-
金线莲Anoectochilus roxburghii为兰科Orchidaceae开唇兰属Anoectochilus多年生草本植物,别名金线草、金线兰等,主要分布在中国福建、浙江、云南和台湾等地区。金线莲因含有黄酮类、多糖类、生物碱类等成分[1],其药用价值日益被重视。由于金线莲对生长环境要求严格,且野生资源被过度采摘,导致金线莲已濒临灭绝[2],因此有必要开展金线莲资源的保护。种质资源收集和遗传多样性评估是金线莲资源保护的基础研究工作。分子标记可以在DNA水平上揭示植物的遗传变异,是一种稳定可靠的遗传分析方法[3]。简单重复序列间扩增多态性(inter-simple sequence repeat,ISSR)和序列相关扩增多态性(sequence-related amplified polymorphism,SRAP)主要针对非特异性序列进行扩增,对于基因组序列信息匮乏的物种较为有效[4]。目前,分别利用ISSR、SRAP等单一分子标记对金线莲遗传多样性的评价[5-11]已取得了一定的进展,但单一的分子标记技术经常受到扩增区域限制、引物扩增能力差异、模糊显性标记主观计入等因素影响,不能完全评价生物遗传多样性[12-13],而综合运用多种分子标记,能最大程度优化聚类分析结果[14-15]。目前,结合ISSR与SRAP分子标记技术已经应用于薄荷Mentha haplocalyx[12]、烟草 Nicotiana tabacum[16-17]、韭菜Allium tuberosum [18]、栀子Gardenia jasminoides [19]等研究。本研究采用ISSR与SRAP相结合的方法对浙江与福建等地引种、杂交与野生的金线莲样品进行研究,揭示金线莲个体间与种源间的遗传分化,为金线莲资源的保护和利用提供参考。
-
48份金线莲新鲜叶片样品详见表1。
编号 样品 来源地 编号 样品 来源地 编号 样品 来源地 1 健君1号 浙江温州 17 尖叶自交辐射选育种 福建福州 33 福建网纹种 福建厦门 2 健君2号 浙江温州 18 红霞变异种 福建厦门 34 福建无网纹种 福建厦门 3 金康1号 浙江金华 19 小叶金线莲 福建福州 35 大福星 福建厦门 4 大圆叶 福建厦门 20 温州文成野生种 浙江温州 36 大圆宝 福建厦门 5 健君1号原种 浙江温州 21 尖叶金线莲 福建泉州 37 红霞 福建厦门 6 戴云山野生种 福建泉州 22 大叶金线莲 福建三明 38 健君1号 浙江温州 7 福建永春黄带种 福建泉州 23 云南金线莲 云南昆明 39 尖叶金线莲 福建三明 8 台湾金线莲 台湾高雄 24 野生无纹 福建厦门 40 庆元无网纹 浙江庆元 9 台湾金线莲 台湾高雄 25 尖叶红杆种 福建福州 41 福建金草繁育种 福建厦门 10 无纹(G) 福建三明 26 尖叶变异筛选种 福建福州 42 小圆叶 福建三明 11 福建金线莲 福建福州 27 尖叶变异筛选种 福建福州 43 银圆宝 台湾高雄 12 大叶(H) 福建三明 28 大叶金线莲 福建三明 44 尖叶自交选育种 福建福州 13 野生种子繁育种 福建三明 29 野生种子繁育种 福建三明 45 尖叶杂交种 福建福州 14 林下尖叶(台州) 浙江台州 30 福建本地银线莲 福建厦门 46 大叶红霞 福建三明 15 林下沙畈本地无纹 浙江金华 31 福建小圆叶 福建厦门 47 尖叶变异筛选种 福建福州 16 野生金线莲 江西萍乡 32 金华本地种 浙江金华 48 无纹金线莲 福建三明 Table 1. Tested samples of A. roxburghii
-
参照DNA提取试剂盒说明书提取金线莲的DNA,得到的DNA用50 μL双蒸水(ddH2O)溶解。以ddH2O为对照,使用超微量分光光度计检测DNA溶解液的浓度以及纯度。
-
采用哥伦比亚大学公布的第9套100个ISSR通用引物序列,由北京擎科生物科技有限公司合成。使用3个已提取的DNA对100个ISSR引物进行筛选,反应体系总体积为20 μL,其中2×Taq Plus MasterMix 10 μL、ddH2O 8 μL、ISSR引物 1 μL、DNA溶解液1 μL,PCR扩增程序为98 ℃预变性30 s;94 ℃变性10 s,退火(不同引物退火温度不同) 15 s,72 ℃延伸15 s,循环35次;最后72 ℃延伸1 min,4 ℃保存。对产物进行琼脂糖凝胶电泳检测,挑选出多态性好、扩增条带清晰的引物对48个DNA样品进行扩增。
-
参照FERRIOL等[20]的设计原理建立SRAP标记分析体系,由北京擎科生物科技有限公司合成,包含14条上游引物(Me1~Me14)和17条下游引物(Em1~Em17),上下游引物可随机组成238对SRAP引物组合。使用2个已提取的DNA对238对SRAP引物组合进行筛选,PCR体系及扩增程序同上。将挑选出的多态性好、扩增条带清晰的引物对48个DNA样品进行扩增。
-
对PCR扩增产物电泳胶图进行人工读带,对扩增条带按有(1)或无(0)进行统计,形成“0, 1”数据矩阵,分别得到ISSR、SRAP及两者综合的数据。利用Excel统计每个(对)引物的总扩增条带数、多态性条带数和多态位点百分率(PPB)。使用POPGENE 32.0软件计算等位基因数(Na)、有效等位基因数(Ne)、Nei’s基因多样性指数(H)、Shannon’s多态性信息指数(I)、居群总基因多样性(Ht)、居群内基因多样性(Hs)、基因分化系数(Gst=1−Hs/Ht)和基因流(Nm)等遗传多样性相关参数,并计算不同种源间的遗传距离和遗传一致度,使用OmicStudio工具对遗传距离进行主坐标分析(PCoA),使用NTSYS-PC 2.1软件对所采集的金线莲样品进行聚类分析并绘制非加权组平均法(UPGMA)树状图。
-
经过2次筛选,共获得11条条带较为清楚的引物(表2)。11条引物共扩增出86条条带,其中多态性条带有84条,PPB平均值为97.67%,平均扩增条带数是7.82个,平均多态性条带数为7.64个。扩增条带数最多的引物是UBC880 (13条),其次是UBC861 (12条),扩增条带最少的是UBC810 (4条)。PPB为83.33%~100%,其中,UBC807、UBC810、UBC826、UBC834、UBC841、UBC842、UBC865、UBC68和UBC861的PPB为100%,均表现出极高的多态性,占总引物的81.82%;UBC856的PPB最低,为83.33%。
编号 ISSR引物 序列
(5′→3′)扩增条
带数多态性
条带数PPB/% 1 UBC807 (AG)8T 6 6 100 2 UBC810 (GA)8T 4 4 100 3 UBC826 (AC)8C 6 6 100 4 UBC834 (AG)8YT 8 8 100 5 UBC841 (GA)8YC 10 10 100 6 UBC842 (GA)8YG 8 8 100 7 UBC856 (AC)8YA 6 5 83.33 8 UBC865 (CCG)6 6 6 100 9 UBC868 (GAA)6 7 7 100 10 UBC880 (GGAGA)3 13 12 92.30 11 UBC861 (ACC)6 12 12 100 平均 7.82 7.64 97.67 合计 86 84 说明:Y=(C, T);PPB为多态位点百分率 Table 2. ISSR primer information and amplification results
-
经过2次筛选,共获得11对条带较为清楚的引物组合(表3)。11对引物共扩增出88条条带,其中多态性条带有86条,PPB平均值为97.73%,平均扩增条带数是8个,平均多态性条带数为7.82个。扩增条带数最多的引物组合是Me11-Em4 (12条);其次是组合Me4-Em13、Me2-Em14和Me13-Em10,扩增出9条条带;扩增条带最少的是Me13-Em16组合,只扩增出6条条带。PPB为88.89%~100%,其中,Me11-Em4、Me8-Em7、Me13-Em7、Me13-Em16、Me4-Em14、Me5-Em11、Me13-Em10、Me14-Em14和Me3-Em2组合的PPB为100%,均表现出极高的多态性,占总引物的81.82%;Me4-Em13和Me2-Em14组合的PPB最低,为88.89%。
编号 SRAP引物 正向引物
(5′→3′)反向引物
(5′→3′)扩增条
带数多态性
条带数PPB/% 1 Me11-Em4 BACG DTGA 12 12 100 2 Me8-Em7 BTGC DCAA 7 7 100 3 Me13-Em7 BAAC DCAA 8 8 100 4 Me4-Em13 BACC DCTA 9 8 88.89 5 Me13-Em16 BAAC DGAT 6 6 100 6 Me2-Em14 BAGC DCTC 9 8 88.89 7 Me4-Em14 BACC DCTC 7 7 100 8 Me5-Em11 BAAG DCAC 7 7 100 9 Me13-Em10 BAAC DCAG 9 9 100 10 Me14-Em14 BTCC DCTC 7 7 100 11 Me3-Em2 BAAT DTGC 7 7 100 平均 8 7.82 97.73 合计 88 86 说明:B=TGAGTCCAAACCGG;D=GACTGCGTACGAATT;PPB为多态位点百分率 Table 3. SRAP primer information and amplification results
-
ISSR研究中遗传一致度为0.4767~0.9070,遗传距离为0.0976~0.7408。其中,遗传一致度最高的1号与2号、3号与17号、14号与17号,均为0.9070,它们的遗传距离最小,均为0.0976,说明其亲缘关系较近;遗传一致度最低的是7号与22号,为0.4767,其遗传距离最大,为0.7408,说明其亲缘关系较远。在SRAP研究中遗传一致度为0.4659~0.9545,遗传距离为0.0465~0.7638。其中,遗传距离最小的是36号与37号,遗传距离最大的是10号与39号。综合ISSR和SRAP的数据后,遗传一致度为0.5115~0.8793,遗传距离为0.1286~0.6704。其中,遗传距离最小的是34号与37号,遗传距离最大的是10号与39号。
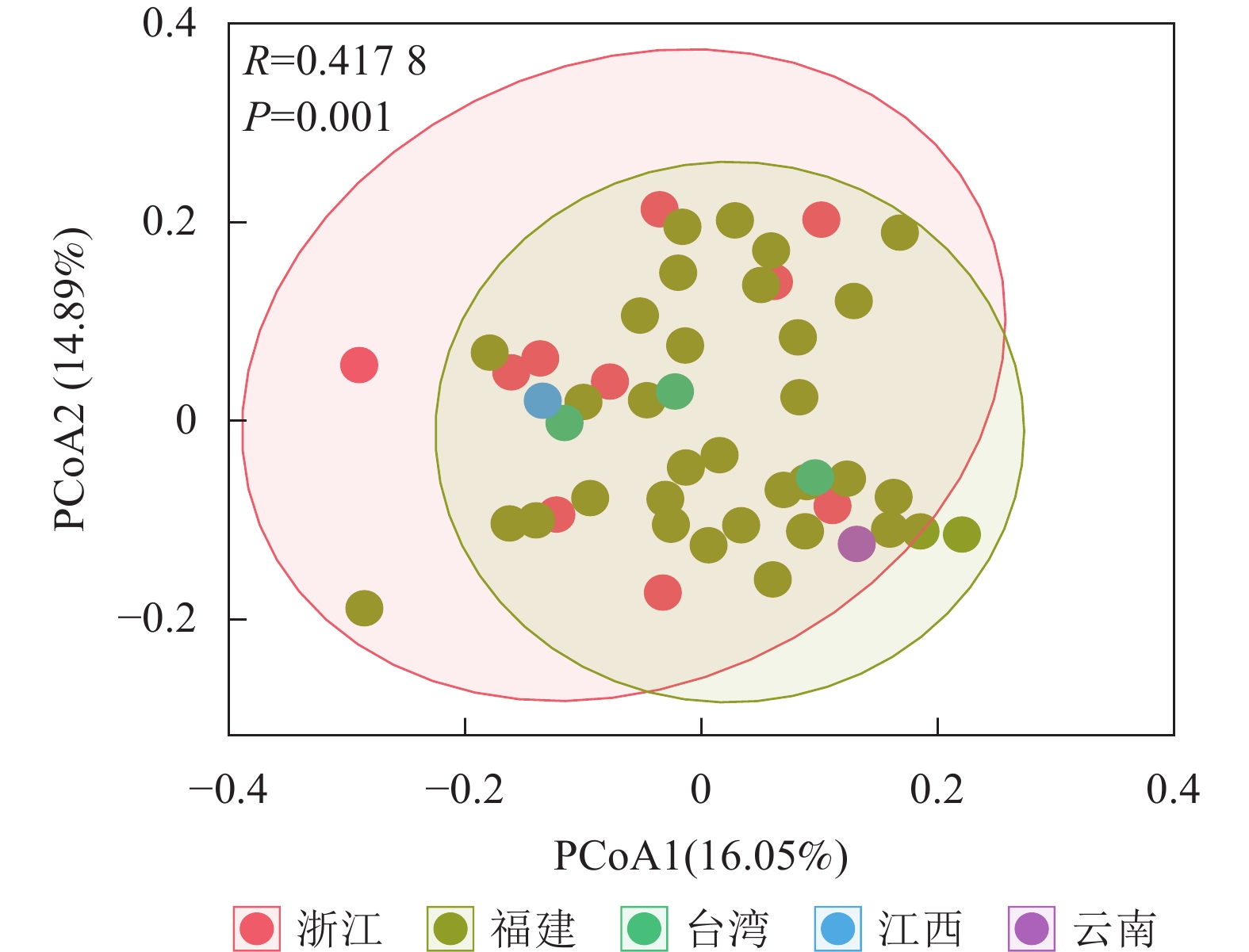
将48份样品按照产地来源分为5个群体(浙江、福建、台湾、江西和云南),利用POPGENE 32软件对其遗传距离和遗传一致度进行计算,结果如表4所示。使用OmicStudio工具将ISSR+SRAP的标记结果进行PCoA分析,结果如图1所示:由于来自台湾(3个)、江西(1个)和云南(1个)的样品数目较少,故不作分析。结合表4与图1可知:浙江省与福建省金线莲种质混杂。
分子标记 产地 浙江 福建 台湾 江西 云南 ISSR 浙江 0.9577 0.8842 0.8416 0.7285 福建 0.0433 0.8916 0.7792 0.7336 台湾 0.1231 0.1148 0.7310 0.6552 江西 0.1724 0.2495 0.3133 0.5814 云南 0.3168 0.3098 0.4228 0.5423 SRAP 浙江 0.9853 0.9480 0.7960 0.8351 福建 0.0148 0.9531 0.7880 0.8233 台湾 0.0534 0.0480 0.7796 0.7978 江西 0.2282 0.2383 0.2490 0.6250 云南 0.1802 0.1945 0.2259 0.4700 ISSR+SRAP 浙江 0.9712 0.9160 0.8187 0.7818 福建 0.0292 0.9229 0.7835 0.7798 台湾 0.0878 0.0802 0.7555 0.7270 江西 0.2000 0.2439 0.2804 0.6034 云南 0.2462 0.2487 0.3188 0.5051 说明:对角线下方为Nei’s遗传距离,对角线上方为Nei’s遗传一致度 Table 4. Genetic agreement and genetic distance among A. roxburghii populations
-
分析浙江与福建种源的样品,结果如表5所示:ISSR分析显示,43个种源在物种水平上,Na为1.9651,Ne为1.4403,H为0.2727,I为0.4247,PPB为96.51%;在群体水平上,Na为1.7093~1.9302,Ne为1.3409~1.4325,H为0.2075~0.2668,I为0.3207~0.4147,PPB为70.93%~93.02%,相对于物种水平而言,群体间的遗传多样性水平较低。SRAP结果与ISSR结果相似。结合ISSR与SRAP的数据分析:43个种源在物种水平上,Na为1.9713,Ne为1.3797,H为0.2429,I为0.3873,PPB为97.13%;在群体水平上,Na为1.7816~1.9425,平均值为1.8621;Ne为1.3578~1.3607,平均值为1.3593;H为0.2239~0.2288,平均值为0.2264;I为0.3488~0.3664,平均值为0.3576;PPB为78.16%~94.25%,平均值为86.21%,也是群体间的遗传多样性水平更低。其中,从ISSR、SRAP及综合研究结果来看,Na、Ne、H、I及PPB中基本上是福建省大于浙江省,说明福建省的金线莲种群遗传多样性更高。
分子标记 产地 等位基
因数 (Na)有效等位
基因数 (Ne)Nei’s基因多样
性指数 (H)Shannon’s多态性
信息指数 (I)多态位点百
分率 (PPB)/%ISSR 浙江 1.709 3 1.340 9 0.207 5 0.320 7 70.93 福建 1.930 2 1.432 5 0.266 8 0.414 7 93.02 群体水平 1.819 8 1.386 7 0.237 2 0.367 7 81.98 物种水平 1.965 1 1.440 3 0.272 7 0.424 7 96.51 SRAP 浙江 1.852 3 1.380 0 0.239 9 0.376 3 85.23 福建 1.954 5 1.284 8 0.191 6 0.319 1 95.45 群体水平 1.903 4 1.332 4 0.215 8 0.347 7 90.34 物种水平 1.977 3 1.320 6 0.213 8 0.350 8 97.73 ISSR+SRAP 浙江 1.781 6 1.360 7 0.223 9 0.348 8 78.16 福建 1.942 5 1.357 8 0.228 8 0.366 4 94.25 群体水平 1.862 1 1.359 3 0.226 4 0.357 6 86.21 物种水平 1.971 3 1.379 7 0.242 9 0.387 3 97.13 Table 5. Genetic diversity parameters of A. roxburghii
-
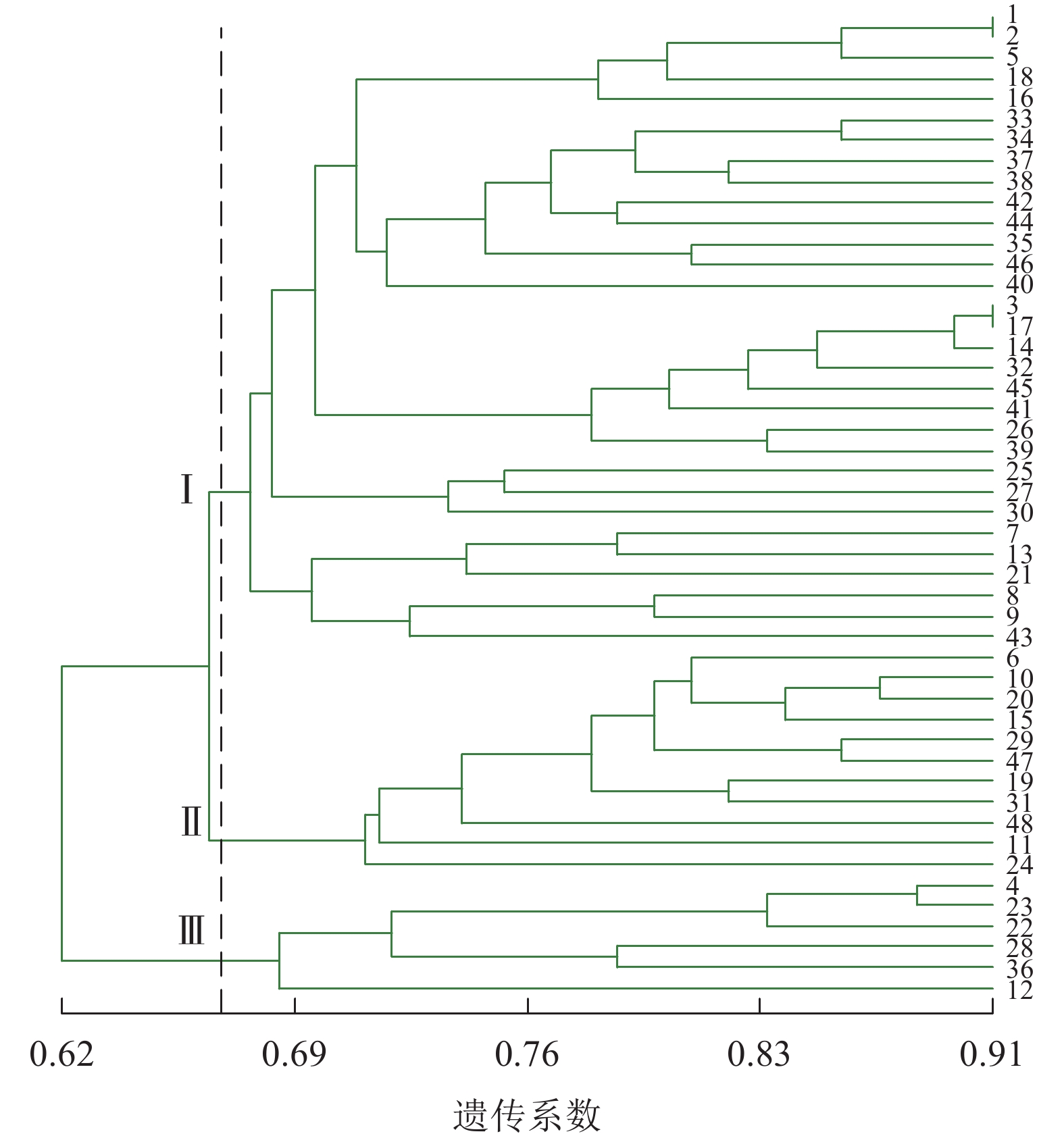
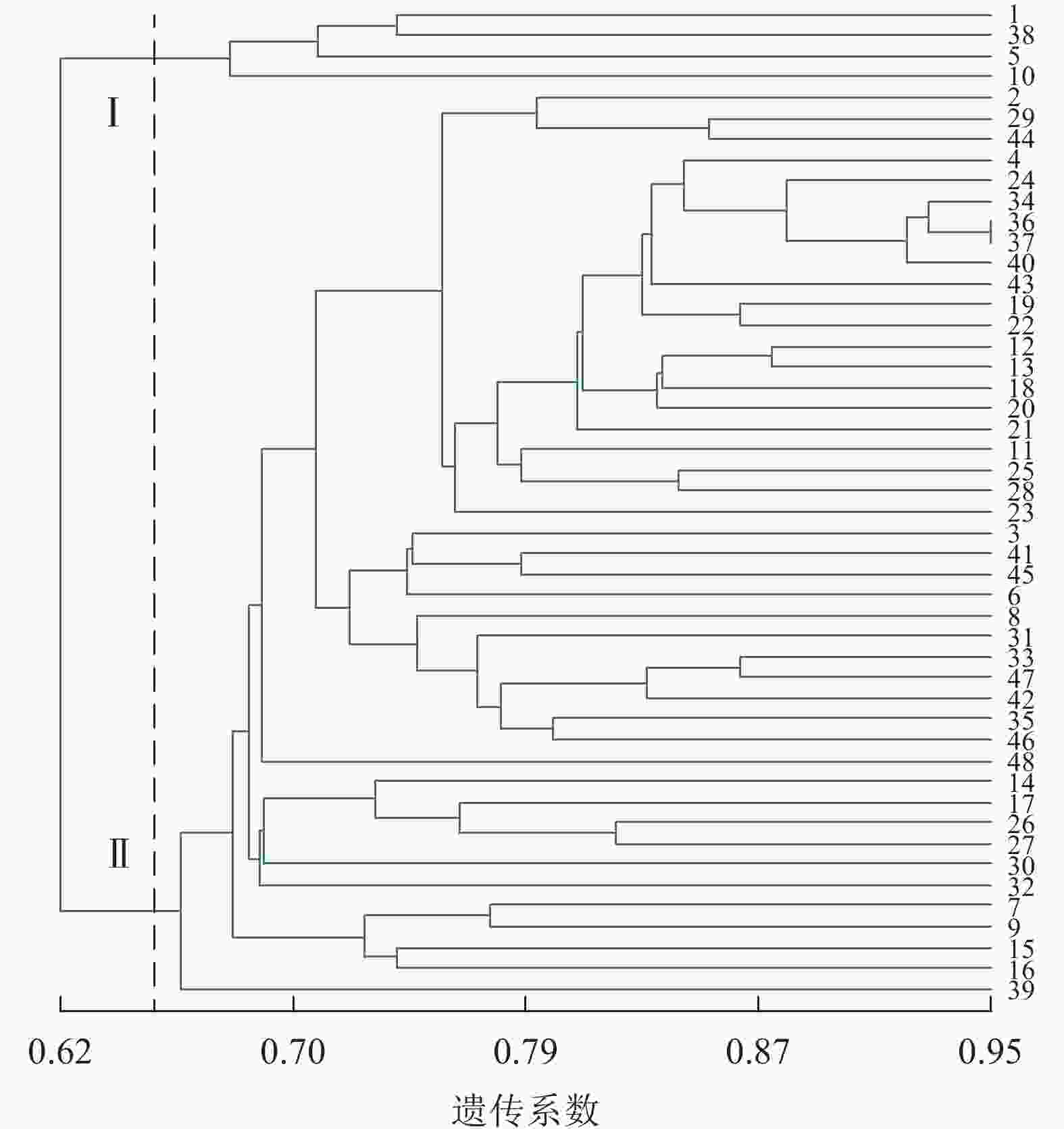
由图2可见:48份金线莲样品的遗传相似性系数为0.62~0.91,变幅为0.29。在遗传相似系数为0.67处,48份金线莲样品被划分为3类:在Ⅰ类中地理位置相同的主要有浙江温州的1、2、5、38号以及福建福州的17、25、26、27、44、45号,两地间亲缘关系相对较近;而Ⅱ类与Ⅲ类中样品的来源组成均较为分散,无明显特征。
-
图3显示:48份金线莲样品的遗传相似性系数为0.62~0.95,变幅为0.33。在遗传相似系数为0.65处,48份金线莲样品被划分为2类,Ⅰ类中1、5、38号均来自浙江温州,且品种相似,故聚为一类;而在Ⅱ类中,各个品种难以明显划分出小类,这也是各地种质较为混乱所带来的结果。与ISSR标记相比,SRAP标记更难划分类别。
-
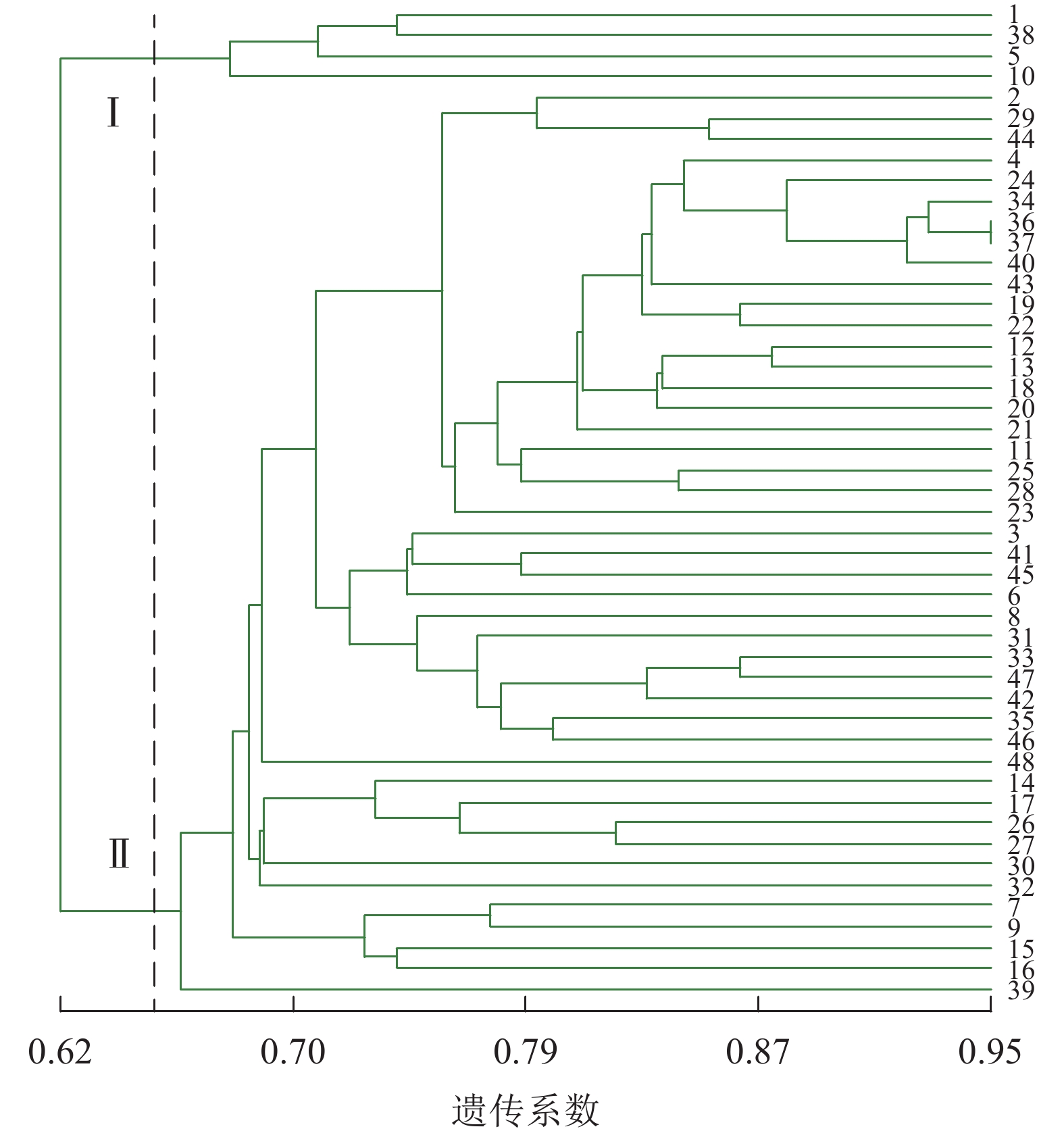
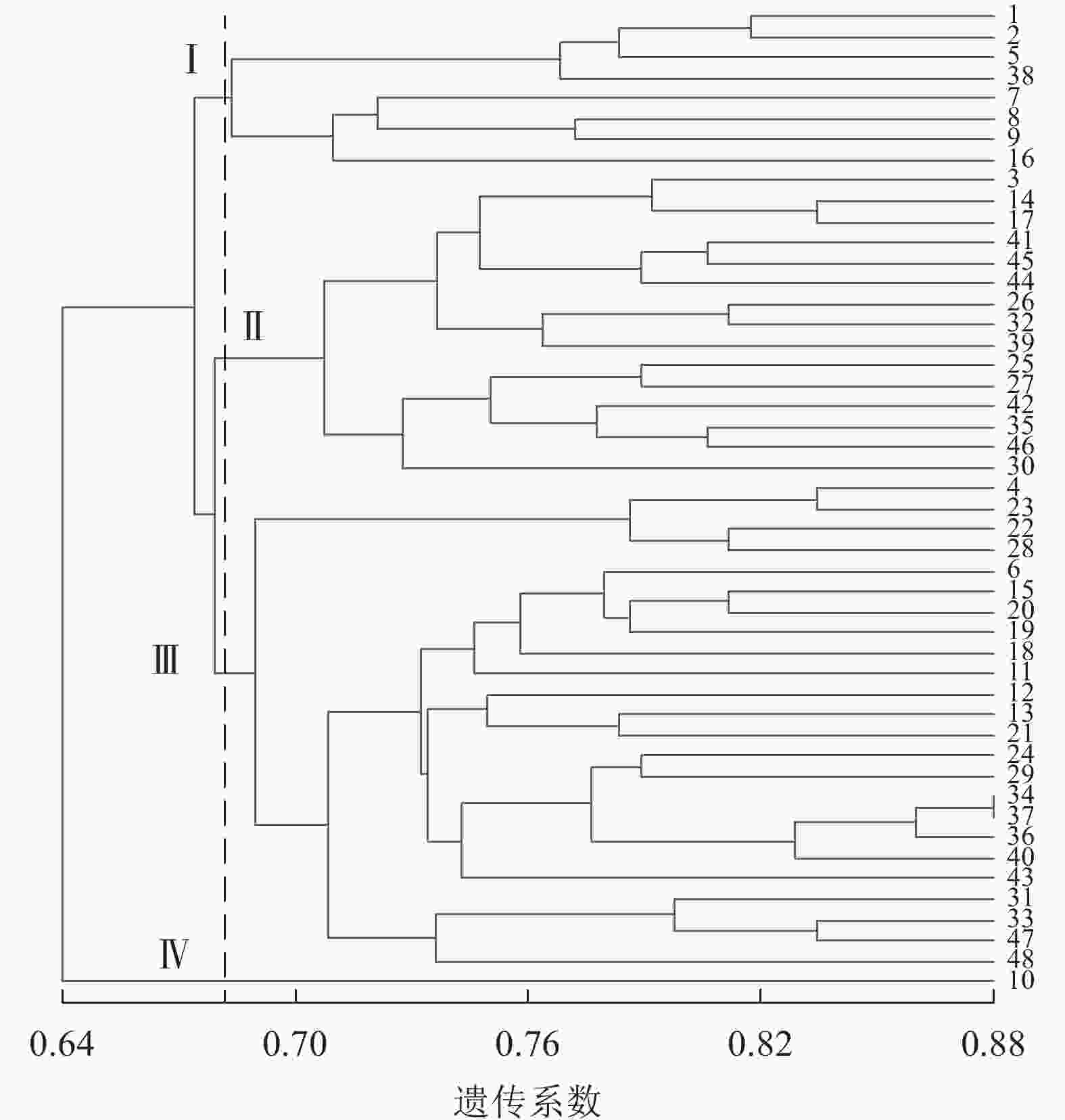
图4显示:48份金线莲样品的遗传相似性系数为0.64~0.88,变幅为0.24。在遗传相似系数为0.68处,48份金线莲样品被划分为4类,Ⅰ类中地理位置相同的主要有浙江温州的1、2、5、38号以及台湾高雄的8、9号,两地间可能品种相互引种;Ⅱ类中主要为来自福建福州的样品,包括17、25、26、27、44、45号;Ⅲ类中地理位置相同的主要有福建厦门的4、18、24、31、33、34、36、37号以及福建三明的12、13、22、28、29、48号,两地间品种互引的可能性较大。2种分子标记结合的方法更易体现样品间亲缘关系、划分类别,更清楚地体现地理位置对亲缘关系的影响。
-
本研究利用ISSR与SRAP对48份不同来源金线莲样品进行遗传特性分析,共筛选获得11条ISSR引物,扩增86条条带,其中多态性条带84条,PPB为97.67%;筛选出的11对SRAP引物共扩增出了88条条带,多态性条带86条,PPB为97.73%。说明供试金线莲样本具有丰富的遗传多样性,且相较于王剑锴等[10]筛选出的用于检测金线莲资源遗传特性的RAPD分子标记,ISSR与SRAP的标记多态性明显增多,进一步验证了ISSR与SRAP在金线莲种源多态性检测方面的高效率。
基于ISSR与SRAP分子标记对金线莲样品的遗传距离与遗传一致度分析发现:遗传距离最小的是34号(福建厦门福建无网纹种)与37号(福建厦门红霞),其亲缘关系较近;遗传距离最大的是10号(福建三明的无纹G)与39号(福建三明的尖叶),其亲缘关系较远。从遗传多样性分析结果来看,来自福建的金线莲遗传多样性更高,同时结合金线莲群体间的遗传一致度与遗传距离结果以及PCoA分析,可以看出各地间金线莲种质资源十分混杂,其中包含了大量的野生种、半野生种和人工驯化的栽培种及杂交种,这种复杂性导致不同种源间的差异性明显,金线莲种质的遗传多样性增加。浙江省市场内流通的栽培种多为省外引入种[21-22],各种质遗传交流频繁,这有利于培育出较优的金线莲品种。
本研究单独使用ISSR或SRAP的UPGMA聚类结果中,均出现各地间种源相互混杂的状况,并未严格按照地理距离的差异进行归类,而2种分子标记结合的聚类结果中,金线莲种质资源的聚类与不同地理位置分布的情况有比较高的一致度,表现出了一定地域性分布规律,说明2种分子标记方法结合相较于单一的分子标记方法能更准确地体现不同地区金线莲的差异性。由于2种分子标记所检测的基因座位以及所用的引物等因素存在差异,故两者得到的遗传距离不同,其聚类图也存在差异[23],且每种分子标记方法均有其优势与不足,结合多种标记技术则能更全面、准确地揭示种质遗传特性。因此,本研究结合ISSR与SRAP能更准确地揭示金线莲资源的遗传多样性和亲缘关系,为金线莲良种培育以及野生金线莲资源保护等方面提供了帮助。此外物种的遗传特性容易受到生态环境、繁殖方式以及各种人为活动的影响,野生金线莲经过长期的自然选择,其遗传背景较为复杂;人工栽培种种源多来自于野生金线莲,品种混杂。同时金线莲在野外萌发所需的自然条件苛刻,加之生境的易碎性对其生存产生很大压力[24],因此金线莲的遗传多样性受到生态环境极大的影响,所以有必要保护当地的生态环境以维护金线莲物种多样性。
Genetic diversity of Anoectochilus roxburghii based on ISSR and SRAP molecular markers
doi: 10.11833/j.issn.2095-0756.20220473
- Received Date: 2022-07-19
- Rev Recd Date: 2022-10-27
- Available Online: 2023-12-19
- Publish Date: 2023-02-20
-
Key words:
- Anoectochilus roxburghii /
- genetic diversity /
- molecular marker /
- germplasm resources /
- ISSR /
- SRAP
Abstract:
| Citation: | HUANG Jinchun, WAN Siqi, CHEN Yang, et al. Genetic diversity of Anoectochilus roxburghii based on ISSR and SRAP molecular markers[J]. Journal of Zhejiang A&F University, 2023, 40(1): 22-29. DOI: 10.11833/j.issn.2095-0756.20220473 |










 DownLoad:
DownLoad: