-
石蒜属Lycoris植物隶属百合目Liliflorae石蒜科Amaryllidaceae,具地下鳞茎,为多年生草本植物;全球约有20多种,中国有15种,主要分布在江苏、浙江和安徽等地[1]。石蒜属植物花型花色变异丰富,种间杂交亲和性高,对石蒜属植物进行遗传育种改良,极有希望选育出有中国特色的切花新品种[2],市场前景和园林应用前景极为广阔。同时,石蒜鳞茎富含生物碱、黄酮及多糖等化学成分,药用价值较高[3-4]。目前,石蒜属植物新品种选育主要通过种间和种内杂交、自然群体选择,对其杂交种真实性的鉴定主要根据开花时花色和花型等农艺性状的变化来判断;但由于石蒜属植物生长周期长,种子播种后5~6 a才开花,早期鉴定及分类较为困难,从而影响销售和生产。
常用的杂种鉴定方法有形态标记法、生化标记法和DNA标记法等。形态标记耗时费力,易受环境条件和检测人员主观判断的影响;生化标记数目少且稳定性差,不能进行多年生植物杂交种的早期鉴定,无法满足生产和市场的需求;相比之下,DNA标记简单序列重复(SSR)、目标起始密码子多态性(SCoT)、内部简单序列重复(ISSR)、相关序列扩增多态性(SRAP)和荧光标记SSR能反映生物个体或种群间某些特异性DNA片段,不受生物自身生长阶段和环境条件的影响,标记数量较多,重复性好,开发成本低,不影响性状表达[5-7],已在冬瓜Benincasa hispida[8]、甜菜Beta vulgaris[9] 、姜黄属Curcum植物 [10]、枇杷Eriobotrya japonica[11]等种质资源遗传多样性、杂交种鉴定、品种鉴定和分子身份证的构建等方面得到广泛应用。其中SSR标记是以PCR技术为基础、较成熟的遗传标记,但由于工作量大、精度低、分析量小,无法完成大批量样品的检测,而荧光标记SSR能准确获得目标DNA片段的大小(精确至1 bp),检测结果稳定、准确且高效适用于大批量品种的检测分析,已被应用于落花生Arachis hypogaea [12]、高山杜鹃Rhododendron lapponicum [13]、百合属Lilium[14]、芒Miscanthus sinensis [15]等F1代杂交种真实性和纯度检测、遗传多样性分析、核心种质的构建等。石艳等[16]开发了石蒜属植物换锦花L. sprengeri的表达序列标签SSR(EST-SSR)标记,用于鉴定换锦花-中国石蒜L. chinensis杂交种,为石蒜属种间杂交种F1的鉴定提供了借鉴。为进一步完善石蒜属植物种间杂交种F1的鉴定技术,本研究筛选了15对EST-SSR引物,通过EST-SSR荧光标记毛细管电泳技术构建亲本换锦花、石蒜L. radiata、中国石蒜以及石蒜-换锦花、石蒜-中国石蒜杂交种等78个样品的分子身份证,并进行杂交种真实性鉴定,为石蒜属植物的亲缘关系分析、品种选育、杂交种早期鉴定及资源保护和利用等提供理论参考。
-
石蒜、中国石蒜、换锦花及石蒜-中国石蒜、石蒜-换锦花杂交种F1共78个样品,均栽植于浙江农林大学石蒜属种质资源圃,于2020年8—9月盛花期采集各材料花瓣。
-
采用改良CTAB法提取供试材料花瓣基因组DNA,用Nanodrop 2000测定DNA的质量和浓度,经质量浓度为1.0%琼脂糖凝胶电泳检测,凝胶成像系统拍照。
分别以亲本(中国石蒜、石蒜和换锦花)基因组DNA为模板,从59对石蒜属植物通用EST-SSR引物中筛选条带清晰、重复性好且具多态性的引物进行扩增[16]。EST-SSR PCR扩增反应体系(10.0 μL)为:Taq DNA聚合酶1.0×16.67 nkat,氯化镁(MgCl2) 1.0 mmol·L−1,dNTPs 0.2 mmol·L−1,引物0.8 μmol·L−1,DNA 40 ng。PCR反应程序为94 ℃预变性3 min;94 ℃变性30 S,退火温度30 S,72 ℃延伸30 S,35次循环;72 ℃延伸10 min;16 ℃保温30 min。用质量浓度10.0%的非变性聚丙烯酰胺凝胶电泳分离银染显色检测扩增反应产物,显色后记录观察拍照。
-
从59对多态性引物中筛选到15对具多态性的EST-SSR引物(表1),在正向引物中分别加注FAM (蓝)、HEX (绿)、TAMRA (黄)和ROX (红)荧光,对78个样品进行多重PCR扩增。利用遗传分析仪(ABI 3730XL,ThermoFisher Scientific,美国)进行毛细管电泳检测分析。根据电泳结果,选择在亲本(中国石蒜、石蒜和换锦花)种内一致、种间具多态性、特异、单一且重复性好的引物用于杂交种鉴定。比较亲本与杂交种F1的 EST-SSR PCR扩增产物,杂交种F1具有父母本互补型或有变异型扩增带型的样品鉴定为真实杂种。计算杂交种鉴定率=真实杂交种样本数/样本总数×100%。
引物 标记荧光 引物序列 (5′→3′) 重复单元 产物长度/bp 退火温度/℃ SSR7 ROX TCATGCATCGCACATGTCAC/AATGTAACCGGTCGCTCCAG (GGA)5 181 52.0 SSR15 FAM GACGCCCAAACAGCCAATTT/TGGAAAGGTTGAGCTTCGGG (CCT)5 247 52.0 SSR20 TAMRA ACAAGTTGGCCCTGTTGTCA/CATTCGATCACTCGGTCCGT (TCT)5 115 53.2 SSR32 TAMRA CCAGTCGTTCCGTTCCATCA/CTGCTGCACTTGTTCCCAAC (TTC)5 165 52.2 SSR85 TAMRA TCAACACAAGTGTCATTTCCAAT/ATGGCCCATTCAAGGTTGGT (TTA)6 108 52.6 SSR96 TAMRA CCGGCCTACAACAAAGGTCT/AAACTGTTGCAGCGACCATG (GGT)5 154 52.0 SSR115 FAM ATTCTGATCGGCGAAGGAGG/ATTTGCAAGGCGGTCAGAGA (GGC)5 235 49.0 SSR138 ROX TCACGAGAGAGGAGGGAGAA/CTCCTTGCGGATCATGGTGT (CAA)5 211 52.0 SSR142 ROX TGTCAGTTGATGGGCTTCGG/TGGTTGCAGTGACAGTTGGT (CCA)5 174 53.0 SSR147 FAM CCAAACAGCAGCTCAAGCAG/TTCGGTTTCGAGATTGGGGG (CCG)5 248 52.8 SSR198 FAM TCAGGGAATAAACCTCCGCC/ACTTGCTATCCTTGGGGCTT (ATC)5 223 51.7 SSR203 ROX ACGTGAGCAGTCCTCCTACT/GACATGCCCACTTCTCCCAA (GAG)5 204 52.0 SSR220 FAM ACTGGTGTCACTTGTGTGCA/GCTGGGCTCCCATCATTTCA (GAT)5 225 51.0 SSR221 HEX ATCTTGAGCTGCGTGTCGAA/CTCATCCACGCCTTCTCCTC (TTG)5 254 52.2 SSR253 HEX CGCCCGTGCAATTTCAAGTT/GCAAGTTGGCAACTCCACAG (AAT)5 277 52.4 Table 1. Sequences and PCR products of 15 pair of SSR primers
-
利用Gene mapper 4.1软件整理分析EST-SSR荧光标记的毛细管电泳数据,根据每个引物对每个样品扩增后有无峰,转化成0/1格式的二元矩阵,无信号或数据缺失赋值“2”。利用POPgene 1.32计算观测等位基因数(Na),有效等位基因数(Ne),Nei’s基因遗传多样性指数(H),Shannon’s指数(In)和多态性信息量(PIC)。使用NTsys2.10e计算石蒜属亲本及杂交种的Nei’s 遗传距离和SM遗传相似系数,并用非加权组平均法(UPGMA)进行聚类分析。
参照徐雷峰等[14]方法将获得的荧光标记EST-SSR多态性数据转换为数字(1~9)表示,当基因型数大于9时,用小写字母(a、b、c
$、\cdots $ )依次表示,无带用0表示。按照引物扩增带型数由少到多的顺序,记录各样品在15对引物上的扩增带型数据,通过个位数字或小写字母编码构建各杂交种的分子身份证。 -
荧光毛细管电泳检测发现:15对引物在25个样品中均能获得大小精确的DNA片段,筛选其中具有种内一致性且种间多态性的4对引物。由表2可知:SSR203和SSR15在3个亲本中均能得到特征条带,SSR32可在石蒜和中国石蒜中得到特征条带,SSR198可在石蒜和换锦花中得到特征条带。因此SSR203、SSR115和SSR198用来鉴定石蒜-换锦花杂交种(包括正交和反交)的真实性,SSR203、SSR115和SSR32可鉴定石蒜-中国石蒜杂交种的真实性。
引物 条带长度/bp 换锦花 石蒜 中国石蒜 SSR203 204 189 195 SSR115 226 230 228 SSR32 175 169 SSR198 228 221 Table 2. Characteristic bands of 4 fluorescent labeled EST-SSR primers on the parents of Lycoris
-
利用SSR203、SSR115和SSR198对27个石蒜-换锦花杂交种F1代进行鉴定。由表3可知:SSR203从石蒜-换锦花杂交F1后代群体中鉴定到88.46%的真实杂交种,其中具有双亲特征条带(189/204 bp)的占69.23%,出现特异条带的有19.23%;SSR115从石蒜-换锦花杂交种F1群体中鉴定到84.61%的真实杂交种,其中具有双亲特征条带的占53.85%,出现特异条带的占30.77%;SSR198只鉴定到53.85%的真实杂交种。分别利用SSR203+SSR115、SSR203+SSR115+SSR198进行多重PCR,从石蒜-换锦花正反交F1杂交群体中均鉴定到96.30%的真实杂交种,大于SSR203+SSR198 (88.89%)和SSR115+SSR198 (88.89%)组合的鉴定结果。因此采用SSR203+SSR115和SSR203+SSR115+SSR198可以早期鉴定石蒜-换锦花杂交种F1的真实性。
同样,利用引物SSR203、SSR115和SSR32对石蒜-中国石蒜杂交种F1进行鉴定。SSR203鉴定石蒜-中国石蒜杂交种F1的真实杂交种占80.77%,其中具有双亲特征条带(175/169 bp)的占42.31%,具变异条带的有38.46%;SSR115和SSR32鉴定石蒜-中国石蒜杂交种F1群体的真实性分别为50.00%和69.23%。采用SSR203+SSR115、SSR203+SSR32和SR203+SSR115+SSR32多重PCR扩增,石蒜-中国石蒜杂交种F1群体的真实杂交种鉴定效率均达96.15%,高于SSR115+SSR32鉴定效率(76.92%)。因此可用SSR203+SSR115、SSR203+SSR32和SSR203+SSR115+SSR32对石蒜和中国石蒜杂交种真实性进行鉴定。
为节省成本,缩短育种周期和减少工作量,本研究采用SSR203+SSR115对石蒜-换锦花、石蒜-中国石蒜杂交种F1代进行早期鉴定。
引物 石蒜-换锦花杂交种的真实鉴定率/% 引物编号 石蒜-中国石蒜杂交种的真实鉴定率/% SSR203 88.46 SSR203 80.77 SSR115 84.61 SSR115 50.00 SSR198 53.85 SSR32 69.23 SSR203+SSR115 96.30 SSR203+SSR115 96.15 SSR203+SSR198 88.89 SSR203+SSR32 96.15 SSR115+SSR198 88.89 SSR115+SSR32 76.92 SSR203+SSR115+SSR198 96.30 SSR203+SSR115+SSR32 96.15 Table 3. Identification of interspecific hybridization of Lycoris by EST-SSR primers
-
筛选出的15对荧光标记EST-SSR引物在78个样品中扩增效果较好(表4),共得到扩增条带92条,扩增条带最多的是SSR32 (10条),最少的是SSR21 (2条),平均6.13条。多态性信息量(PIC)为0.6923~1.0000,其中SSR253最小(0.6923),SSR20和SSR142最高(1.0000),平均为0.9137。所有PIC值均大于0.500 0,说明均为高多态性引物,可反映杂交种间遗传差异和遗传多样性。
引物 Na Ne H In PIC 扩增条带/条 引物 Na Ne H In PIC 扩增条带/条 SSR7 1.974 4 1.291 4 0.218 1 0.369 4 0.974 4 6 SSR142 2.000 0 1.270 5 0.210 7 0.364 2 1.000 0 8 SSR15 1.833 3 1.167 7 0.136 8 0.250 2 0.833 3 7 SSR147 1.987 2 1.678 3 0.387 6 0.570 2 0.987 2 3 SSR20 2.000 0 1.382 5 0.273 1 0.440 4 1.000 0 5 SSR198 1.782 1 1.203 0 0.156 8 0.273 1 0.782 1 6 SSR32 1.987 2 1.193 0 0.159 5 0.293 1 0.987 2 10 SSR203 1.974 4 1.378 7 0.264 2 0.426 8 0.974 4 5 SSR85 1.897 4 1.212 6 0.168 0 0.298 0 0.897 4 6 SSR220 1.974 4 1.294 5 0.219 9 0.371 7 0.974 4 6 SSR96 1.871 8 1.082 1 0.259 2 0.409 8 0.871 8 5 SSR221 1.910 3 1.643 6 0.377 0 0.550 5 0.910 3 2 SSR115 1.846 2 0.214 9 0.167 3 0.292 0 0.846 2 7 SSR253 1.692 3 1.096 4 0.083 7 0.164 6 0.692 3 9 SSR138 1.974 4 1.252 8 0.196 0 0.340 9 0.974 4 7 平均 1.913 7 1.224 1 0.218 5 0.361 0 0.913 7 6.13 Table 4. Number of alleles and polymorphism information of 15 EST-SSR primers
-
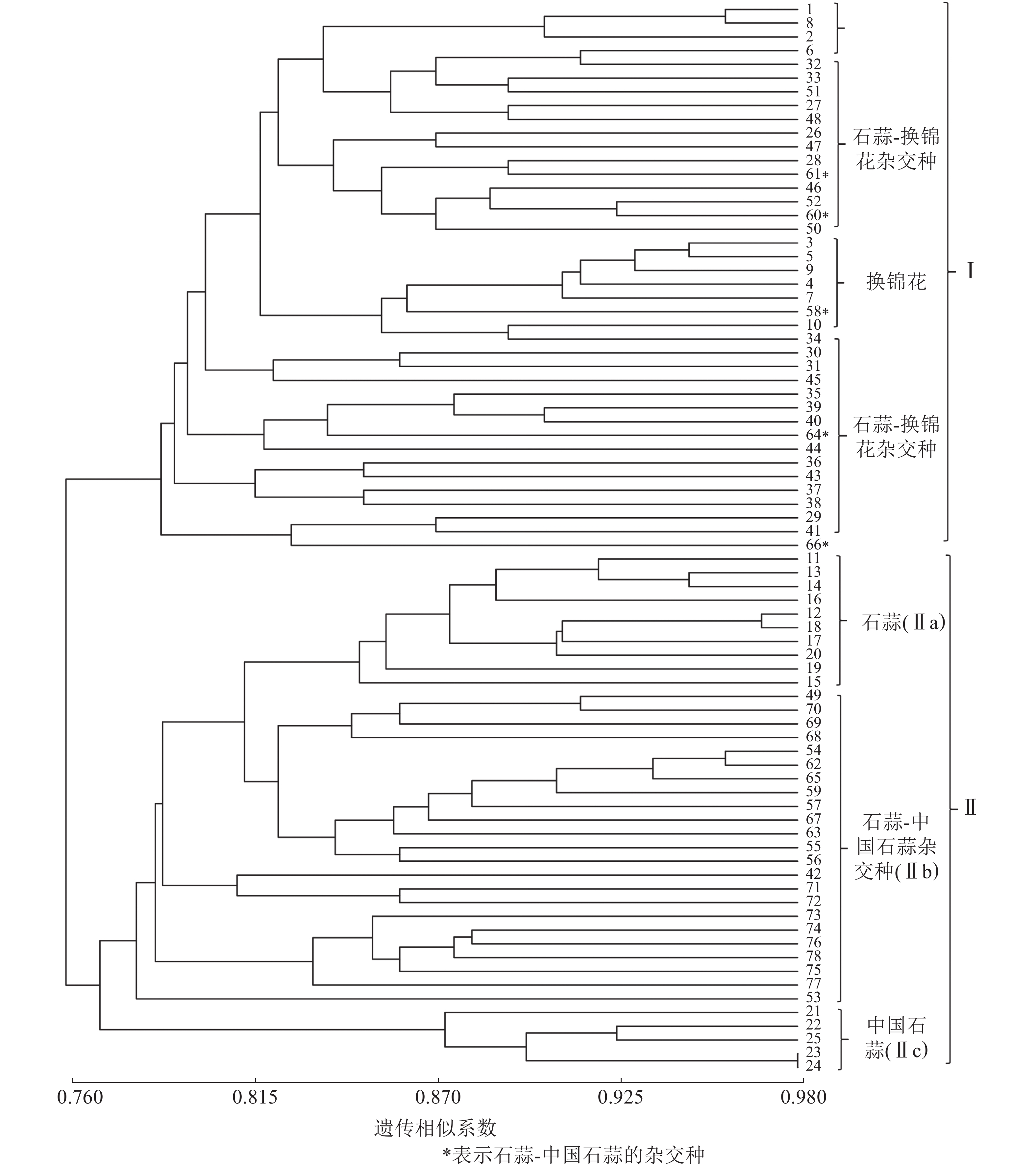
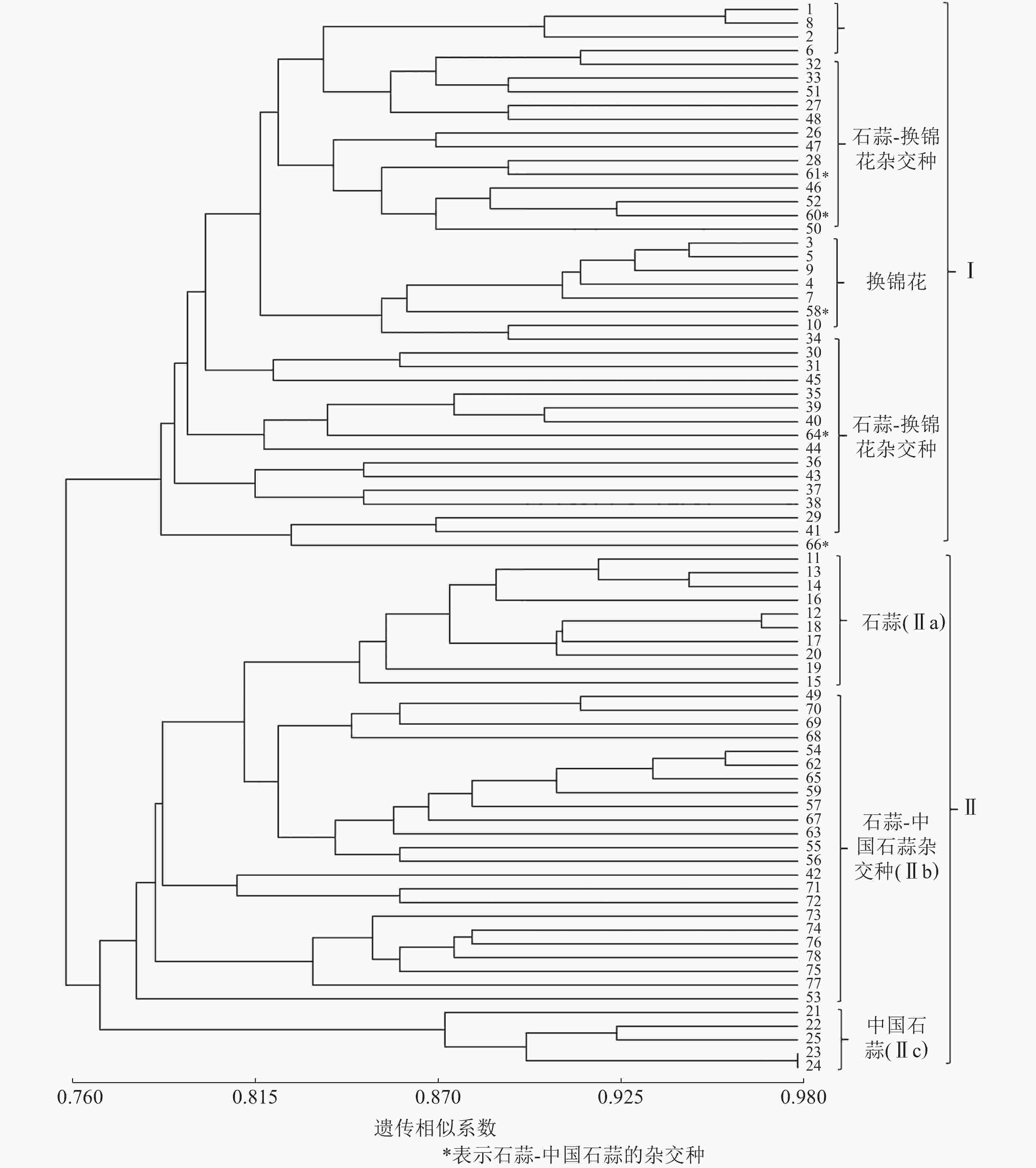
以15对荧光标记EST-SSR标记扩增结果计算遗传相似系数并进行UPGMA聚类分析。由图1可知:各样品遗传相似系数为0.76~0.98,在相似系数为0.77处25个亲本和53个杂交种聚为Ⅰ和Ⅱ两大类,Ⅰ类主要为换锦花、石蒜-换锦花杂交种,但包含了58、60、61、64和66等石蒜-中国石蒜杂交种,且与石蒜-换锦花杂交种聚在一起,推测5个杂交种的母本可能是石蒜。Ⅱ类包括Ⅱa(石蒜)、Ⅱc(中国石蒜)、Ⅱb(中国石蒜-石蒜杂交种)等3个亚类,说明本研究所用的15对荧光标记EST-SSR可用作石蒜、中国石蒜、换锦花及种间杂交种的早期鉴定。
-
由表5可知:15对荧光标记EST-SSR引物扩增各样品,共得到153种带型,平均每条扩增10.2带型,其中SSR221扩增带型最少(2种),SSR32扩增带型最多(30种)。根据表4对15对荧光标记EST-SSR引物扩增得到的基因型或带型赋值并排序,以此对各样品的扩增情况编码分子身份证,扩增条带位置相同则编码符号相同,说明基因型相同。由表6可知:各亲本和各杂交种均具有唯一的分子身份证,15对荧光标记EST-SSR引物可有效鉴别石蒜属亲本和种间杂交种。
基因型数 SSR15 SSR253 SSR138 SSR32 SSR20 SSR142 SSR198 SSR85 SSR147 SSR7 SSR220 SSR221 SSR96 SSR203 SSR115 0 − − − − − − − − − − − − − − − 1 241 263/268 202 166/172 116 247 220/228 83/105 245 173/188 224 249/255 149/154 189 226 2 241/244 269 202/206 166/175 117/120 247/253 221 89/94 245/248 182 224/228 255 151 189/195 226/228 3 242 272/277 202/209 166/181 118 247/254 221/228 100 245/251 182/185 224/234 151/154 189/204 226/230 4 242/250 274 206 166/188 118/121 247/256 221/230 107/110 248 182/188 224/237 151/156 195 226/231 5 244 274/277 206/209 169 171/120 247/259 222/228 108 251 185/188 224/244 151/157 195/204 226/242 6 244/246 274/279 206/212 169/172 121 247/260 228 108/110 188 228 154 201/207 227/231 7 244/250 277 206/215 169/175 247/262 229 110 191 228/237 154/156 204 228 8 245 277/279 206/218 169/178 247/267 230 203 237 154/157 207 228/230 9 246/254 278 209 169/188 247/270 238/244 156 230 a 248 279 209/215 169/191 270 157 230/242 b 250 291 209/212 169/194 231 c 250/254 212 172 237 d 254 212/215 172/175 e 215 172/178 f 215/221 172/181 g 172/191 h 175 i 175/178 j 175/188 k 175/191 l 175/194 m 178 n 178/185 o 178/188 p 178/191 q 185 r 185/188 s 188 t 188/191 u 188/194 说明:基因型数用1~9表示,大于9时用小写字母依次表示,0表示无条带;−表示没有带型 Table 5. Amplification patterns of 15 pairs of fluorescent labeled EST-SSR primers on parents and hybrid F1
编号 亲本 编号 石蒜-换锦花杂交种 编号 中国石蒜-石蒜杂交种 1 6aek3267522161a 26 574n37053312833 53 eb8m31013242120 2 5a4e3467331131a 27 57fi43053412330 54 50b74177124232a 3 7a7k32a7542231a 28 5b4p44653312423 55 a1b737063342300 4 7a7k4267342131a 29 525q4177023230a 56 82b73777335234b 5 2ad14467322241a 30 57bi42363321b43 57 3b7i31071341310 6 7adj4767521131a 31 57bn44051211040 58 544c61051342541 7 ca423767321171a 32 57fi43351312433 59 104737671642328 8 ca4e3467531231a 33 5bbi44161212031 60 507764053212633 9 77fi6467536271a 34 507u42365212b33 61 5b4344053212431 10 5afi3267332271a 35 505662663222733 62 50b737771342328 36 74d427353232333 63 02dc42771342728 11 53am25451212a71 37 52aa22051212252 64 52bf2277335283b 12 537m45451212a71 38 57bt22353212336 65 e07741771342328 13 54ap62251312b71 39 5bbf21353222833 66 2a838703252738 14 542p61451212b71 40 5bbh63361222633 67 0bfc3a77327232a 15 543u63251312b71 41 525841061252275 68 70f731a3367272a 16 55a243251312a71 42 fafg42b53322314 69 d0ek34a73312728 17 50ao63251312871 43 566421b61212734 70 d0d743a7122261c 18 57a225a51212a71 44 0abs17765222533 71 50bd4346326262c 19 557t22251312b71 45 5afu4216122208d 72 00c047a03262328 20 5ae442251312b71 46 075k64653212333 73 00bi33703562080 47 0bbt35353312833 74 5b4r47703312060 21 e00d31062022040 48 50bm43353112334 75 1bfl44704312060 22 00053a042082047 49 40f743a73242738 76 0cf747005312080 23 0015380717a2247 50 504d24251212733 77 8b6d42703572060 24 0015380417a2247 51 50bi41851222433 78 5bb641771342080 25 00c531001682247 52 504l44b53212634 说明:1~10为换锦花;11~20为石蒜;21~25为中国石蒜;26~52为石蒜-换锦花杂交种;53~78为石蒜-中国石蒜杂交种 Table 6. Molecular ID codes of parent L. radiata, L. chinensis, L. sprengeri and interspecific hybrids
-
SSR荧光标记在毛白杨Populus tomentosa[17]和建兰Cymbidium ensifolium [18]等核心种质构建、落花生[12]和山茶属Camellia植物[19]等分子身份证构建以及三叶青Tetrastigma hemsleyanum [20]、杂交兰[21]等种质资源遗传多样性研究中得到验证。本研究表明:利用荧光标记EST-SSR检测石蒜属各亲本及杂交种的真实性、分析其间遗传关系,精准高效,结果可靠。
本研究利用15对荧光标记EST-SSR引物从石蒜、中国石蒜、换锦花和种间杂交种共78个样品中扩增到92个条带,平均每对引物6.13条;15对ESR-SSR引物多态性信息量均大于0.5000,平均为0.9137,较好地反映了杂交种间的遗传差异和遗传多样性,与茶Camellia sinensis[22]、百合[14, 23]等的研究结果类似。利用荧光标记EST-SSR引物鉴定石蒜-换锦花、石蒜-中国石蒜杂交种F1代,发现SSR203+SSR115等引物对杂交种鉴定的真实性分别达96.30%和96.15%,与SSR203+SSR115+SSR198和SSR203+SSR115+SSR32等引物鉴定结果相同,因此用SSR203+SSR115引物对石蒜-换锦花、石蒜-中国石蒜杂交种F1进行鉴定,可大大缩短育种周期,减少工作量并节约成本。
分子身份证构建以 DNA 指纹图谱为基础,是识别种质资源的标志,可以更加简单明了地识别和检索种质资源[24]。本研究利用15对荧光标记EST-SSR引物共扩增出92个条多态性条带,153种带型,按照统计方便、书写简洁、字符串长短适中、易于检索及充分利用引物的原则,采用基因型赋值编码法[14]对石蒜属亲本和杂交种单一位点带型或杂合带型进行编码,构建石蒜属植物亲本和杂交种的分子身份证,较好地区分了78份石蒜属植物材料,说明该方法适用于石蒜属植物的鉴定。
以RAPD、ISSR和SCoT等分子标记技术分析石蒜属植物种质资源的遗传多样性,建立遗传距离和遗传相似系数矩阵,构建相应分子树状图和指纹图谱[2, 25-26],其目的主要是为了杂交种的亲本配制、野生资源的高效利用等。本研究利用15对荧光标记EST-SSR引物扩增了78个石蒜属植物的遗传物质,发现各样本遗传相似系数为0.76~0.98;UPGMA聚类分析发现:相似系数为0.77时,25个亲本和53个杂交种聚为两大类,Ⅰ类包括亲本换锦花、石蒜-换锦花的杂交种,Ⅱ类又分为Ⅱa、Ⅱb和Ⅱc 等3个亚类,包括石蒜、中国石蒜-石蒜杂交种和中国石蒜,说明15对荧光标记EST-SSR可将石蒜、中国石蒜、换锦花及种间杂交种有效聚类,结合杂交种的遗传多样性认为:荧光标记EST-SSR可有效鉴定石蒜属杂交种F1,并厘清其间的遗传关系。
Construction of molecular identification card of Lycoris interspecific hybrids
doi: 10.11833/j.issn.2095-0756.20210296
- Received Date: 2021-04-18
- Accepted Date: 2022-01-31
- Rev Recd Date: 2022-01-19
- Publish Date: 2022-05-23
-
Key words:
- Lycoris /
- hybrid identification /
- EST-SSR /
- molecular identification card
Abstract:
| Citation: | WANG Li, ZHOU Qi, GAO Yanhui. Construction of molecular identification card of Lycoris interspecific hybrids[J]. Journal of Zhejiang A&F University, 2022, 39(3): 562-570. DOI: 10.11833/j.issn.2095-0756.20210296 |










 DownLoad:
DownLoad: