-
黄精Polygonatum spp.的干燥根茎是常用的一味中药[1],具有补气养阴、健脾、益肾等功效,临床常用于脾胃气虚、胃阴不足、精血不足、腰膝酸软等证[2−3]。现代研究发现:黄精含有多糖、甾体皂苷、木质素等多种化合物,其中黄精多糖被认为是黄精主要的活性成分,可清除自由基,延缓衰老,降低血乳酸,缓解疲劳等。黄精多糖的降血糖和抗炎等多种作用相互协作,为研究治疗糖尿病提供了较好的思路。黄精药性平和,不仅具有多种药理作用,还有一定滋补作用,故在复方汤剂以及中成药的守正创新中具有较好的应用前景[4−5]。另外,黄精因具有独特风味和较好的口感,已被开发出多种保健食品,如黄精药膳、黄精酒、黄精饮料、黄精酸奶等[6]。随着大健康产业的发展,黄精保健食品的复合开发具有极大的市场潜力。
黄精品种多,适应能力强,分布广[7]。黄精遗传多样性保护与优良品种选育可以提高黄精品质与产量,促进黄精产业高质量发展。叶钱等[8]对多花黄精P. cyrtonema有效成分与主要环境因子的相关性进行研究,发现不同种源的多花黄精化学成分存在差异,这为进一步探讨黄精品质与环境因素之间关系提供了理论依据。
相关序列扩增多态性(SRAP)分子标记技术在植物遗传多样性方面研究应用较多,但在药用植物应用较少,如何首乌[9]。另有研究表明:SRAP分子技术能较好地用于铁皮石斛Dendrobium officinale品种亲缘关系的研究[10],但此技术尚未见用于黄精遗传多样性分析。与之前研究使用的简单重复序列SSR多态位点分析方法[11]、多态性分子标记(SCoT)方法[12]和简单序列间重复(ISSR)分子标记方法[13]、叶绿体基因联合序列分析[14]等技术相比,SRAP分子标记技术以基因的开放阅读框 (open reading frames,ORFs)的特定区域进行扩增,具有简单、可靠、共显性高等特点[15]。本研究采用SRAP分析技术对华东、西北、华中、西南4个居群47份黄精种质资源的遗传多样性进行研究,为黄精的遗传多样性与优良品种选育研究提供依据,以更好地保护和开发黄精种质资源。
-
收集华东居群(浙江省、福建省、安徽省、江西省),西北居群(河北省、陕西省),华中居群(湖南省),西南居群(四川省、贵州省、云南省)的黄精干燥叶片,共47份种质资源,各种质资源位置信息如表1所示。样品采集后置于密封袋中干燥保存,经浙江省亚热带作物研究所陶正明研究员鉴定后为多花黄精、长梗黄精P. filipes、黄精P. sibiricum、滇黄精P. kingianum。
编号 采样点 样品数 基源 北纬(N) 东经(E) 海拔/m 居群 ZJTS1 浙江省温州市泰顺县竹里畲族乡 1 多花黄精P. cyrtonema 27.70° 119.79° 229 华东 ZJCA2~3 浙江省杭州市淳安县里商乡 2 多花黄精P. cyrtonema 29.50° 118.95° 169 华东 ZJYJ4 浙江省温州市永嘉县岩头镇 1 多花黄精P. cyrtonema 28.44° 120.79° 919 华东 ZJJD5 浙江省杭州市建德市寿昌镇 1 多花黄精P. cyrtonema 29.43° 119.15° 300 华东 ZJOH6 浙江省温州市瓯海区景山街道 1 多花黄精P. cyrtonema 28.00° 120.64° 10 华东 ZJYQ7 浙江省温州市乐清雁荡山风景区 1 多花黄精P. cyrtonema 28.39° 121.06° 123 华东 ZJQY8~14 浙江省丽水市庆元县荷地镇 7 多花黄精P. cyrtonema 27.59° 119.24° 943 华东 ZJSY15 浙江省丽水市松阳县玉岩镇 1 长梗黄精P. filipes 28.41° 119.25° 606 华东 ZJSY16~17 浙江省丽水市松阳县玉岩镇 2 多花黄精P. cyrtonema 28.41° 119.25° 145 华东 ZJLQ18~20 浙江省丽水市龙泉市城北乡 3 多花黄精P. cyrtonema 28.30° 119.08° 671 华东 ZJJN21~22 浙江省丽水市景宁畲族自治县东坑镇 2 多花黄精P. cyrtonema 27.89° 119.69° 1190 华东 HBBD23~24 河北省保定市易县塘湖镇 2 黄精P. sibiricum 39.16° 115.36° 103 西北 FJZH25~26 福建省南平市政和县杨源乡 2 多花黄精P. cyrtonema 27.16° 119.02° 519 华东 FJFZ27 福建省福州市鼓岭旅游度假区 1 多花黄精P. cyrtonema 26.06° 119.40° 789 华东 SXLT28 陕西省西安市临潼区斜口街道 1 多花黄精P. cyrtonema 34.36° 109.19° 454 西北 HNAH29~32 湖南省益阳市安化县江南镇 4 多花黄精P. cyrtonema 28.35° 111.43° 149 华中 SCBZ33~34 四川省巴中市南江县南江镇 2 多花黄精P. cyrtonema 31.25° 106.45° 441 西南 GZTR35~36 贵州省铜仁市印江自治县杨柳镇 2 多花黄精P. cyrtonema 27.74° 108.49° 1394 西南 YNDG37~40 云南省昭通市大关县翠华镇 4 多花黄精P. cyrtonema 27.75° 103.89° 2611 西南 AHQM41~42 安徽省黄山市祁门县芦溪乡 2 多花黄精P. cyrtonema 29.68° 117.57° 487 华东 JXWY43~44 江西省上饶市婺源县珍珠山乡 2 多花黄精P. cyrtonema 29.19° 117.51° 183 华东 YNPE45~47 云南省普洱市镇沅自治县者东镇 3 滇黄精P. kingianum 24.00° 101.30° 1280 西南 Table 1. Collected information of Polygonatum spp.
-
将150~450 mg叶片于液氮下研成粉末,采用杭州莱枫生物科技有限公司的plant DNAzol试剂,逐一提取DNA后用核酸蛋白分析仪测定其质量浓度及质量分数,均稀释至20 mg·L−1,于−20 ℃保存待用。
-
PCR反应体系参照文献[16]。具体为:12.5 μL 2×预混型Taq酶,1.0 μL浓度为10 mmol·L−1的引物,模板DNA 40~60 ng,补水至25.0 μL。扩增程序为:预变性94 ℃ 5 min;变性94 ℃ 30 s,退火52 ℃ 30 s,延伸72 ℃ 2 min,35个循环后结束。72 ℃延伸7 min,4 ℃保存。电泳方法:反应产物5.0 μL,质量分数为1.2%聚丙烯酰胺凝胶电泳,5×TBE 缓冲液,150 V 跑胶 40 min,结束后凝胶成像系统拍照留存。
-
由SRAP-PCR电泳结果获得0/1矩阵。使用PopGene version 1.32进行数据分析,计算多态位点百分率(PPB)、等位基因数(Na)、有效等位基因数(Ne)、Nei’s基因多样性指数(H)、Shannon’s多态性信息指数(I)、Nei’s基因分化系数(Gst)、居群总基因多样性(Ht)、居群内基因多样性(Hs)、基因流(Nm);使用Ntsys-PC 2.1进行非加权组平均法(UPGMA)聚类分析。
-
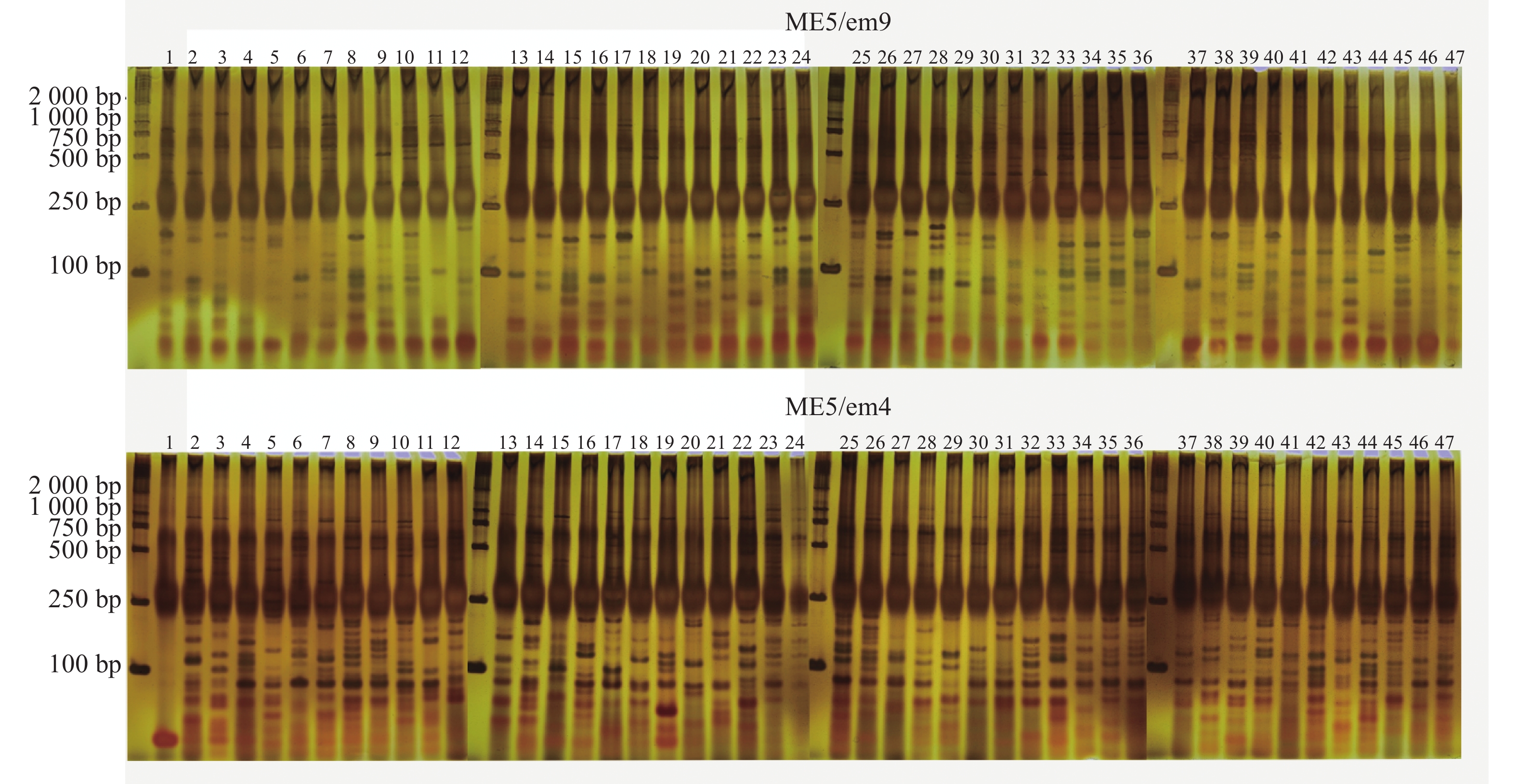
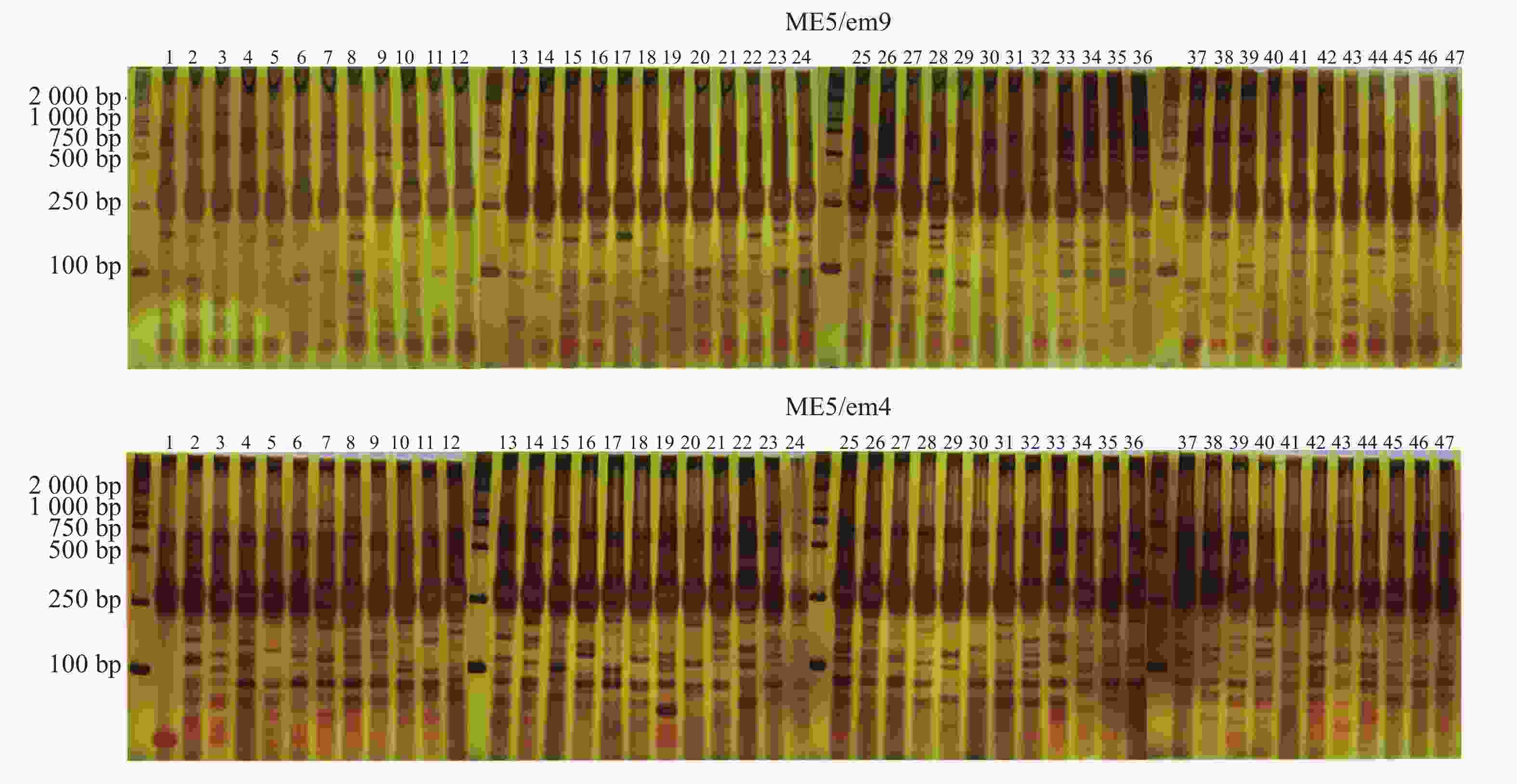
88对SRAP引物中筛选出7对扩增条带清晰且多态性明显的SRAP引物。引物序列见表2。47份样品共检测到159条扩增条带,其中140条为多态性条带,PPB为88.05%。引物ME4/em3扩增条带数最多,但其多态率最低,为68.29%。而引物ME5/em9、ME6/em4两者多态性条带数与扩增条带数量一致,多态率为100.00%(表3,图1)。
引物名称 ME引物序列(5′→3′) em引物序列(5′→3′) ME2/em9 TGAGTCCAAACCGGAGC GACTGCGTACGAATTCGA ME4/em3 TGAGTCCAAACCGGACC GACTGCGTACGAATTGAC ME4/em11 TGAGTCCAAACCGGACC GACTGCGTACGAATTCCA ME5/em9 TGAGTCCAAACCGGAAG GACTGCGTACGAATTCGA ME6/em4 TGAGTCCAAACCGGTAA GACTGCGTACGAATTTGA ME8/em4 TGAGTCCAAACCGGTGC GACTGCGTACGAATTTGA ME8/em9 TGAGTCCAAACCGGTGC GACTGCGTACGAATTCGA Table 2. Primer sequences for SRAP
引物名称 扩增条带数/条 多态性条带数/条 PPB/% 条带大小/bp ME2/em9 23 23 100.00 100~2 000 ME4/em3 41 28 68.29 100~2 000 ME4/em11 17 16 94.12 100~2 000 ME5/em9 19 19 100.00 100~2 000 ME6/em4 19 19 100.00 100~2 000 ME8/em4 17 14 82.35 100~2 000 ME8/em9 23 21 91.30 100~2 000 总计 159 140 88.05 Table 3. Amplification results of combinated SRAP primers
-
物种水平上黄精PPB为88.05%,H为0.259 8±0.166 9,I为0.401 0±0.227 2 (表4),表明黄精有较高的遗传多样性和遗传变异度。居群水平上, Na为1.421 4~1.861 6,平均值为1.600 6;Ne为1.276 1~1.421 2,平均值为1.343 6;H为0.188 1~0.259 1,平均值为0.204 1;I为0.238 2~0.399 4,平均值为0.308 0;PPB为42.14%~86.16%(表4),从高到低依次是华东居群、西南居群、华中居群、西北居群,表明华东居群遗传多样性丰富,遗传变异度较高。4个黄精居群之间的Ht为0.253 3±0.029 4, Hs为0.204 1±0.020 1,基因分化系数Gst为0.194 1,即居群间遗传变异占居群遗传变异的19.41%,80.59%的遗传变异在居群内进行;种群间的遗传分化系数基因流Nm为2.075 4,表明居群间存在一定的基因流动。
居群 Na Ne H I PPB/% 华东(HD) 1.861 6±0.346 4 1.421 2±0.325 1 0.259 1±0.167 1 0.3994±0.2291 86.16 西北(XB) 1.421 4±0.495 3 1.276 1±0.366 2 0.160 6±0.198 4 0.2382±0.2876 42.14 华中(HZ) 1.522 0±0.500 1 1.318 2±0.363 8 0.188 1±0.197 1 0.2819±0.2850 52.20 西南(XN) 1.597 5±0.492 0 1.358 8±0.381 1 0.208 7±0.201 3 0.3123±0.2860 59.75 居群水平 1.600 6 1.343 6 0.204 1 0.3080 60.06 物种水平 1.880 5±0.325 4 1.422 6±0.325 7 0.259 8±0.166 9 0.4010±0.2272 88.05 说明:华东种质资源数29个;西北种质资源数3个;华中种质资源数4个;西南种质资源数11个。 Table 4. Genetic diversity of Polygonatum spp.
-
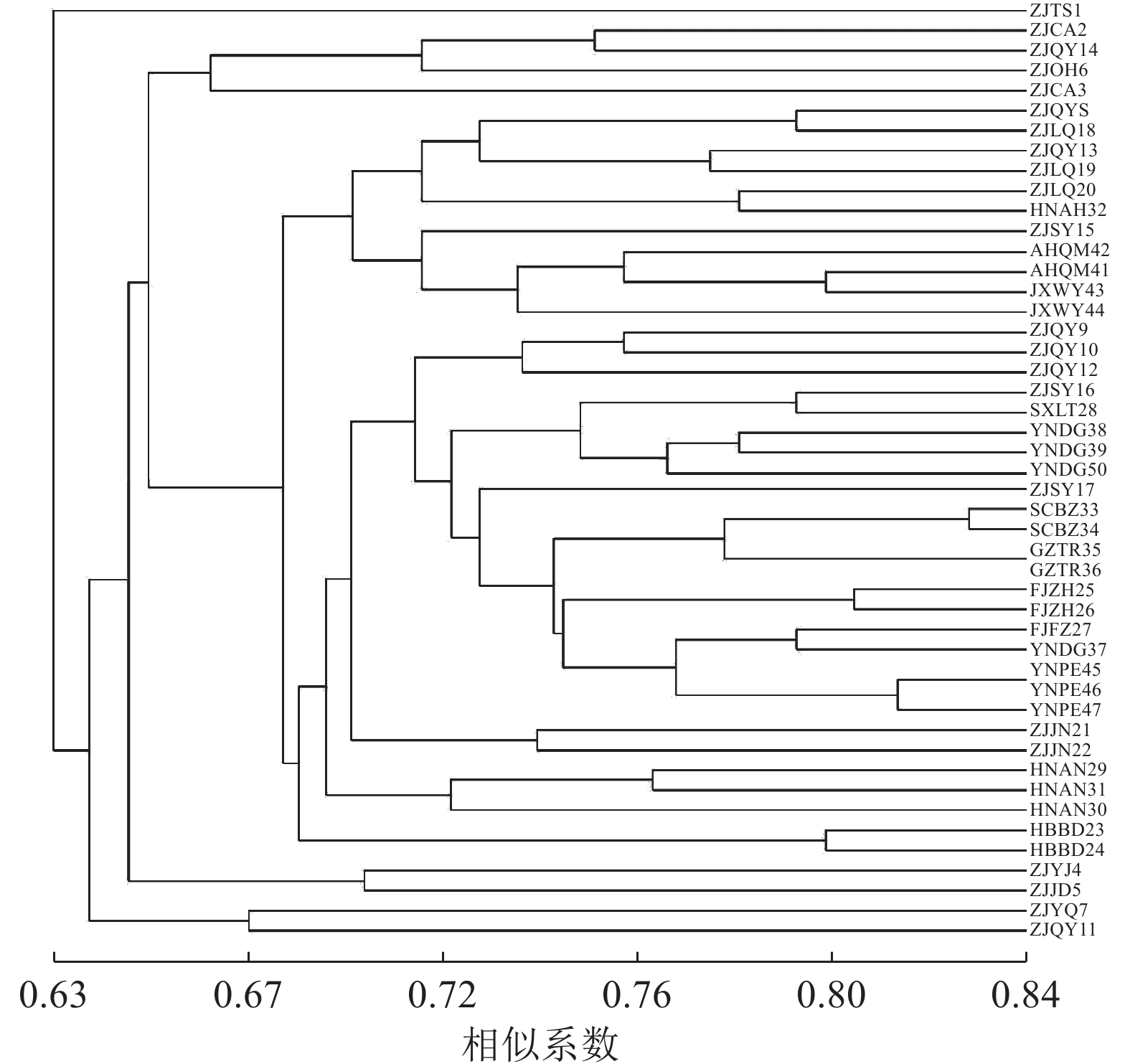
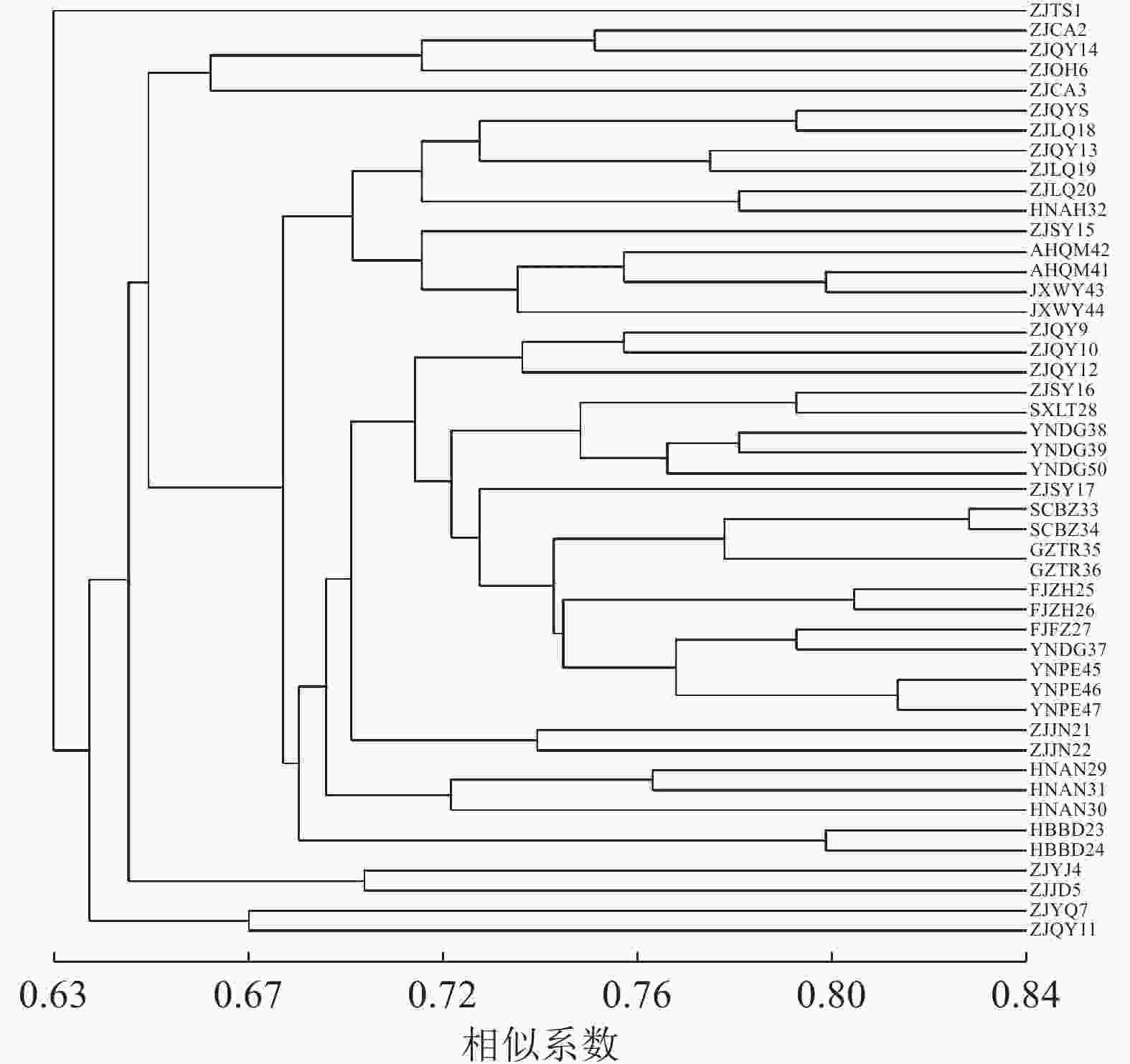
对样品个体进行聚类分析(图2),47份黄精种质资源的遗传相似系数为0.63~0.84。当相似系数为0.63,浙江泰顺种质资源单独聚为一类,表明此种质资源与其他黄精种质资源的遗传距离较远。当相似系数为0.66,聚为4类,Ⅰ、Ⅲ、Ⅳ 类为浙江部分种质资源单独聚类,表明这些地区的黄精种质资源在亲缘关系上比较接近,可能与引种有关。当相似系数为0.68时,Ⅱ类分聚为2支,华东地区的安徽、江西与浙江龙泉、庆元、松阳部分种质资源以及少数华中居群聚为一支,另一支为华东地区的浙江景宁、庆元和松阳部分种质资源与多数华中、西南、西北居群聚类,西北居群与西南居群虽然地理差异大,但黄精种质资源之间的遗传距离相差并不大,种质资源之间亲缘关系较近,可能与黄精引种栽培有关。庆元地区是黄精种植开始较早的地方,因此种质引进种植比较多样,所以采集数量最多。由图2可以看出,庆元7份种质资源分别聚为4类:ZJQY8和ZJQY13与ZJSY15、ZJLQ18~20、AHQM41~42、JXWY43~44以及HNAH32聚为一类;ZJQY11与ZJYQ7聚为一类;ZJQY14与ZJCA2~3、ZJOH6聚为一类;ZJQY9~10和ZJQY 12与部分华东、华中以及西北、西南种质资源聚为一大类,遗传多样性丰富。
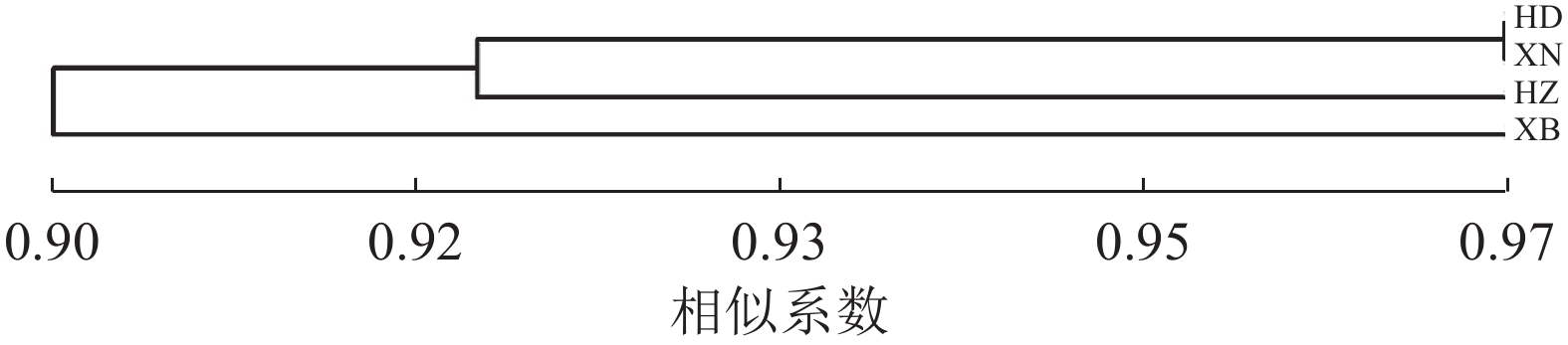
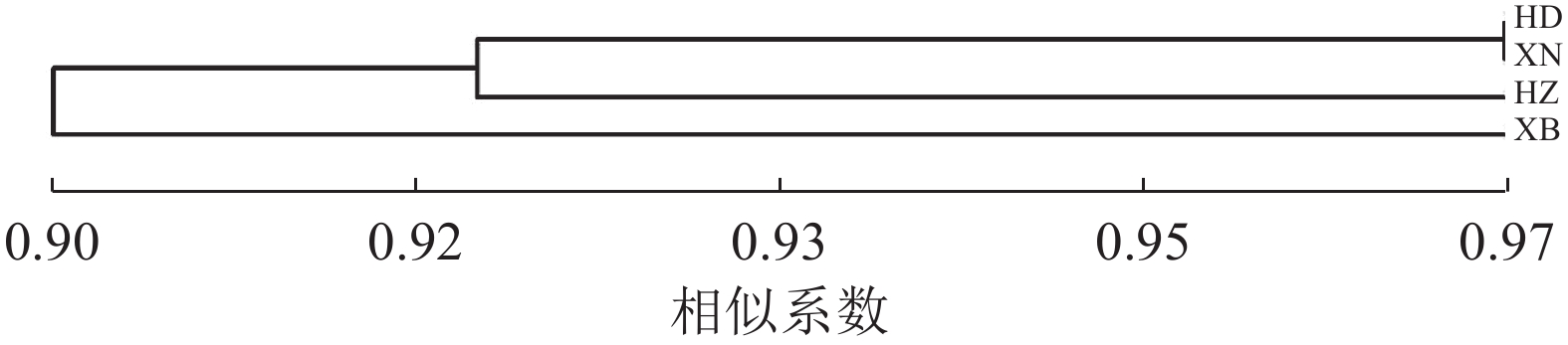
对4个居群的种质资源进行聚类,遗传相似系数为0.90~0.97 (图3)。相似系数为0.90时,可分为2类,华东居群、西南居群与华中居群聚为一类,西北居群单独为一类。
-
遗传多样性是生物多样性的核心,是物种保持进化潜能的基本条件,也是优良品种选育的主要依据[17]。本研究采用SRAP分子标记技术对47份黄精种质资源进行分析,多态性比率为88.05%,表明黄精遗传多样性丰富,其中华东居群遗传多样性相对其他地区更丰富一些。推测有2种原因:一是与地区采样量有关,二是华东地区黄精种质资源以多花黄精为主。多花黄精遗传多样性高于黄精[18]。相对于SRAP分子标记方法,使用ISSR方法,对福建多花黄精与长梗黄精多态性比率可达100%,使用SCoT分子标记技术,多态性比率可达为99.6%[12−13]。从多态位点来看,另外2种标记方法优于SRAP技术。物种或者居群的遗传多样性越高,对环境变化的适应能力就越强。在本研究中,华东居群整体上遗传多样性高,与朱巧等[19]的研究结果相反。他们对60份黄精种质资源进行SSR分子标记分析,其结果为西部地区黄精种质资源遗传多样性最高,与黄精属起源中心为西南地区染色体基数较高有关,但结合居群聚类结果,华东与西南遗传相似系数最近,且聚为一类。这可能是因为黄精自西南引种之后,在华东地区对环境变化的适应力较强,保留了一定的遗传多样性,所以华东地区在黄精遗传多样性保护以及优良品种选育方面有一定的地理优势,可充分利用以促进黄精产业高质量发展。
遗传分化是指遗传多样性在居群内和居群间的分布[20]。本研究47份黄精资源的Gst 为0.194 1,表明80.59%的遗传分化发生在居群内,19.41%的遗传分化存在于居群间,居群内有较高的遗传分化。这一结果与籍蓉蓉[21]对安徽省多花黄精遗传变异研究结果一致,居群内变异要大于居群间变异。基因流大于1时,表明居群间有一定程度的基因交流。本研究中黄精基因流Nm 为 2.075 4,表明不同居群间存在一定的基因交流。黄精可自花授粉,也可借助昆虫等媒介进行异花授粉。对各种质资源的聚类分析发现:华东居群有较高的遗传多样性,这可能与引种其他地区黄精种质资源有关,引种在同一地区增加了不同居群间的基因交流,从而提高了黄精遗传多样性。
遗传距离能判断种质资源间遗传关系远近。云南普洱和贵州铜仁采集的黄精种质资源都有最小的遗传距离,遗传相似度高,亲缘关系较近,与刘新等[22]研究结果一致,相同或相近地区种质资源基本聚在一起,可能与黄精无性繁殖有关。浙江庆元黄精种质资源则不同,遗传距离远近不一,具有复杂的亲缘关系,这与庆元地区为早期种植黄精的地区,种质资源引进种植多样有关。浙江一带黄精种质资源有较为复杂的亲缘关系,这种复杂性增加了黄精的遗传多样性。浙江一带引种栽培较多,为各种质资源之间遗传交流提供基础,有助于优质黄精品种选育。
Genetic diversity of Polygonatum spp. from different production areas based on SRAP markers
doi: 10.11833/j.issn.2095-0756.20220436
- Received Date: 2022-06-28
- Accepted Date: 2023-02-25
- Rev Recd Date: 2023-02-20
- Publish Date: 2023-05-20
-
Key words:
- Polygonatum spp. /
- SRAP molecular markers /
- genetic diversity /
- production areas
Abstract:
| Citation: | LI Yaping, DAI Huiming, JIANG Wu, et al. Genetic diversity of Polygonatum spp. from different production areas based on SRAP markers[J]. Journal of Zhejiang A&F University, 2023, 40(3): 658-664. DOI: 10.11833/j.issn.2095-0756.20220436 |









 DownLoad:
DownLoad: