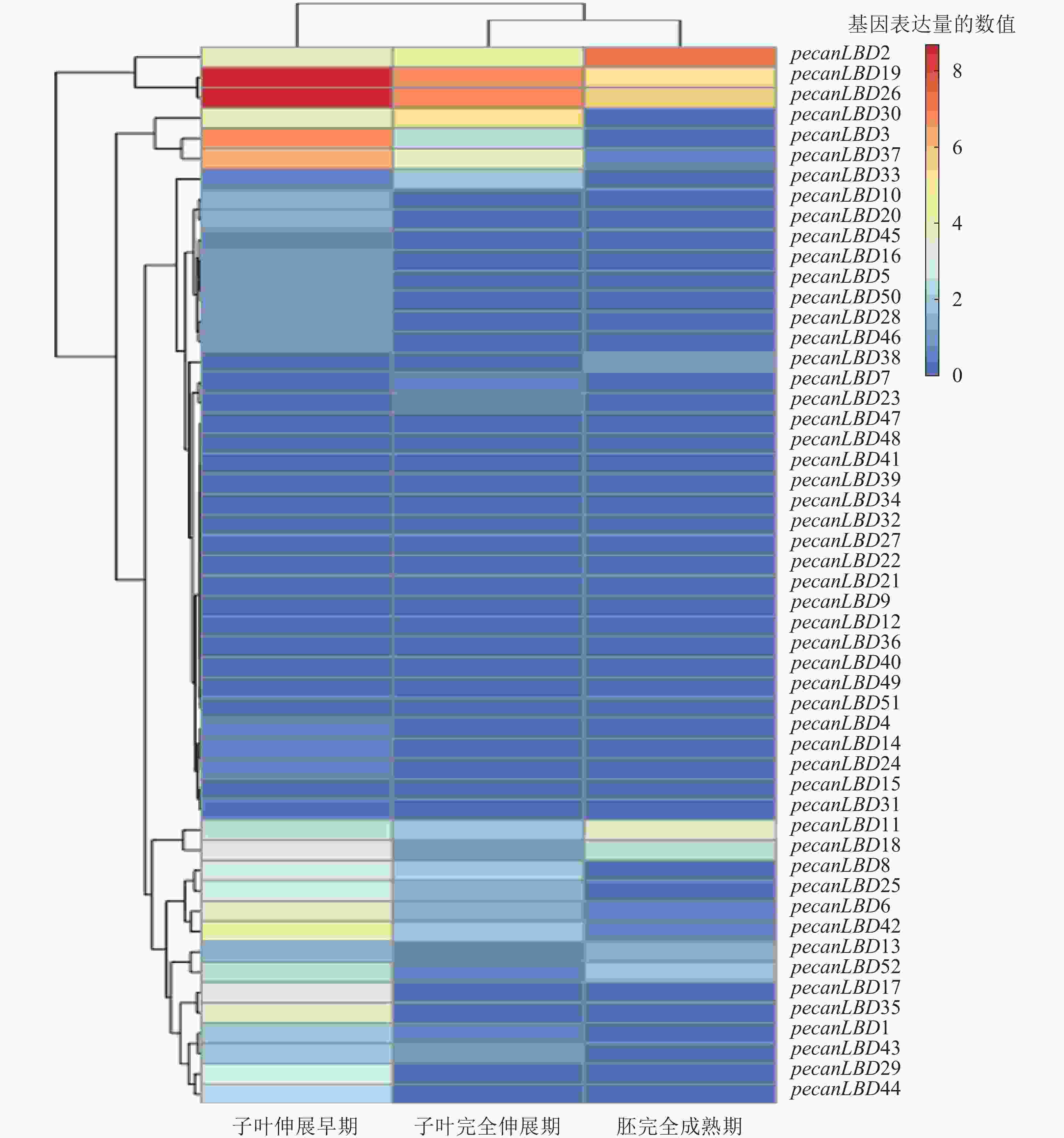
为了探究LBD转录因子家族在胚发育的关键过程中的作用,从NCBI网站的SRA原始测序数据库中下载了薄壳山核桃胚成熟过程中3个关键时期的原始测序数据:子叶伸展早期(early extended stage of cotyledon development)、子叶完全伸展期(fully extended stage of cotyledon development)、胚完全成熟期(fully matured stage of embryo),并进行分析和绘制聚类热图(图5)。不同LBD基因和不同发育阶段之间的表达模式有显著差异。基因表达量可以聚为4类:①pecanLBD2、pecanLBD19和pecanLBD26在3个阶段的表达量比较高;②pecanLBD30、pecanLBD3和pecanLBD37在子叶伸展早期和子叶完全伸展期表达;③pecanLBD11、pecanLBD18、pecanLBD8、pecanLBD25、pecanLBD6、pecanLBD42、pecanLBD13、pecanLBD52、pecanLBD17、pecanLBD35、pecanLBD1、pecanLBD43、pecanLBD29和pecanLBD44仅在子叶伸展早期有表达;④其余的LBD基因在3个阶段都不表达。从2.2中系统发育分析的结果来看:③类表达的14个LBD基因(78.6%),属于较为保守的Group I,因此它们在胚发育过程中主要参与经典的器官分化过程,加快子叶边界新生的细胞分化。而①类表达的基因都是进化枝较长、变异程度较大的GroupⅡ中Sub GroupⅡ的成员,可见GroupⅡ中Sub Group Ⅱ的LBD成员已经发生了较大程度的变异,它们参与到胚发育的全程,可能与胚中营养物质的积累过程和胚组织分生发育的过程密切相关。②类中pecanLBD30和pecanLBD3属于GroupⅡ, pecanLBD 37属于GroupⅠ,它们在功能上可能介于①类和③类,即既参与了器官分化、细胞分裂的过程调控,又和胚中营养物质积累的过程相关。总体上,薄壳山核桃胚发育过程中LBD基因功能较为保守,主要参与新生细胞分裂分化、子叶形态建成等,但GroupⅡLBD基因的变异程度较大,可能进化出新的功能,在胚发育过程中发挥更加关键的作用。
2.1. LBD基因鉴定与蛋白质理化性质分析
2.2. 薄壳山核桃LBD基因系统发育、基因结构和基序分析
2.3. 薄壳山核桃LOB结构域多序列比对
2.4. 显花植物LBD转录因子家族系统发育分析
2.5. 薄壳山核桃LBD基因组织表达模式分析
-
本研究分别在银杏、无油樟、蓝星睡莲、水稻、拟南芥、葡萄、大豆、核桃、山核桃和薄壳山核桃全基因组蛋白质序列中系统鉴定了551个LBD基因。这些LBD基因包含典型的LOB结构域,即1个含有DNA结合活性所需的 (CX2CX6CX3C)锌指结构、1个Gly-Ala-Ser(GAS)结构和1个负责蛋白质二聚体化的亮氨酸拉链 (LX6LX3LX6L)结构。根据LOB结构域的组成模式和系统发育学分析结果。薄壳山核桃52个LBD基因可分为GroupⅠ、GroupⅡ和GroupⅢ 3类。33个LBD基因被划分为GroupⅠ,15个被划分为GroupⅡ,4个为GroupⅢ。根据进化枝的长度,GroupⅠ是数量较多同时也比较保守的一支,而GroupⅡ和 GroupⅢ都发生了一定程度的变异。
新基因功能产生或分化的一大动力是基因重复,基因重复对物种提高环境适应性至关重要[40]。在进化过程中,重复的基因可能会经历功能分化、亚功能分化或功能丢失等多种过程[41]。基因重复通常会导致基因家族扩张[42]。为了揭示薄壳山核桃LBD基因家族的复制机制,利用MCScanX对薄壳山核桃的基因组共线性区块内成对的LBD基因进行筛选。根据目前的基因组内LBD基因位置分析结果,薄壳山核桃基因组内只存在1个连续3个LBD基因的串联重复事件,但是存在13对23个由全基因组加倍产生的LBD基因共线性基因对。这说明全基因组加倍在LBD基因家族进化中起到了主导性的推动作用。
本研究构建了以裸子植物(银杏)、早期被子植物(无油樟、睡莲)、单子叶植物(水稻)、双子叶植物(拟南芥、葡萄、大豆)、近缘种植物(核桃、山核桃)和薄壳山核桃为显花植物中主要类群的LBD基因系统发育树。显花植物LBD系统发育树与与薄壳山核桃LBD相似,同样可以聚为3类。从分布来看,GroupⅠ、GroupⅡ和GroupⅢ中每个植物都存在LBD基因,因此LBD转录因子家族可能在裸子植物和被子植物未分化前已经分化为3类。从变异程度看,GroupⅠ和GroupⅡ相对较为保守,而GroupⅢ内的所有LBD基因共享一支较长的分支,说明已发生了较大的变异,可能已经分化出新的功能。
LBD蛋白质协调了很多植物发育过程,并对环境刺激作出反应,特别是在调控侧根器官发育和代谢过程中起着至关重要的作用。例如LBD18通过抑制LBD18 DNA-结合活性控制侧根发育[43]。一般来说,主根受到侧根过度生长的影响,会降低植物提取水分和养分的能力。在土壤中蔓延的侧根过多,将增加被病原体攻击的可能性。以前的一些工作已经证明了这些预测。如LBD1通过抑制主根生长和维持侧根萌发来控制盐胁迫下的根结构[44]。LBD基因由于其对一系列下游基因的转录调控,成为植物病原体入侵的关键分子靶基因[7]。
在薄壳山核桃研究中胚的发育是非常重要的阶段[45],特别是胚内蛋白质、不饱和脂肪酸、淀粉等重要营养物质累积过程是薄壳山核桃研究的热点[46-47]。本研究分析LBD转录因子家族在薄壳山核桃子叶伸展早期、子叶完全伸展期和胚完全成熟期3个时期的转录数据发现:薄壳山核桃LBD基因存在4种表达模式。可见LBD基因具有参与调控胚发育过程的功能,通常基于在侧生器官底部的边界新生的细胞中调控细胞分裂分化来控制子叶的发育和形态建成。薄壳山核桃LBD基因中有在整个胚发育过程中都高表达的一簇基因,这些基因在胚发育过程中发挥了更加重要的作用,因此有必要对这一簇LBD基因进行更为深入的基因功能研究,可为进一步改良薄壳山核桃遗传性状提供基础。










 DownLoad:
DownLoad: