-
花榈木Ormosia henryi为豆科Leguminosae红豆属Ormosia重要木本药用植物,主要产于东南亚和南美洲,属于国家二级保护植物,在野外处于中度濒危状态[1]。花榈木常以根、根皮及茎、叶入药。《全国中草药汇编》记述,其性味归经为辛、温,有毒,具有活血化瘀、祛风、消肿之功。研究发现:红豆属植物含有生物碱、黄酮、苯丙素、萜类和挥发油等多种化学成分,其中萜类化合物是一类重要的活性物质[2]。迄今为止,已从红豆属不同植物中发现大量的萜类化合物,如丁香酚、苯乙醇、榄香素和棕榈酸等化合物,并发现多种物质具有消炎、抗氧化、抗菌的作用[3−4]。萜类物质是多种药用植物的主要活性化合物,并且是天然化合物中规模最大种类最多的一类物质[5−6],了解花榈木中萜类物质能够提高对花榈木的药用认知。
植物萜类化合物主要由甲基赤藓糖磷酸酯 (MEP) 途径或甲羟戊酸 (MVA) 途径产生的二甲基烯丙基二磷酸酯 (DMAPP) 和异戊烯基二磷酸酯 (IPP) 生成[7]。萜类化合物分子骨架是基于异戊二烯或其异构体二甲基烯丙基焦磷酸C5单元构成的,根据骨架碳链长度的不同,主要分为半萜、单萜、二萜、三萜等几种类型的萜类化合物[8]。MEP途径仅发生在质体中,MVA途径却分布在细胞质、内质网和过氧化物酶体之间[9]。IPP与DMAPP通过戊烯基转移酶的催化,缩合产生萜类化合物前体物质香叶酰二磷酸 (GPP)、法呢基二磷酸 (FPP) 和香叶基香叶基焦磷酸 (GGPP)。其中GPP是合成单萜类化合物前体物质,GGPP是合成二萜前体物质,FPP则是合成倍半萜和三萜化合物的前体物质[10−11]。随后通过萜类合成酶 (TPS) 和氧角鲨烯环化酶 (OSCs) 催化前体形成不同种萜类化合物,这2种酶的多样性使得萜类化合物的种类有很多[12−13]。
TPS基因家族主要分为7个亚家族,分别命名为TPS-a、TPS-b、TPS-c、TPS-d、TPS-e/f、TPS-g 和 TPS-h。其中TPS-a、TPS-b和TPS-g属于被子植物特有分支,这3个分支完全由专门的单萜、倍半萜或二萜生物合成基因组成,主要在植物生态相互作用中起作用。TPS-d是裸子植物特异性亚家族,主要包含特异性代谢的裸子植物TPSs[14]。TPS-c和TPS-e/f是被子植物与裸子植物所共有的亚家族,TPS-c分支成员只包含“DXDD”序类,是单功能柯巴基焦磷酸合酶 (CPS)。与TPS-c类似,TPS-e/f分支成员只包含“DDXXD”序类,是单功能的贝壳杉烯合成酶 (KS)[15−16]。植物OSC家族是一个超基因家族,主要包含达玛烯二醇合成酶 (DS) [17]、β-香树脂醇合成酶 (β-AS) [18]、α香树脂醇合成酶 (α-AS) [19]和羽扇豆醇合成酶 (LUS) [20]。近些年来对萜类的研究越来越广泛,但是豆科植物花榈木中的萜类化合物种类以及生物合成情况却未见报道。
本研究利用代谢组学测序数据首次对花榈木中的萜类物质进行鉴定,并结合转录组测序结果,利用生物信息学方法分析花榈木中萜类的主要合成情况,分析相关基因在花榈木不同组织部位中的表达水平,为研究花榈木的药用活性物质以及花榈木中萜类的代谢调控途径奠定基础。
-
2021年4月23日于华南农业大学 (中国广州) 苗圃中采集3年生花榈木新鲜的根、茎、树皮、老叶和幼叶5个组织部位样品,每样品重复4次,纯净水冲洗干净,立即在液氮中冷冻,并分成2个部分,一部分立即在−80 ℃下保存用于总RNA提取,另一部分在真空下冷冻干燥用于代谢物提取。
-
将采摘的根、茎、树皮、老叶和幼叶样品冻干磨粉后,精确称量各个样品粉末0.5 g,分别用5 mL体积分数为80%的HPLC级甲醇提取过夜,其中内标为1 μmol·L−1的白杨素。4 ℃下12 000 g离心30 min,取上清液装入样品瓶中,进行超高效液相色谱质谱 (UHPLC Q-TOF/MS) 分析。为了分离花榈木不同器官的代谢物,采用超高效液相色谱串联质谱法(Q Exactive Plus)进行测定。为保证平行试验每个样品平行4针。将1 μL样品进入2.1 mm×100.0 mm、1.9 μm粒径的Hypersil GOLD色谱柱,柱温30 ℃,流速0.3 mL·min−1。流动相A为体积分数0.1%的HPLC级甲酸,流动相B是HPLC级乙腈。梯度洗脱:0~2.0 min,0~10%流动相B;2.0~10.0 min,10%~55%流动相B;10.0~10.1 min,55%~80%流动相B;10.1~13.0 min,80%流动相B;13.0~14.0 min,80%~95%流动相B;14.0~18.0 min,95%~10%流动相B。全扫描质谱和数据关联扫描(dd-MS2)的分辨率分别设置为70 000和17 500,在正负离子模式下均使用加热的ESI源。在正负模式下,喷雾电压被设定为3.5和3.2 kV。毛细管温度被设定为320 ℃,辅助气体加热器的温度被设定为350 ℃。
使用Compound Discoverer 3.2对原始数据进行分析,然后根据综合分子量、质荷比 (m/z)、保留时间 (RT) 和二级光谱,与代谢物数据库mzCloud、mzVault和Chemspider进行对比。
-
总RNA由RNAprep Pure Assay试剂盒提取,RNA质量用NanoPhotometer®分光光度计测定。为了进行转录组测序,样品被送到百迈客生物技术公司。库的制备在Illumina Hiseq 2000平台上进行测序,并产生成对的读长 (reads)。首先,使用内部的perl脚本处理fastq原始数据,以去除含适配器的读数、含ploy-N的读数和低质量读数。经过Trinity软件组装获得一个非冗余的基因数据库 (universal gene, unigene),并进一步使用HMMER软件与Pfam数据库比对,获得unigene的注释信息。
-
以常用的基因表达水平估算方法中每千个碱基转录每百万映射读取的片断(fragments per kilobase million,FPKM) 值进行表达量统计。使用错误发现率(false discovery rate, FDR) ≤0.01 和 |log2CF| ≥2 (CF为差异倍数, fold change) 的阈值,写入R中的DESeq 2包,对花榈木不同部位样本进行差异基因表达分析[21]。使用TBtools构建差异显著基因 (DEGs) 的热图,京都基因和基因组百科全书(KEGG)对DEGs的富集分析是使用R中的Cluster Profiler包进行的,加权基因共表达网络(WGCNA)是在R包WGCNA的帮助下进行的,并使用相关P值建立统计学意义。
-
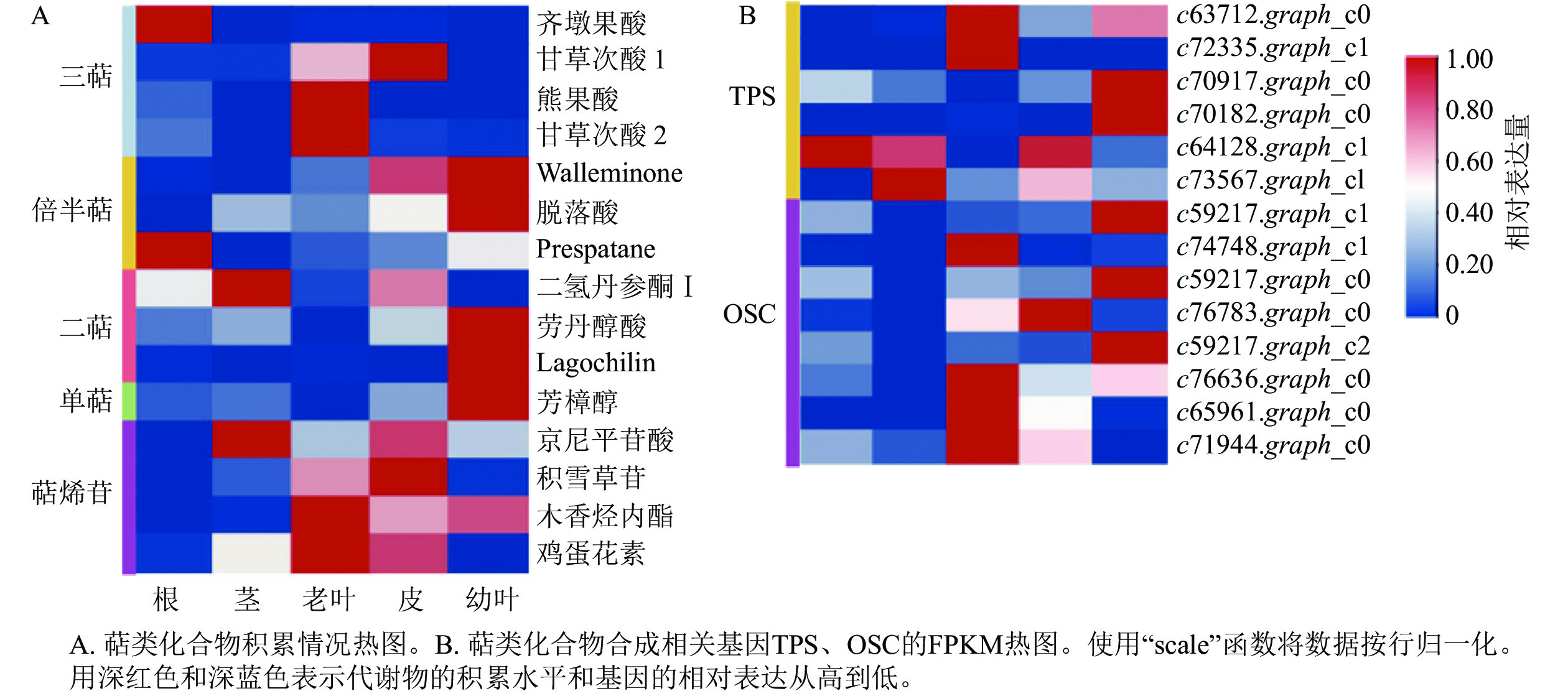
根据总离子流图所得到的化合物精确相对分子质量、出峰时间、二级碎片信息,与mzCloud和mzVault数据库比对,共鉴定出15种萜类化合物(表1)。为了分析花榈木不同组织部位中萜类化合物的分布情况,对5个部位的萜类化合物的相对含量进行了热图分析(图1A),发现不同部位的萜类相对含量差别较大。能明显看出幼叶、老叶与根中萜类相对含量较高,与之相反的是茎与皮中相对含量比较少,并发现甘草次酸 (enoxolone)、齐墩果酸 (oleanolic acid)、芳樟醇 (linalool) 和二氢丹参酮Ⅰ (dihydrotanshinone Ⅰ) 等具有抗氧化、抗炎免疫调节、抗心血管疾病和镇定作用的萜类化合物[22],表明萜类化合物可能是花榈木中主要的药用活性物质。
名称 分子式 质荷比 保留时间/min 峰面积 根 茎 老叶 皮 幼叶 齐墩果酸 C30H48O3 456.360 87 13.411 264 762 138 689 614 4 392 055 4 501 446 1 185 143 二氢丹参酮Ⅰ C18H14O3 278.093 90 7.125 293 469 237 564 453 110 77 252 839 415 872 759 45 119 942 Walleminone C15H24O3 252.172 21 12.877 27 142 310 25 925 485 38 139 793 90 127 128 102 273 216 甘草次酸1 C30H46O4 470.338 67 9.011 10 782 521 10 749 752 19 745 477 26 015 732 10 142 324 熊果酸 C30H48 O3 456.359 91 13.631 40 193 042 1 258 439 318 763 532 2 194 119 2 055 746 京尼平苷酸 C16H22O10 374.121 60 1.932 16 460 987 112 195 727 49 129 697 97 661 105 52 007 928 甘草次酸2 C30H46 O4 470.338 61 13.353 6 685 919 1 285 086 34 589 990 2 917 822 2 213 863 劳丹醇酸 C20H36O3 324.266 84 12.857 1 810 075 2 772 437 427 160 3 608 483 8 629 568 芳樟醇 C10H18O 154.135 49 12.167 2 423 647 2 992 083 1 182 137 4 399 836 13 100 226 脱落酸 C15H20O4 264.136 42 8.304 1 324 567 8 782 319 6 127 101 13 051 696 24 808 273 积雪草苷 C48H78O19 958.511 41 8.844 4 835 086 22 449 131 116 659 465 169 326 989 9 393 498 Lagochilin C20H36O5 356.255 49 12.385 1 032 268 511 236 837 844 660 680 27 515 252 木香烃内酯 C15H20O2 232.146 14 9.727 600 726 697 407 6 046 546 4 143 536 4 973 618 鸡蛋花素 C15H14O6 290.078 91 9.824 1 179 709 9 187 944 17 465 820 14 830 113 618 679 Prespatane C15H24 204.187 43 12.689 15 917 262 1 479 579 2 907 710 4 359 663 8 310 627 Table 1. Mass spectral information of terpenoids in O. henryi
-
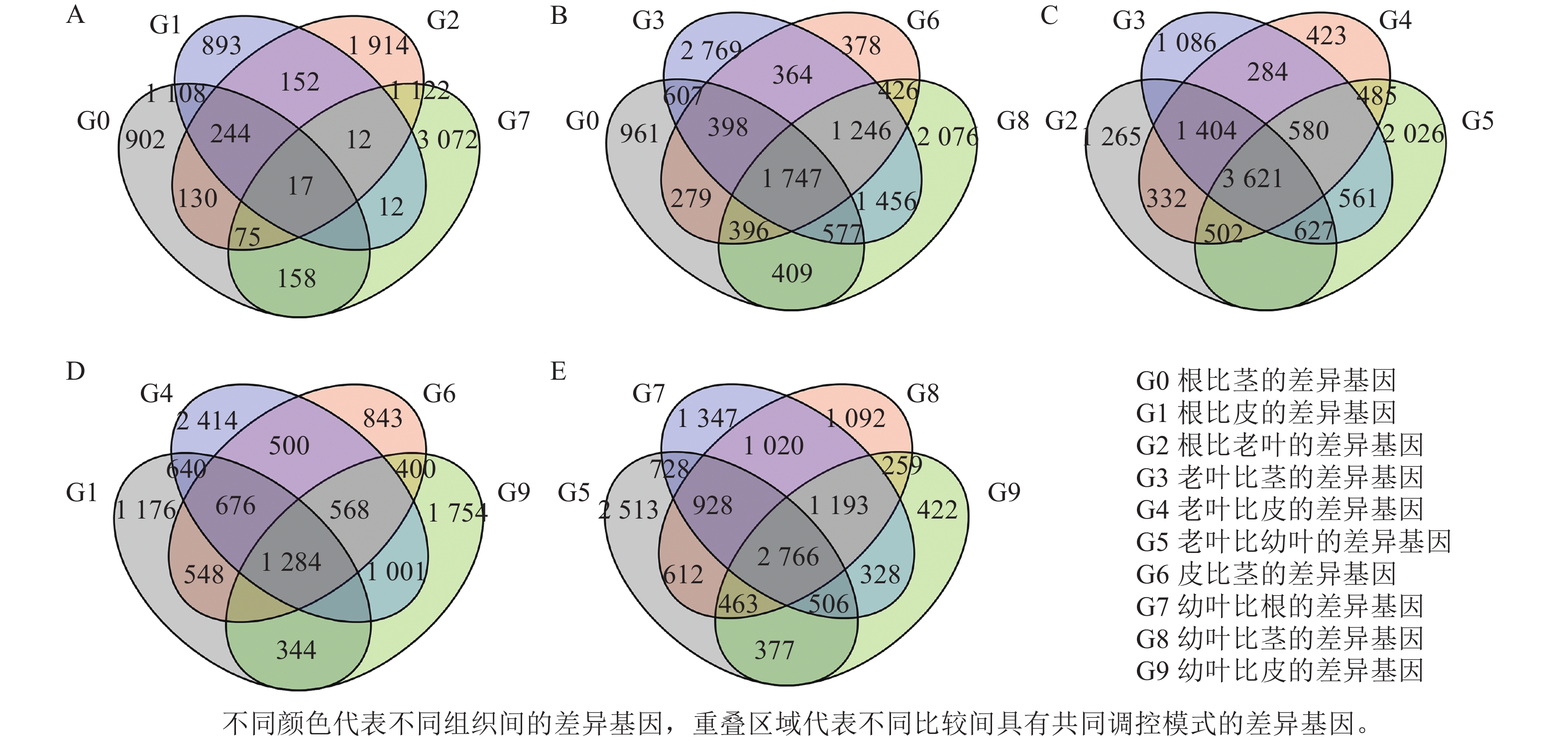
为了筛选花榈木中与萜类生物合成相关的潜在基因,利用RNA-seq对花榈木5个部位的所有转录组进行分析。使用Illumina Hiseq 2000平台进行测序,在去除低质量和短的读数后,总共获得了1.9~2.4 M的高质量待分析数据,共获得96 302个最长转录本,并与NR、eggNOG、TrEMBL、Pfam、SwissProt、KEGG、COG、KOG及GO等9个数据库比对,确定编码序类有47 809条,并对其进行功能注释。计算出FPKM值,代表每个基因的表达水平。对花榈木5个部位的转录组进行差异基因分析,共得到13 840个差异显著基因 (DEGs)。在差异基因分析中发现,与其他3个部位样本相比,叶的特异差异基因更多,茎中的特异差异基因最少(图2)。
-
根据其他物种萜类生物合成信息,花榈木萜类物质合成途径大致可分为萜类前体物质的合成、萜骨架的合成和后修饰3个阶段,其中萜类化合物的前体GPP、FPP和GGPP主要来自植物的MVA和MEP途径。MVA通路共鉴定到8个基因,包括2个乙酰辅酶A乙酰转移酶 (AACT, c68943.graph_c2, c73242.graph_c0)、2个羟甲基戊二酰辅酶A合成酶 (HMGS, c57495.graph_c0, c70166.graph_c1)、1个羟甲基戊二酰辅酶A还原酶 (HMGR, c68938.graph_c1)、1个甲羟戊酸激酶 (MK, c70988.graph_c0)、1个磷酸甲羟戊酸激酶 (PMK, c71400.graph_c0)、1个甲羟戊酸焦磷酸脱羧酶 (MVD, c76163.graph_c2);MEP通路共鉴定到10个基因,包括4个1-脱氧-D-木酮糖-5-磷酸合酶 (DXS, c62607.graph_c0, c70494.graph_c2, c75298.graph_c5, c75577.graph_c0)、1个1-脱氧-D-木酮糖-5-磷酸还原酶 (DXR, c71705.graph_c2),1个4-焦磷酸胞苷-2-甲基-D-赤藓醇激酶 (CMK, c73279.graph_c4)、1个2-甲基-D-赤藓醇-2,4-环焦磷酸合酶 (MDS, c62387.graph_c0)、1个羟甲基丁烯基-4-焦磷酸合酶 (HDS, c56719.graph_c0)、2个羟甲基丁烯基-4-磷酸还原酶 (HDR, c29394.graph_c0, c70095.graph_c0)。为了更直观地比较萜类生物合成途径主要基因在不同组织中的表达情况,采用FPKM值对花榈木不同组织部位的合成途径基因表达量作图。发现参与MVA途径的基因在幼叶中表达量普遍较高,MEP途径相关基因在老叶中表达量较高,与萜类相对含量比较对应,可能与叶中的萜类化合物的积累有关(图3)。还检测出2个异戊烯基焦磷酸异构酶 (IPPI, c31804.graph_c0, c89450.graph_c0),它们在叶中的表达量并不高,可能这些基因与萜类化合物相对含量并不密切相关。
MVA和MEP这2个途径后生成萜类化合物的前体,还需要3个比较重要的酶参与:香叶基二磷酸合成酶 (GPPS)、法尼基二磷酸合成酶 (FPPS) 和香叶基香叶基焦磷酸合成酶 (GGPPS)。在花榈木的转录组数据中共检测到2个FPPS基因 (c86328.graph_c0, c75342.graph_c0),2个GPPS基因 (c36059.graph_c0, c76294.graph_c0),5个GGPPS基因 (c55157.graph_c0, c71336.graph_c0, c65845.graph_c2, c63610.graph_c0, c69470.graph_c0),表达情况如图3所示。2个GGPPS基因c55157.graph_c0和c69470.graph_c0在幼叶中表达量比较高,多数二萜在幼叶中的积累量也是最多的(图1),可能这2个基因是花榈木二萜物质合成的关键酶基因。TPSs是催化合成不同种单帖、二萜和倍半萜类化合物的酶,OSCs可以催化合成三萜类化合物,在花榈木转录组中还鉴定出6个TPSs和8个OSCs基因,其表达谱如图1B所示。这些基因的表达在花榈木中出现了组织差异性,每个组织中都有其表达量高的基因,其中与MVA和MEP途径基因表达情况相似:在幼叶和老叶中表达量高的基因占比较大。这些TPSs和OSCs的组织差异表达可能是导致花榈木中萜类多样性和具有不同积累模式的主要原因。
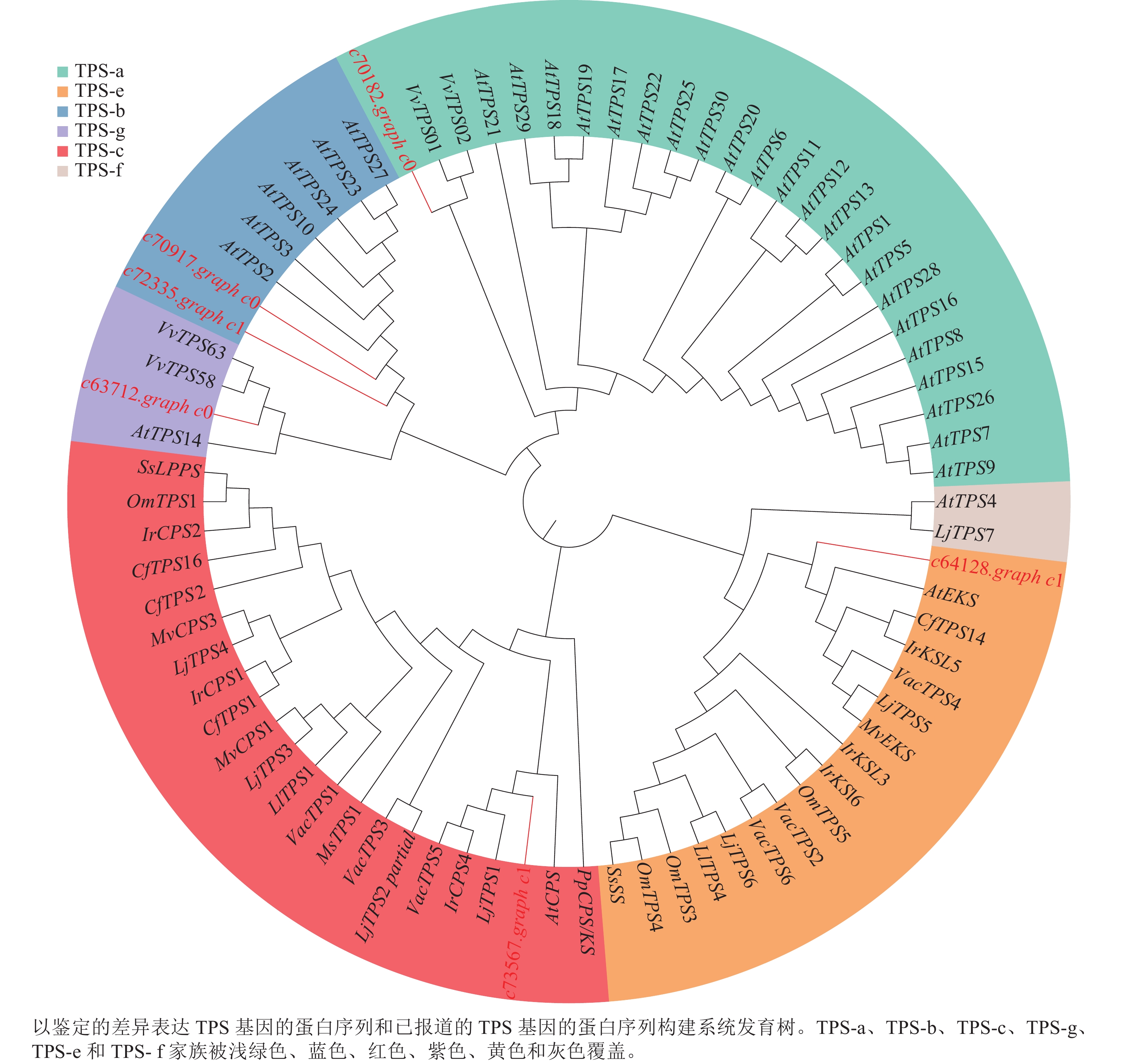
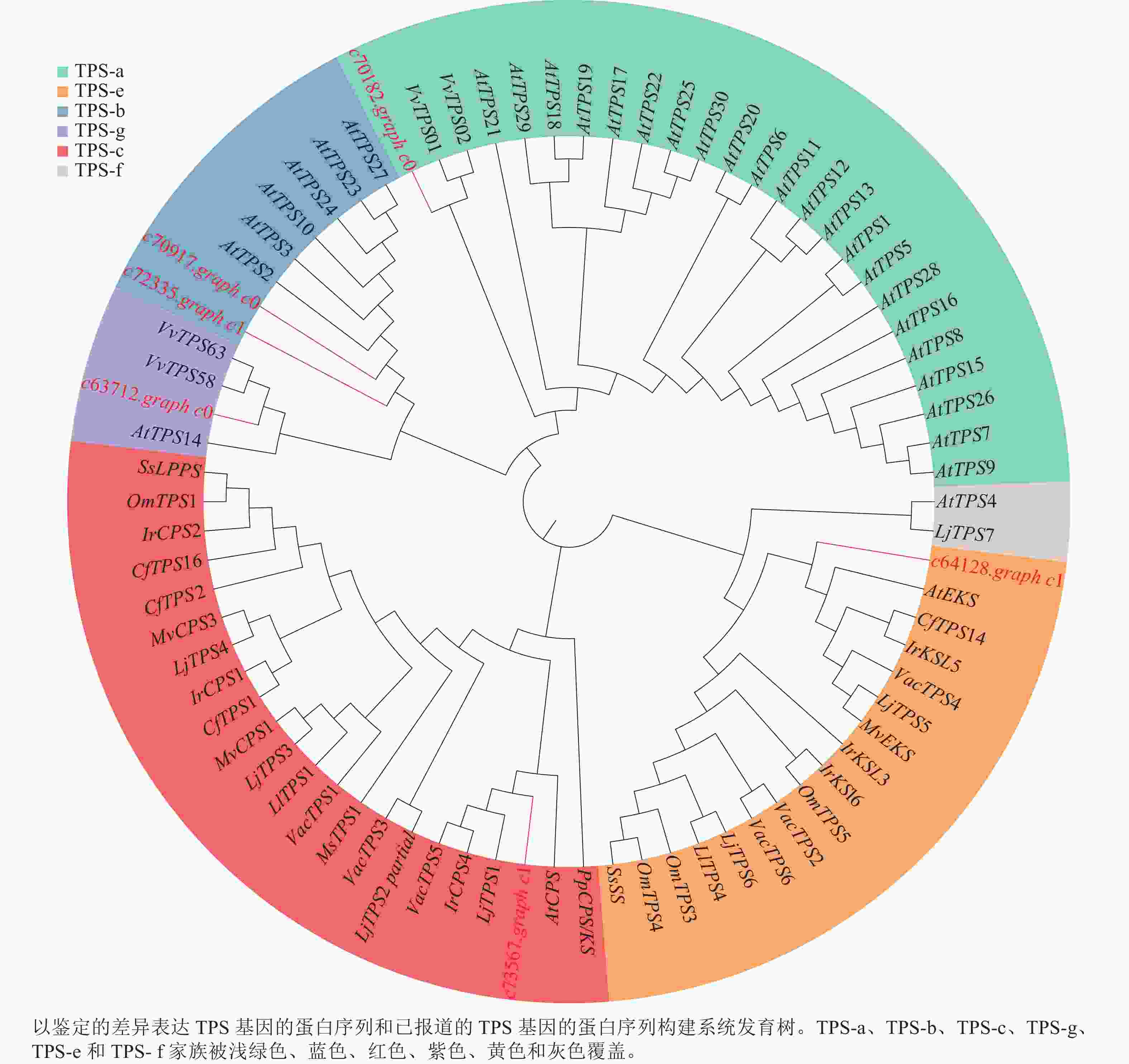
为了进一步了解研究中6个TPSs的假设功能,根据其蛋白序列,将其与已经在拟南芥Arabidopsis thaliana、葡萄Vitis vinifera和番茄Lycopersicon esculentum等物种中确定的TPS进行系统发育分析(图4)。发现与之前报道相似,花榈木中的TPSs主要分布在TPS-a、TPS-b、TPS-c、TPS-e和TPS-g亚家族。c70917.graph_c0和c72335.graph_c1被分类到TPS-b中,可能这2个基因在功能上比较类似;c64128.graph_c1被分到TPS-e亚家族,说明c64128.graph_c1可能在花榈木中行使着KS酶功能;c73567.graph_c1则被分到TPS-c亚家族中,表明它可能有着CPS酶的功能。为了进一步推测花榈木OSCs的功能,对每个基因再次进行了美国国家生物技术信息中心数据库的比对注释,发现除c71944.graph_c0基因是编码LUC的基因外,其他7个都是编码β-AS的基因。
-
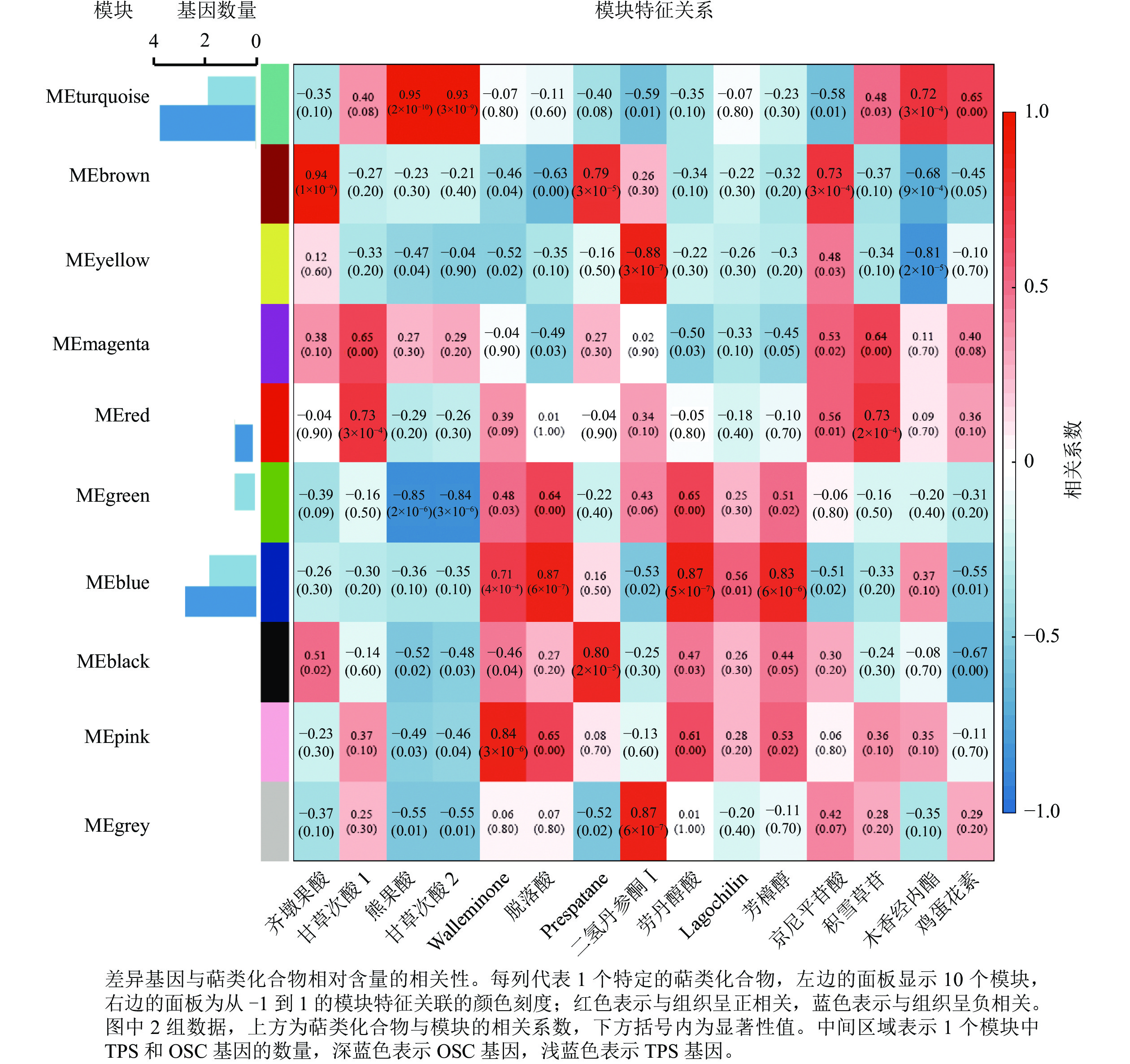
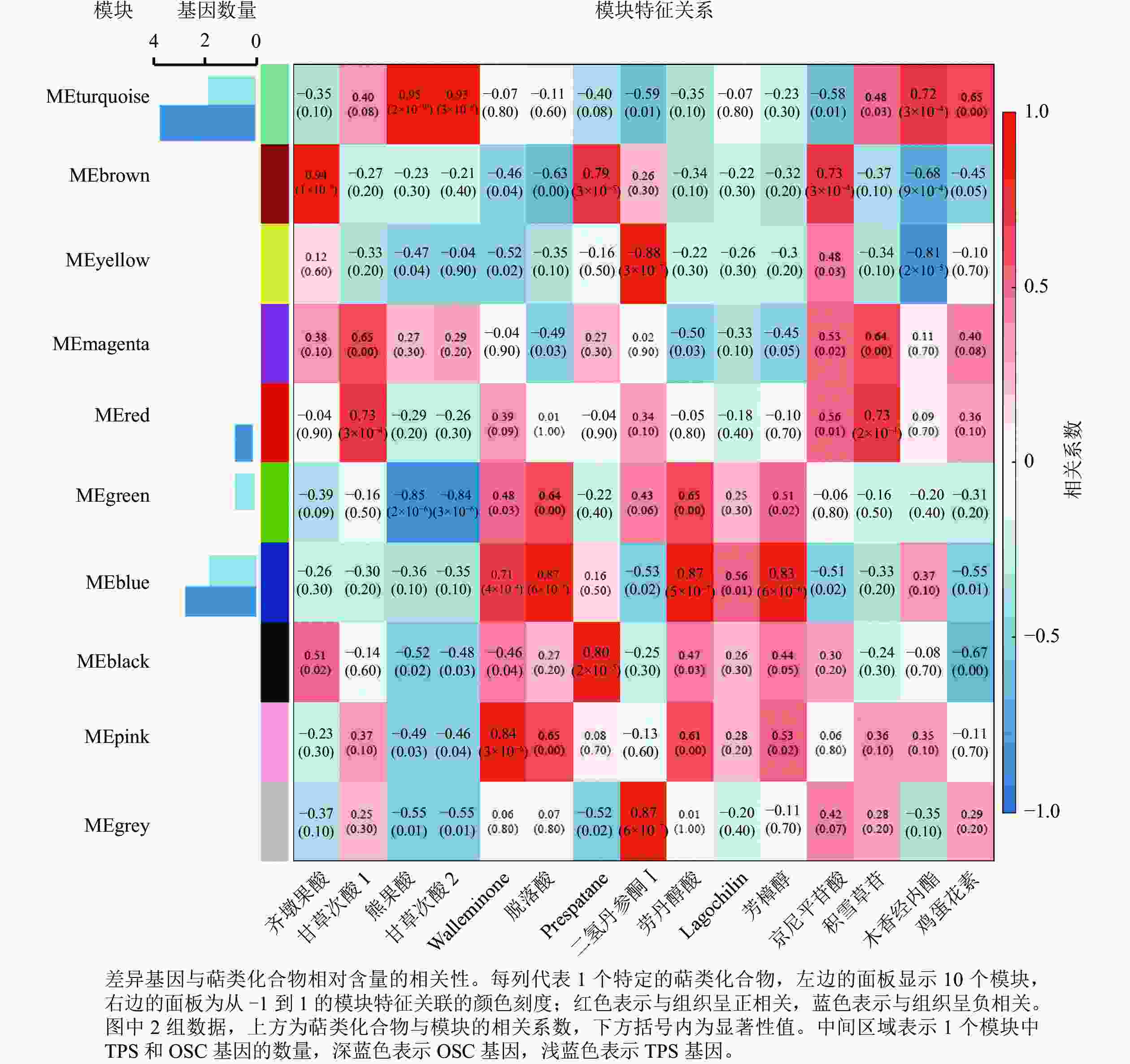
为进一步分析萜类化合物合成相关基因,利用WGCNA对确定的全部DEGs进行了共表达分析。这些DEGs被聚类为10个分支,每个不同颜色标记的模块代表了1个分支(图5)。模块是由具有相似表达模式的基因簇组成的,MEturquoise模块包含的基因数量最多(3 761个基因),而MEmagenta模块的基因数量最少(104个基因)。其中有7个模块与一些特定的萜类化合物表现出明显的正相关(相关系数>0.80)(图5)。除了c63712.graph_c0基因外,共有5个TPSs 和8个OSCs基因表达差异明显,推测这13个差异表达的TPSs和OSCs可能是导致花榈木5个组织部位萜类化合物组成差异的重要候选基因。其中, MEturquoise模块中共有2个TPSs和4个OSCs基因;MEblue模块有2个TPSs和3个OSCs基因。MEturquoise和MEbrown与三萜类化合物的相关性较强(相关性系数>0.90),而MEblue、MEblack和MEpink模块与倍半萜化合物有相关性,其中MEblue模块也与二萜和单萜具有相关性。二氢丹参酮Ⅰ这个二萜化合物与MEyellow和MEgrey 2个模块都具有正相关性。这些正相关的模块表明:这些基因在调节花榈木萜类化合物的生物合成中具有潜在的作用。
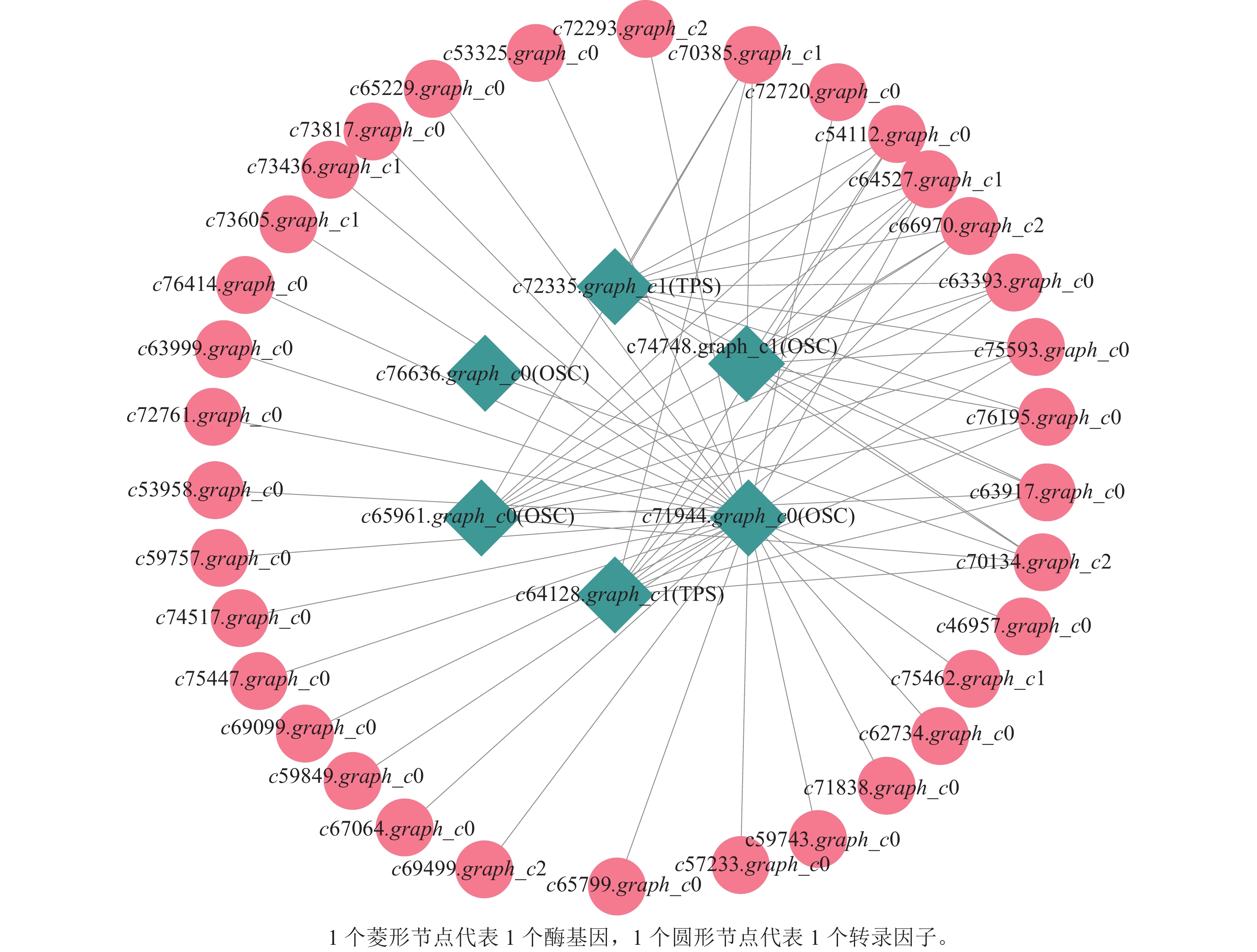
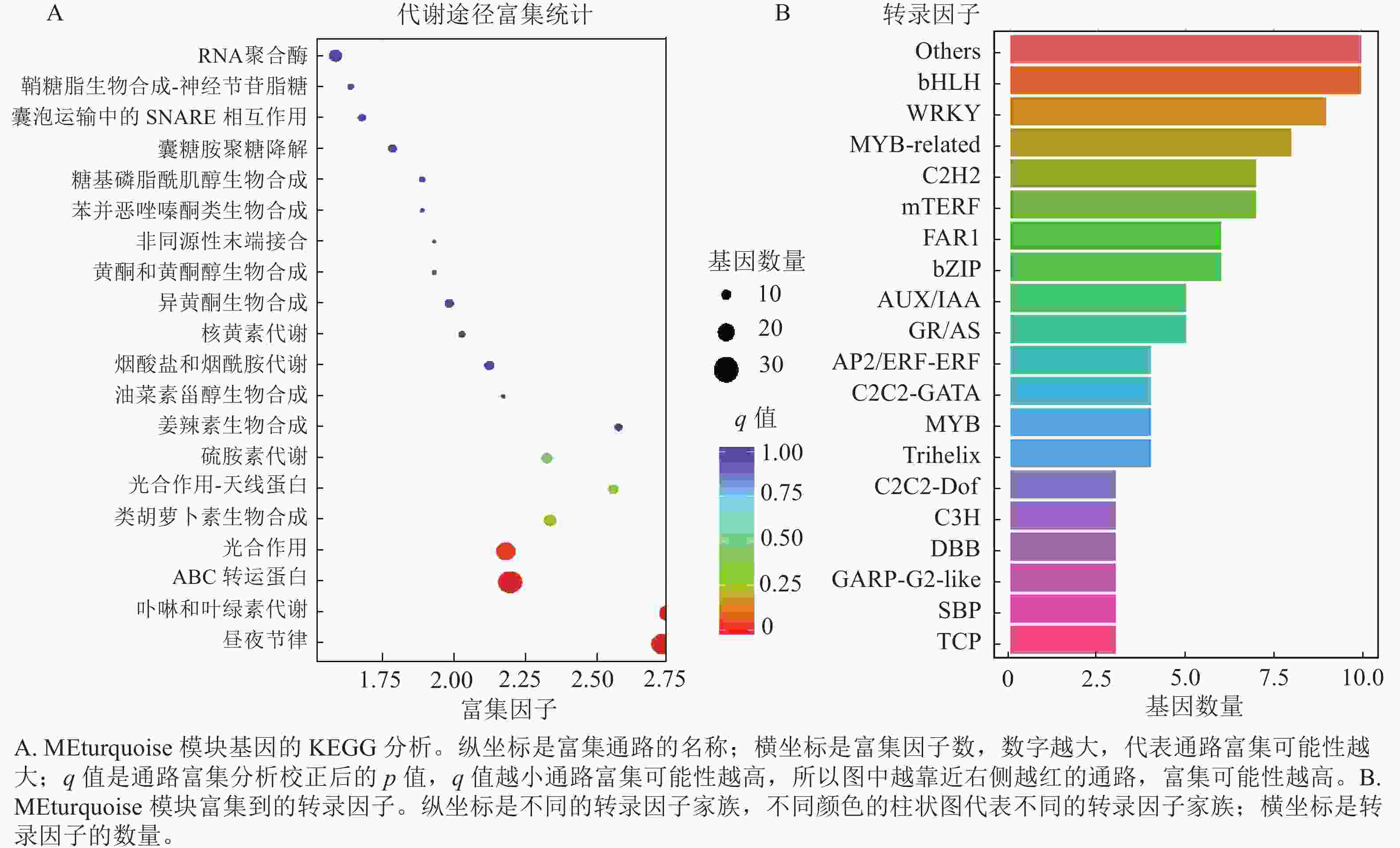
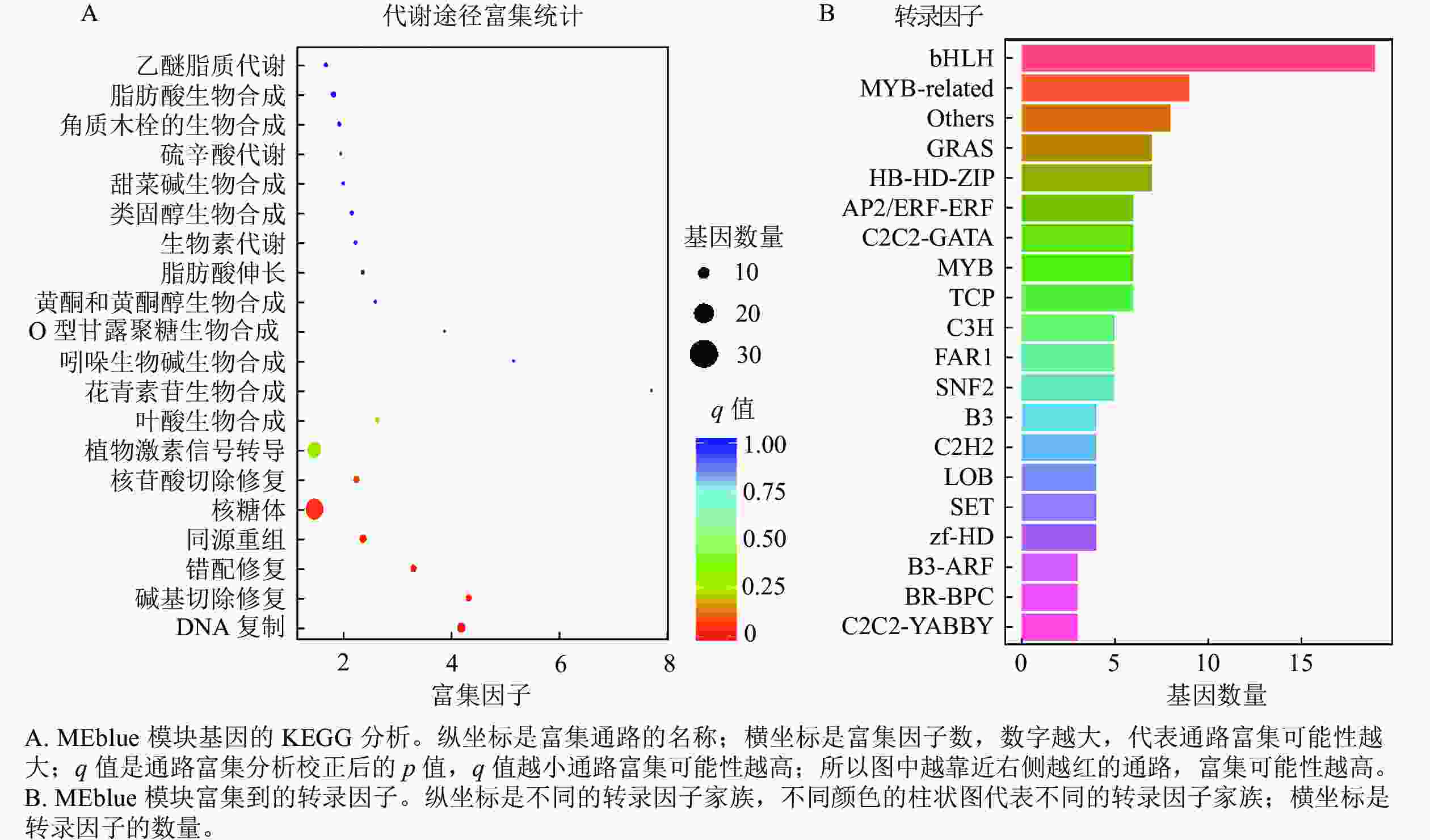
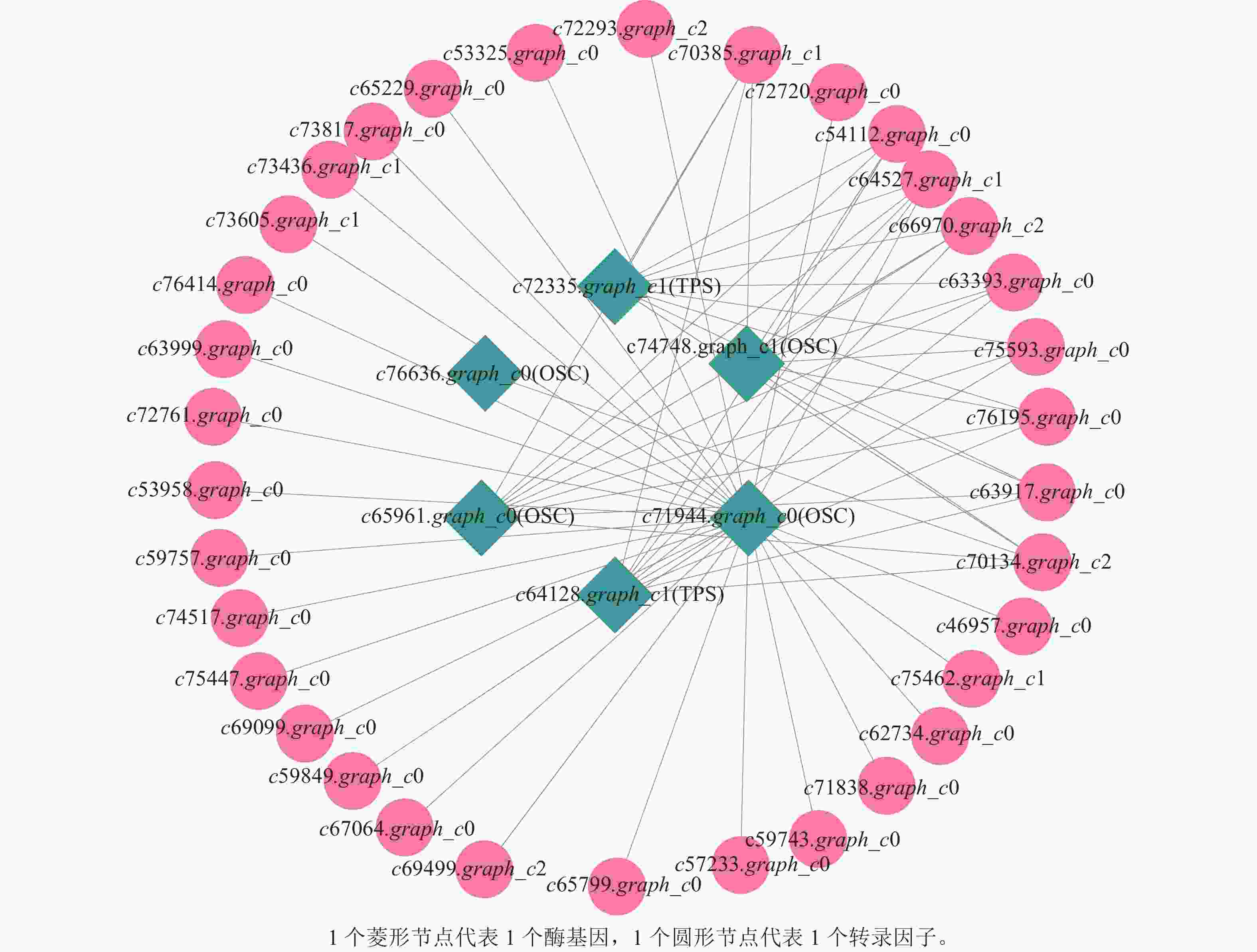
上述模块的分析结果表明:MEturquoise模块富集了最多的TPSs和OSCs,并且该模块与乌苏酸和甘草次酸呈正相关,相关性分别为0.95和0.93(图5);MEblue也富集到了较多的TPSs和OSCs基因,并且与一些倍半萜和二萜成正相关,所以对这2个模块开展进一步的分析。在MEturquoise模块中发现:基因主要富集在次生代谢和信号转导通路,共检测到105个转录因子可能参与TPSs的调控(图6 A和图6 B):bHLH、WRKY和MYB家族数量最多,分别有10、9和8个,此外C2H2、mTERF、FAR1和bZIP家族数量也达到了6~7个。通过对MEblue模块基因进行KEGG分析,发现基因主要富集在DNA复制、同源重组和修复通路,并检测到152个转录因子,数量最多的是bHLH和MYB家族转录因子 (图7 A和图7 B)。为了进一步研究MEturquoise模块中转录因子与相应TPSs和OSCs基因的关系,构建了一个共表达的网络图,选择了与MEturquoise模块中TPSs和OSCs基因相关性较强的34个转录因子 (边缘权重≥0.4),包括MYB、WRKY、bHLH、C3H、DBB、HB-HD-ZIP和一些其他家族的转录因子(图8)。最终利用Cytoscape插件CytoHubba的Degree算法分析识别到了6个转录因子,即HB-HD-ZIP (c64527.graph_c1)、GRF (c76195.graph_c0)、DBB (c66970.graph_c2)、DBB (c75593.graph_c0)、HB-HD-ZIP (c63393.graph_c0)和C3H (c70385.graph_c1)。这些转录因子在花榈木中可能与萜类化合物的合成基因有着密切的关系。
-
目前,天然植物次生代谢物已被广泛应用于抗癌药和治疗感染性疾病的药物[23]。根据合成起始分子不同,植物次生代谢物可以分为生物碱、萜类、苯丙烷类三大类化合物[24]。萜类一直是天然产物中重要的药用化合物,具有多种药理活性,如紫杉醇可以抗肿瘤,青蒿素属于抗疟疾特效药物,雷公藤内酯能够抗炎等[25−26],但药用木本植物花榈木中的萜类还没有过报道。本研究利用代谢组学技术分析了花榈木中萜类物质在不同部位的积累情况,共检测出比较确定的15种萜类化合物,并发现了甘草次酸、齐墩果、芳樟醇和二氢丹参酮Ⅰ等具有抗氧化、抗炎免疫调节和镇定等药用活性的萜类化合物[27]。通过对花榈木不同组织部位的萜类代谢物热图分析发现,萜类化合物积累具有明显组织特异性,主要积累在幼叶和老叶中,其他部位中积累较少,可能为了保护叶片免遭危害[28]。今后在提取和分析花榈木药用物质时应该更多利用它的叶片。
近些年来,RNA-Seq 高通量测序技术被越来越多地应用于药用植物基因信息解读、新基因发掘与基因功能研究中。人们已对药用植物连翘 Forsythia suspense、银杏 Ginkgo biloba、款冬 Tussilago farfara等进行了转录组的研究,获得了大量有用的基因信息[29−30]。这使得阐明药用植物中活性物质的合成及积累规律成为可能,为增加次生代谢物积累、改善药用植物品质提供更多途径。本研究采用RNA-Seq对花榈木5个不同组织部位进行无参转录组分析,共获得96 302个,在经过与数据库比对后共注释了47 809条转录本。利用FPKM值对基因表达量进行分析比较,共获得显著差异基因13 840个。韦恩图分析发现大量基因表达具有组织特异性,并通过注释信息在差异显著的基因中共鉴定出49个与萜类化合物生物合成相关的基因,包括29个萜类骨架生物合成途径的酶基因,6个单帖、倍半萜和二萜生物合成酶基因,8个三萜生物合成酶基因以及6个可能参与萜类生物合成调控的转录因子,对花榈木的萜类合成有了初步的认识,为后续研究提供了信息资源。
先前的研究表明:MVA途径在许多植物的三萜生物合成中起主导作用,如人参Panax ginseng、三七P. notoginseng和茶树Camellia sinensis等植物[31−32],MEP途径通常有助于单萜类化合物和二萜类化合物的生物合成[33]。但是本研究利用热图对萜类生物合成基因的表达分析发现:MVA途径相关酶基因大多数在幼叶中表达量较高,MEP途径多数相关基因则集中在老叶中表达。根据相应FPPS、GPPS和GGPPS酶基因的表达情况,以及花榈木中三萜主要在老叶中积累,而二萜、倍半萜和单萜在幼叶中积累较多的情况,推测在花榈木中三萜类化合物前体可能主要由MEP途径生成,而二萜、单帖和倍半萜的前体物质则主要由MVA途径提供。由图4看出:花榈木的TPS基因在进化上相对于各种模式植物来说是相对分离的,也进一步印证了其萜类的合成具有特殊性。也有可能由于转录组学的限制,部分基因没有检测到,且研究中还没有检测到所有的萜类物质,从而导致判断出现误差,所以后续还需要大量的试验来验证其功能,判断花榈木中萜类的具体合成情况。
基因的转录调控一直是植物代谢研究领域的热点。对不同植物的研究表明:参与萜类生物合成的转录因子主要分布在bHLH、AP2/ERF、bZIP和WRKY家族中,如在西洋参P. quinquefolius中转录因子PqWRKY1是三萜人参皂苷生物合成相关的正调节因子[34],在艾叶Artemisia argyi中AarbHLHs的基因表达与1, 8-桉树烯或β-石竹烯的含量变化呈显著相关[35]。本研究利用WGCNA对差异基因和萜类化合物进行了相关性分析,筛选出一些可能对萜类生物合成的关键酶基因表现重要调控作用的转录因子家族,如MYB、WRKY、bHLH和HB-HD-ZIP转录因子。有研究预测传统中药走马胎Ardisia kteniophylla中AP2/ERF、MYB、WRKY和bHLH转录因子可能调控萜类合成,预测赤桉Eucalyptus camaldulensis中WRKY、MYB、NAC和bHLH转录因子对萜类生物合成中的关键酶基因表现出重要的调控作用[36]。这和本研究预测的结果基本相同。经过分析进一步筛选出6个转录因子处于共表达网络的中心位置,推测这些候选转录因子可能调控了萜类化合物的生物合成,后续还需要进一步的试验证明。
-
本研究通过代谢组与转录组学的分析,发现花榈木叶中萜类相对含量最高,并鉴定出49个与萜类化合物生物合成相关的基因。预测了可能调控萜类化合物生物合成的上游转录因子。本研究为花榈木资源活性成分萜类化合物的积累状况、生物合成及调控提供了大量的信息,弥补了花榈木萜类合成研究中的空白,为进一步开展花榈木的主要药用活性物质研究提供基础。
Analysis of candidate genes for terpene synthesis in Ormosia henryi based on metabolome and transcriptome
doi: 10.11833/j.issn.2095-0756.20220737
- Received Date: 2022-11-28
- Accepted Date: 2023-03-20
- Rev Recd Date: 2023-03-09
- Available Online: 2023-09-26
- Publish Date: 2023-09-26
-
Key words:
- Ormosia henryi /
- metabolome /
- transcriptome /
- terpenoids /
- biosynthesis
Abstract:
| Citation: | WANG Jiaqi, WANG Xin, DENG Xiaomei, et al. Analysis of candidate genes for terpene synthesis in Ormosia henryi based on metabolome and transcriptome[J]. Journal of Zhejiang A&F University, 2023, 40(5): 970-981. DOI: 10.11833/j.issn.2095-0756.20220737 |












 DownLoad:
DownLoad: