-
磷脂酸(phosphatidic acid,PA)是一种常见的细胞甘油磷脂,也是最简单的膜脂之一[1]。几乎所有生物体内都会产生PA,包括酵母[2]、动物[3]和植物[4]等。在植物中PA的合成主要包括2种方式:第1种是从头合成途径,3-磷酸甘油先后在3-磷酸甘油酰基转移酶和溶血磷脂酸脂酰基转移酶的作用下发生连续2步酰化反应生成PA,这也是PA合成的主要途径[5];第2种是磷脂的降解途径,磷脂降解又可分为2种,一种是磷脂酶D通过水解某些结构磷脂直接生成PA[6−9],另一种是磷脂酶C通过水解磷脂酰肌醇生成二脂酰甘油[10],继而经二脂酰甘油激酶磷酸化生成PA[11]。PA作为一种结构膜脂,磷酸基团头部能够以“静电/氢键开关模型”的方式与蛋白质结合[12]。在该模型中,PA结合蛋白中带正电荷的碱性氨基酸残基[13−17],可以吸引磷脂双分子层上的负电荷,并通过静电作用影响膜环境,继而与PA头部的磷酸基团形成氢键,产生一个稳定的蛋白质结合位点。PA这种携带负电荷并且在生理条件下呈圆锥体的独特分子构型[18],使得PA结合蛋白的结构域能够特异性的与其结合,保证了生物膜结构的稳定和功能的发挥。除了作为膜脂的一个结构成分以及甘油脂合成的前体物质,PA还是一个关键信号分子,在多种生物学过程中发挥重要作用。正常条件下,行使信号功能的PA含量甚微;在植物应答各种生物与非生物胁迫过程中PA水平则会迅速升高,但这种积累往往是短暂的。这说明生物体拥有一套严格控制信号分子PA含量的机制,这对调节PA信号传递强度、维持细胞物质与能量代谢的动态平衡至关重要[19−22]。
尽管PA拥有多种功能,但目前对于细胞内PA含量的动态变化仍知之甚少,这主要受PA检测手段的限制。过去主要采用诸如薄层层析、高效液相色谱和放射性同位素标记等理化手段对其含量进行测定[23]。近年来,质谱分析技术的应用使得对包括PA在内的各种脂质的测定更准确和高效[24−25]。然而,这些理化手段都只能对组织器官中PA总量与种类进行定量或定性分析,无法捕捉细胞内PA的动态与瞬时变化信息。通常认为,内质网上的PA含量相对较高,这有助于促进磷脂和三酰甘油的合成[26−27];而质膜和细胞器中行使信号功能的PA含量甚微。显然,采用理化测定会掩盖胞内PA的真实变化。因此,亟需开发一种能可视化分析细胞内PA的工具。
大量研究表明:生物体中PA结合蛋白种类繁多,这些蛋白中的PA结合域(phosphatidic acid binding domains,PABD)结构亦存在差异。其中一些PABD因与PA结合专一性强,被用来开发多种PA探针 [28−33]。酵母蛋白Spo20p中的PA结合域已被证实具有极强的专一性[33],只结合PA而不结合其他磷脂。目前基于这一PABD所开发的PA探针在医学研究中应用广泛,但在植物中的应用却有限[34]。因此,本研究旨在构建基于Spo20p中的PABD的植物PA荧光探针,为剖析植物早期应答逆境胁迫的细胞分子机制提供新工具。
-
将拟南芥Arabidopsis thaliana种子置于含有湿润滤纸的培养皿中,4 ℃冰箱内避光放置3 d后将其点播在湿润的土壤基质上(V营养土∶V蛭石∶V珍珠岩=3∶1∶1)。将播种后的盆栽用透明塑料盖覆盖,置于24~25 ℃室温、50%~60%湿度、14 h光照/10 h黑暗的植物培养箱中培养,7 d后揭去塑料盖间苗。
-
将拟南芥种子进行消毒,加入适量灭菌过的蒸馏水,在4 ℃冰箱内避光放置3 d,点播在1/2 MS培养基上,置于植物培养间中培养。
-
酵母蛋白Spo20p中存在一个高度专一的PA结构域(PABD),该PABD由40个氨基酸残基构成,只结合PA而不结合其他磷脂[33−34],因此,本研究选择该PABD与荧光蛋白融合,构建PA荧光探针。根据PAPD核苷酸序列[33]设计引物(表1)进行PCR扩增,获得与该PABD相对应的DNA片段;同时PCR扩增获得GFP基因编码区序列。随后,利用无缝克隆试剂盒(生工),将GFP和PABD片段克隆至植物表达载体pSY06的限制性内切酶位点Kpn I和Pst I之间,获得重组质粒pSY06-GFP-PABD。PABD连接至GFP基因编码区的3′端,构成GFP-PABD融合基因,该融合基因的表达由组成型启动子UBQ10驱动。重组质粒中的插入片段经菌落PCR、Kpn I和Pst I双酶切以及测序验证正确后,采用热激法将质粒转化至农杆菌Agrobacterium tumefaciens GV3101,用于后续拟南芥遗传转化。
用途 引物名称 引物序列(5′→3′) PABD基因合成 ZYP301 CCTGCTAATTTCAAGGCTAATTCATTGCTGTTCGCTCGAGATCTGAGTCCGGACTGCAGGTTGGATCCGGTACCGAGCT ZYP302 GAAGACGTGATAGGCTACATGTGAAGCTTAAATCCTTGAGGAATAAAATCCACAAACAACTTCACCCAAA ZYP303 TGCTTCCTGAACAATTGTCCATTCTAGATTCTGTCTTGTTGATATCAAGACCTGCTAATTTCAAGGCTAA ZYP304 GTACCCAAGCTTCACATGTAGCCTATCACGTCTTCTGCTTCCTGAACAATTGTCC ZYP305 CACAAACAACTTCACCCAAACTGTCGGTTCGATGACGCCACTAAGACTAGTTAAGGTACCGGCGTAATCATGGTC PABD基因克隆 ZYP308 TCCGGACTCAGATCTCGAGC ZYP309 AGCTATAGTTCTAGATCTAGATTAACTAGTCTTAGTGGCGTC GFP基因克隆 ZYP306 TATCGATGGCGCCAGCTGAGGATGGTGAGCAAGGGCGA ZYP307 AGATCTGAGTCCGGACTTGTACAGCTCGTCCATCCGGACTCAGATCTCGAGC 转基因拟南芥鉴定 MSY01 TGGTGTTAGTTTCTAGTTTGTGCG MSY02 TCATCATGACAGATCTGCGC 表达量分析 ZYP389 GACCACTACCAGCAGAACACC ZYP390 CTTGTACAGCTCGTCCATGC Table 1. Sequence of related primers
-
以哥伦比亚野生型拟南芥(WT)为植物材料,利用农杆菌花序侵染法转化拟南芥。将获得的T1代拟南芥种子进行表面消毒,均匀铺在含有草铵膦(10 mg·L−1)和特美汀(50 mg·L−1)的1/2 MS筛选培养基上进行筛选。将具有除草剂抗性的阳性苗移栽到土壤中继续培养,3周后提取叶片DNA并设计引物(表1)进行PCR鉴定,对T2和T3代转基因株系进行除草剂抗性的遗传分析,最终筛选得到纯合单插入位点转基因株系。
-
以在1/2 MS培养基中生长7 d的转基因拟南芥根系作为测定材料,使用高纯度RNA提取试剂盒(全式金)提取RNA并对其进行纯度检测和浓度测定。取等量RNA,使用反转录试剂盒(翌圣)进行单链cDNA的合成。选择拟南芥肌动蛋白基因Actin为内参基因,设计引物(表1)进行实时荧光定量PCR (RT-qPCR),并对数据采用2−∆∆Ct法进行分析。
-
将野生型和转基因株系种子点播在1/2 MS方形培养基上,竖直培养6 d后,挑选长势均匀一致的幼苗(每个基因型,20株),分别转移至含有0和50 mmol·L−1 NaCl的1/2 MS固体培养基中继续竖直培养,7 d后观察表型。
-
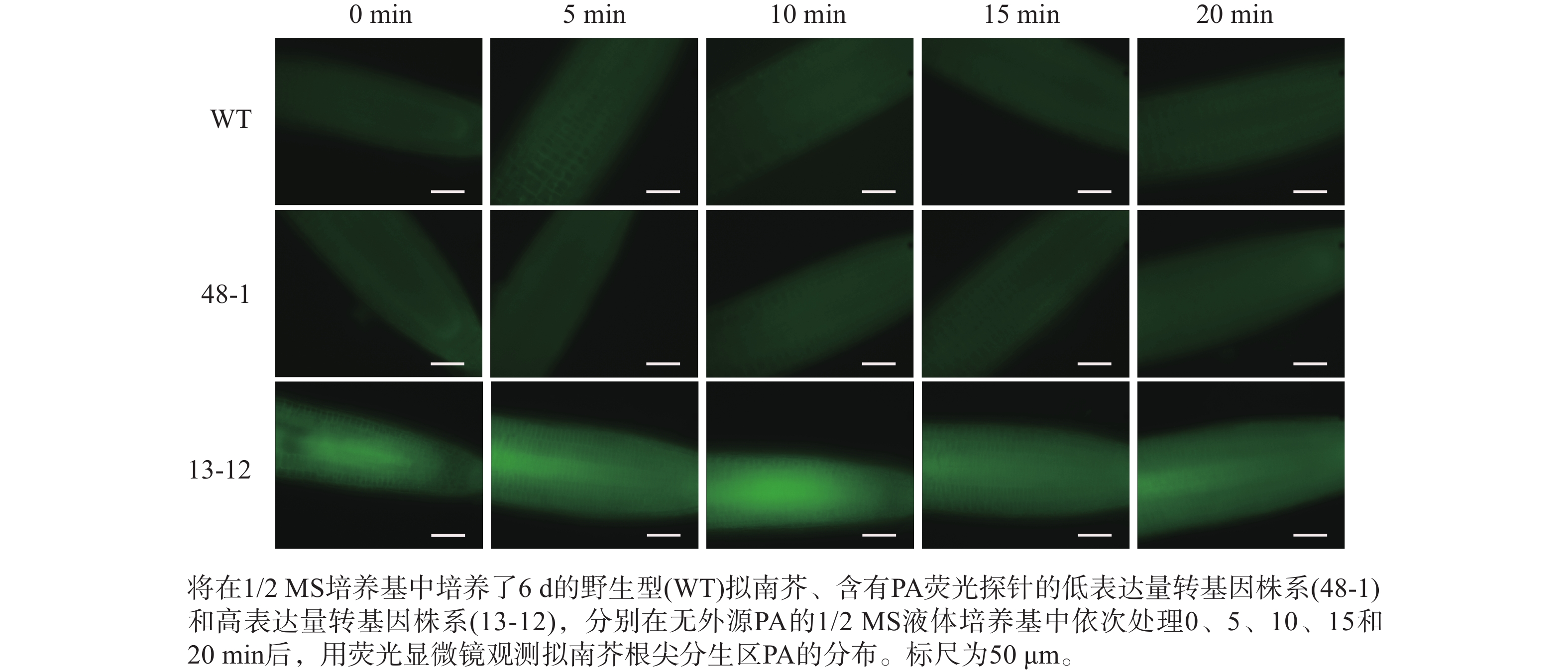
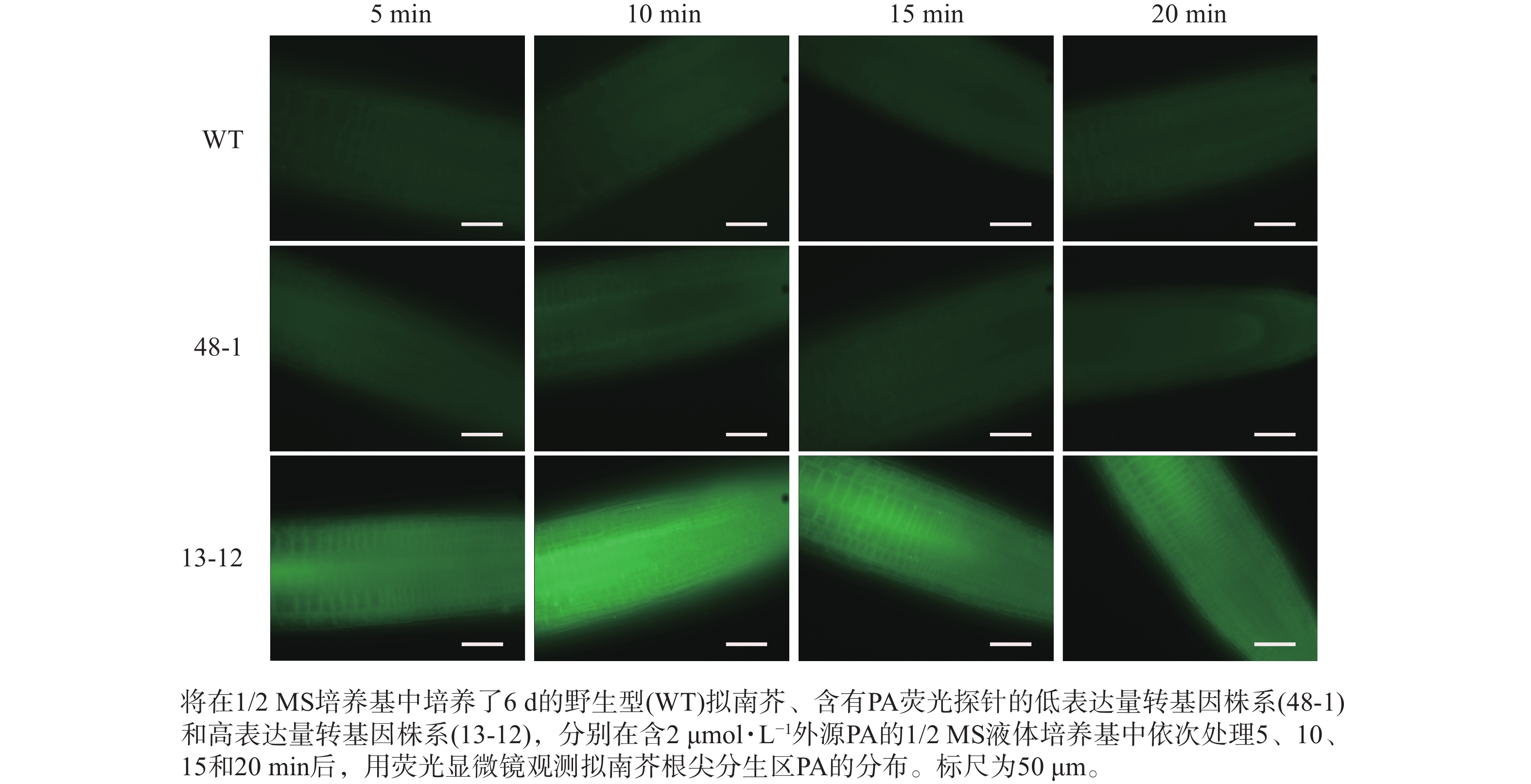
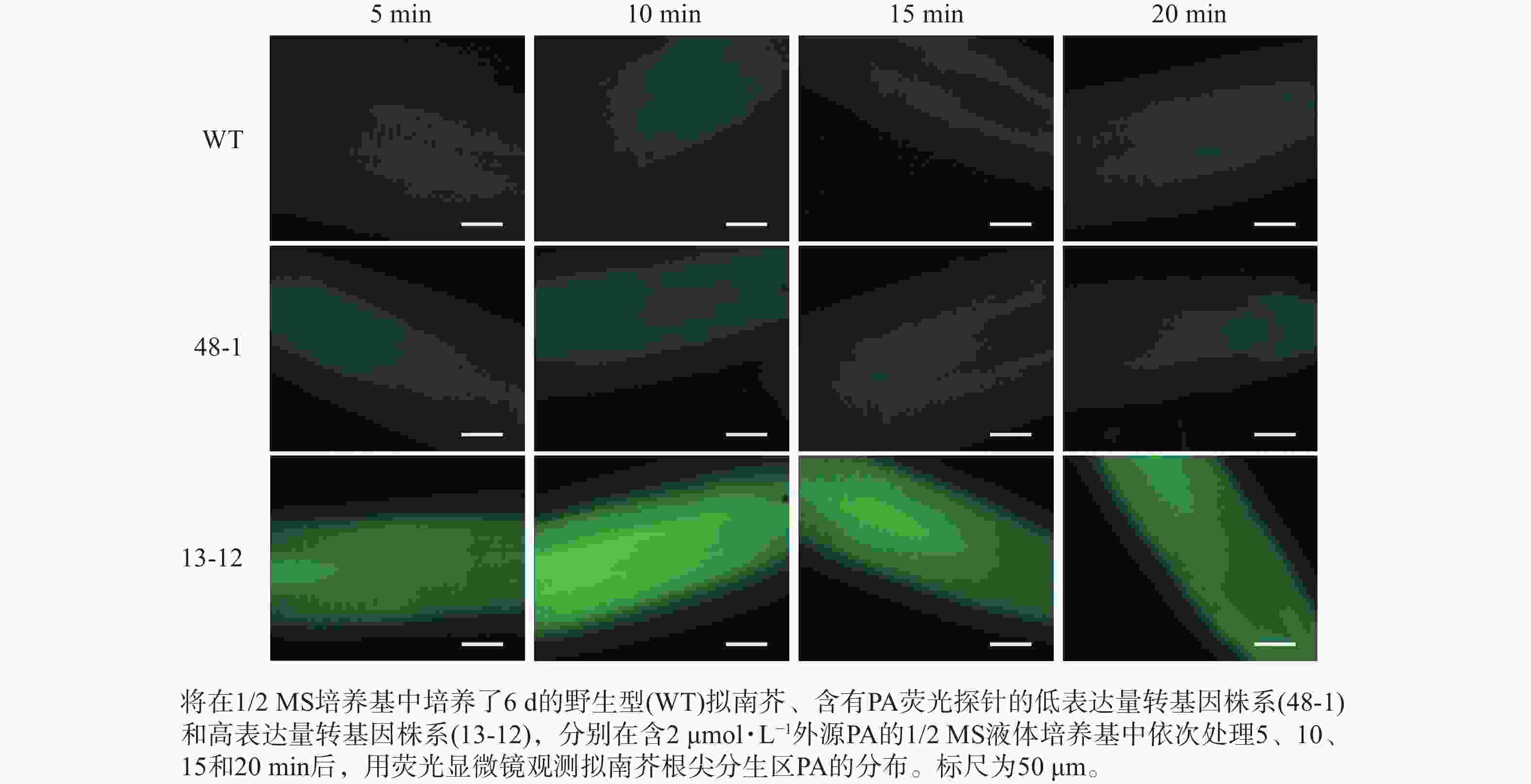
将在1/2 MS方形培养基上竖直培养6 d的幼苗分别浸泡在含有0、2和10 μmol·L−1 PA的1/2 MS液体培养基中,依次处理5、10、15和20 min后,利用荧光显微镜(ZEISS,Axio Imager 2)对拟南芥根尖分生区PA的分布进行观察并拍照。
-
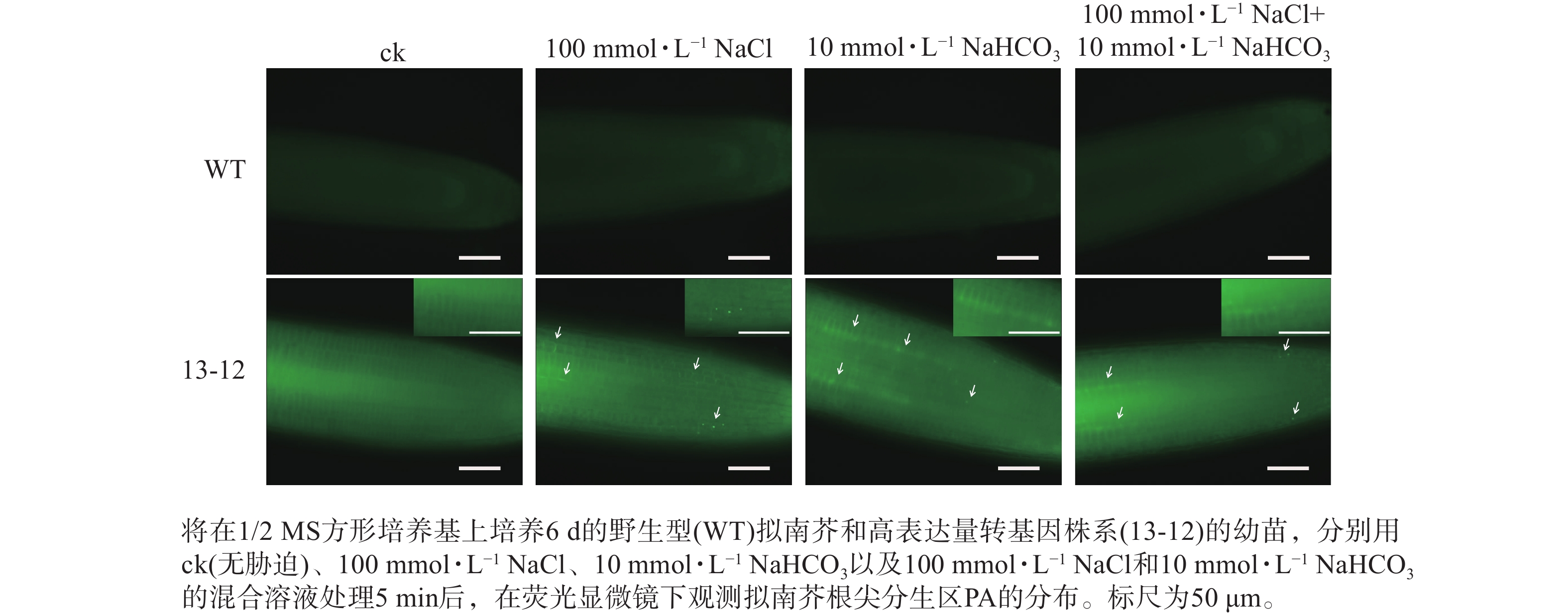
将含PA荧光探针的拟南芥转基因株系种子点播在1/2 MS方形培养基上,竖直培养6 d,选择均匀一致的幼苗分别浸泡在含有100 mmol·L−1 NaCl、10 mmol·L−1 NaHCO3和兼含两者的1/2 MS液体培养基中,处理5、10和15 min后用荧光显微镜观察根尖分生区PA的分布。
-
如图1所示:该载体以pSY06为骨架,由35S 启动子驱动植物选择标记基因——草铵膦抗性基因(bar)的表达,而GFP-PABD融合基因的表达则由UBQ10启动子驱动。
采用农杆菌花序侵染法将GFP-PABD融合基因转入拟南芥,获得T1代转基因种子。对T1代幼苗进行草铵膦(10 mg·L−1)抗性筛选与PCR鉴定,获得56个含融合基因的独立转基因株系。随后,对相应株系的T2代幼苗(每个株系约300株)继续进行草铵膦除草剂抗性筛选。卡方测验显示:符合3(阳性苗数)∶1(阴性苗数)分离规律的株系共8个,这些株系为含单插入位点的转基因株系。再对T2代各个株系不同植株的自交后代(T3代)进行除草剂抗性筛选,最终获得7个纯合、单插入位点转基因拟南芥株系,分别为8-1、11-1、13-12、25-5、42-4、48-1和53-1。
-
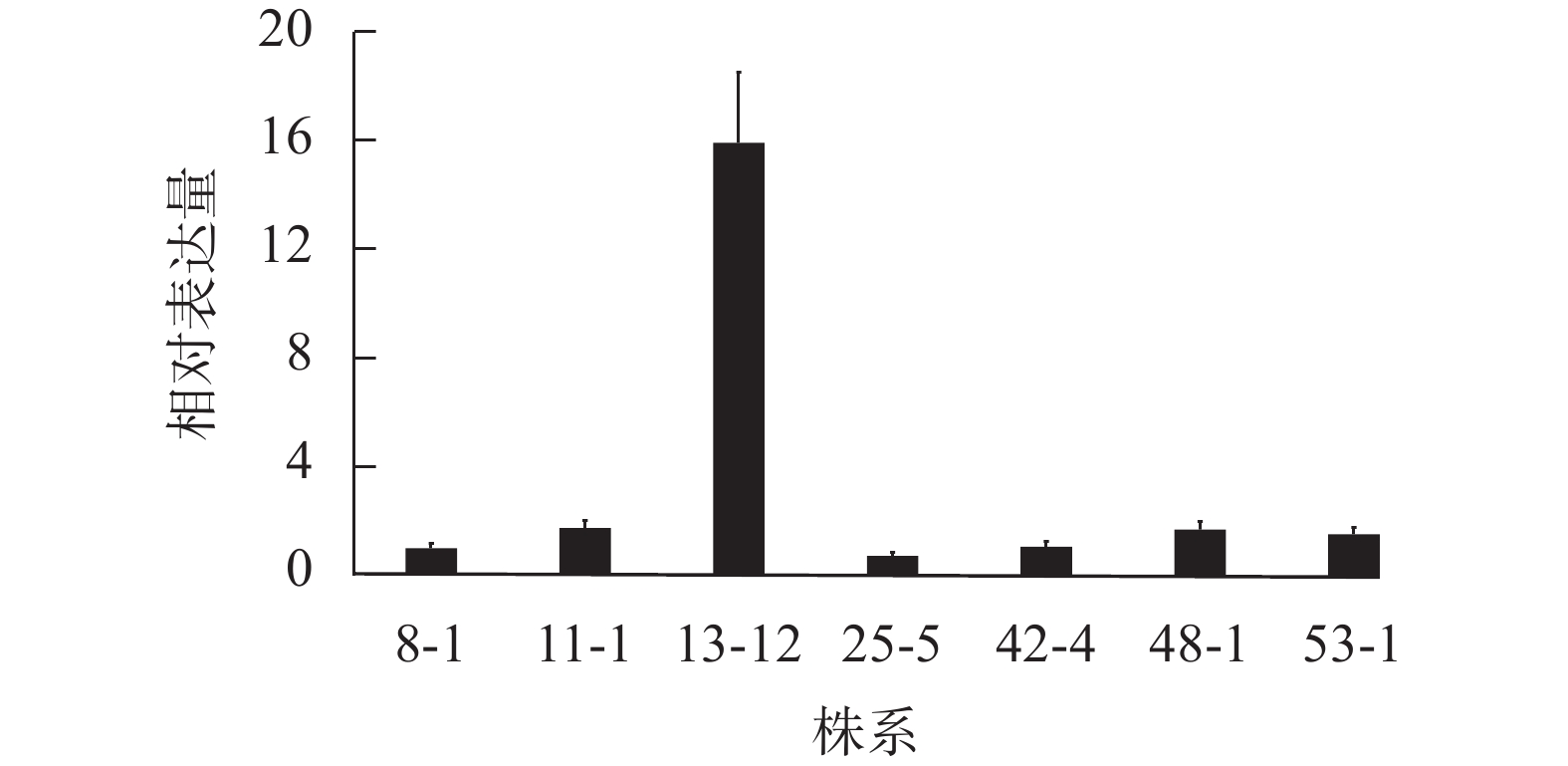
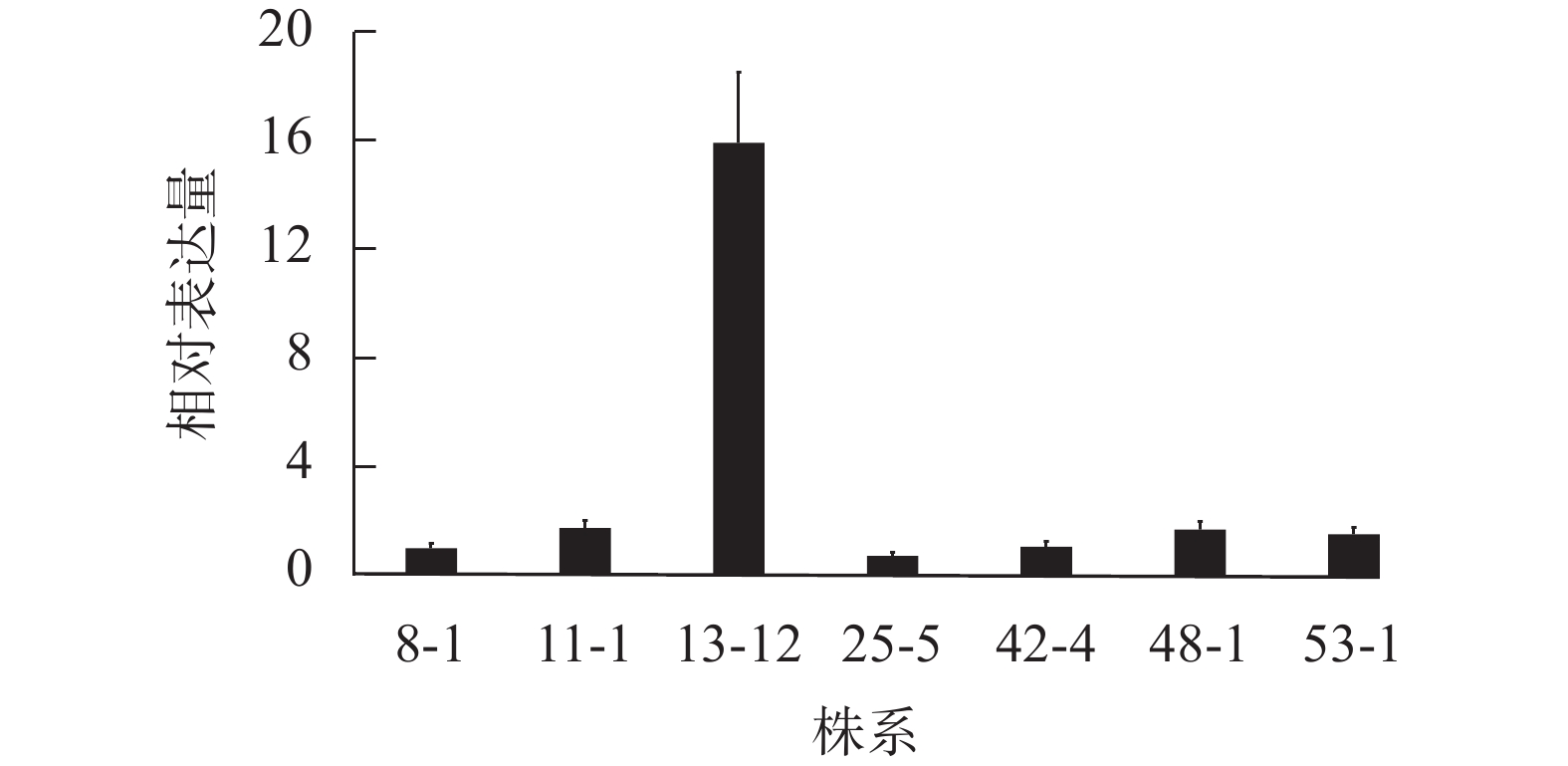
为了明确不同转基因株系PA荧光探针表达量的差异,对上述7个转基因株系进行了目的基因的表达量测定。RT-qPCR结果(图2)显示:当将转基因株系8-1中融合基因的相对表达量设为参考值“1”,其余6个株系(11-1、13-12、25-5、42-4、48-1和53-1)的相对表达量分别为1.74、15.91、0.72、1.06、1.69和1.52。株系13-12的PA探针的表达量最高,是其他株系的9~22倍,为PA探针高表达转基因株系;相应地,将株系48-1作为PA探针低表达材料。
-
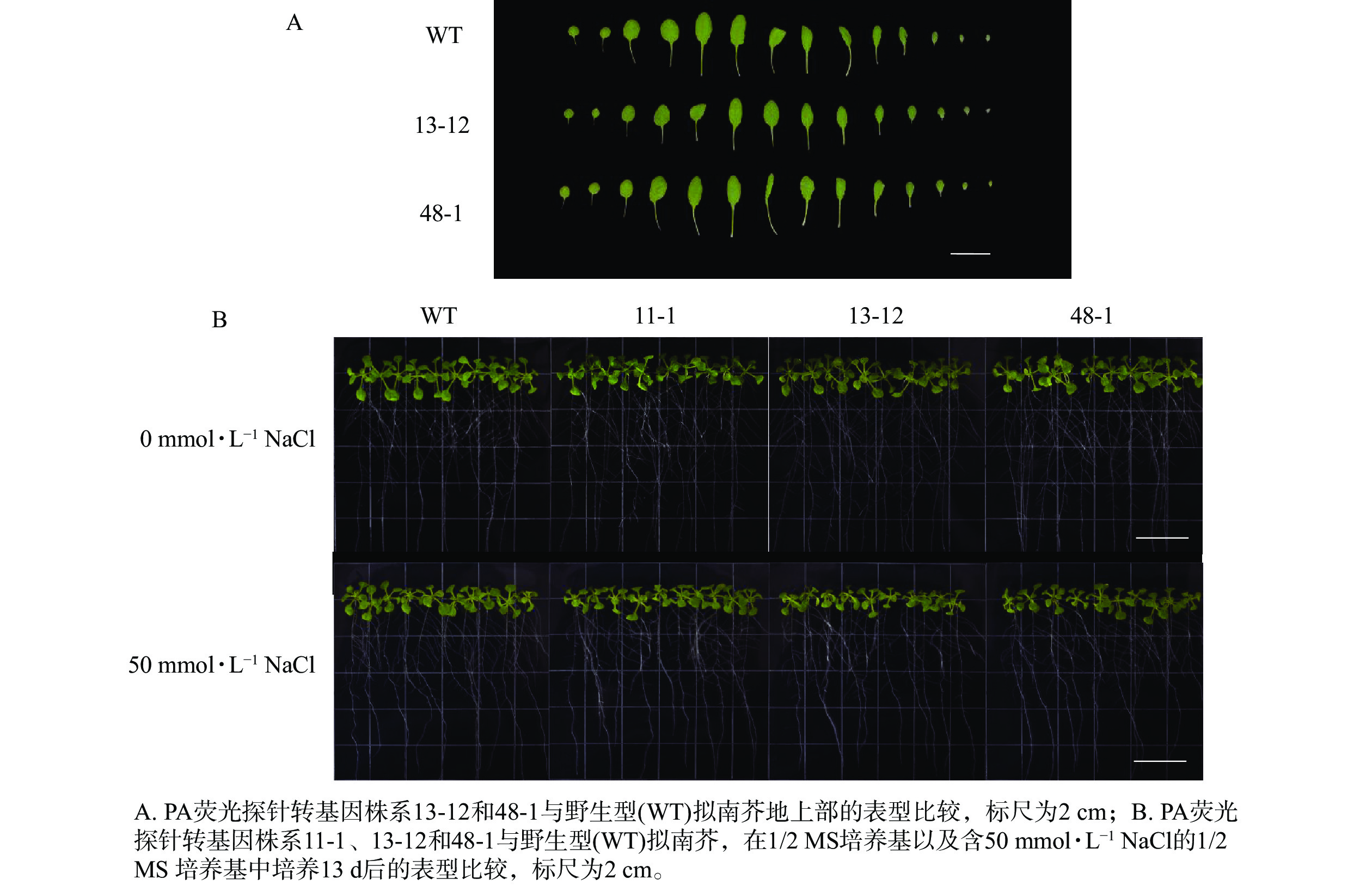
为了明确PA荧光探针的表达是否会对植物生长发育产生影响,比较分析了野生型拟南芥和不同转基因株系在整个生长发育周期的表型差异。土培生长实验显示:野生型与转基因拟南芥株系的叶片生长无明显差异,高表达转基因株系(13-12)与低表达转基因株系(48-1)之间亦无可见表型差异(图3A)。另外,在含有0和50 mmol·L−1 NaCl的1/2 MS培养基中生长时,野生型与不同转基因株系之间也未表现出生长速率的差异(图3B)。这些结果说明:在一定范围内PA荧光探针的表达不会影响拟南芥的正常生长发育,这一特性有助于合理解释特定条件下胞内PA变化对植物生长发育产生的影响。
-
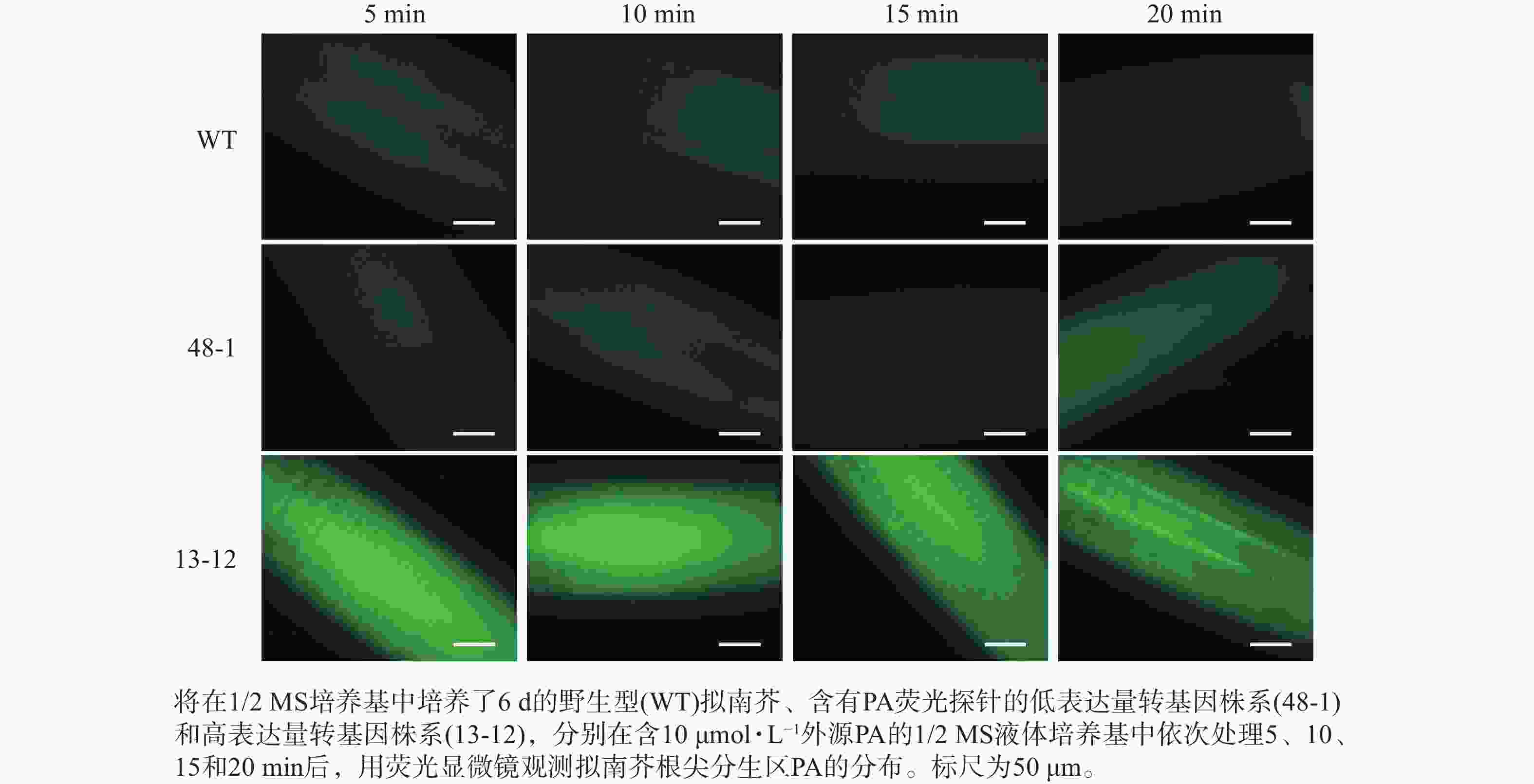
随着荧光探针表达量的增加,细胞内的荧光强度也会增强,这会导致PA检测的信噪比下降;相反,荧光探针表达量过低则会影响PA检测的灵敏度。为了获得检测灵敏度和信噪比都较高的PA荧光探针,用不同浓度的外源PA处理荧光探针表达量存在差异的转基因株系,进而在荧光显微镜下观察根尖PA的积累情况。结果(图4~6)显示:野生型拟南芥在有、无外源PA处理下,根尖细胞自发荧光的强度无明显差别。另外,在不同浓度外源PA处理下,荧光探针低表达的转基因株系(48-1)的根尖分生区的PA含量变化未能被捕捉到。相反,对于荧光探针高表达的转基因株系(13-12),2和10 μmol·L−1 PA分别处理10 和5 min后,其根尖分生区细胞的胞内或质膜上的PA积累清晰可见(图5)。当外源PA浓度低时,随着处理时间延长,细胞中PA积累减少(图5),这很可能是因为作为甘油脂合成前体物质的PA已被转化为其他物质。而当外源PA浓度升至10 μmol·L−1时,处理10、20 min后依然可见细胞中的PA呈点状分布(图6)。这些结果说明:荧光探针表达量较高的转基因株系(13-12)可被用于监测细胞中PA含量的变化。
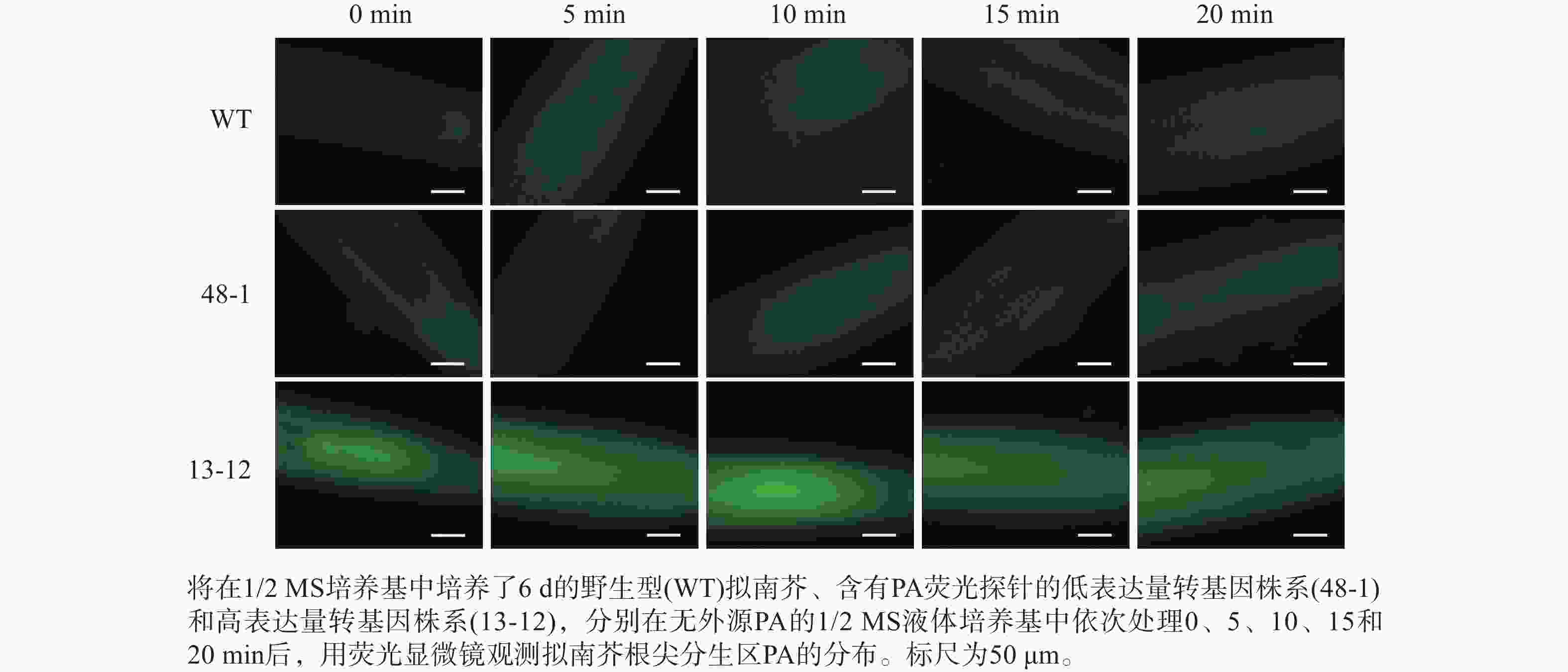
Figure 4. Analysis of discrepancy in fluorescent intensity of PA probe between wild type and two transgenic lines of A. thaliana
-
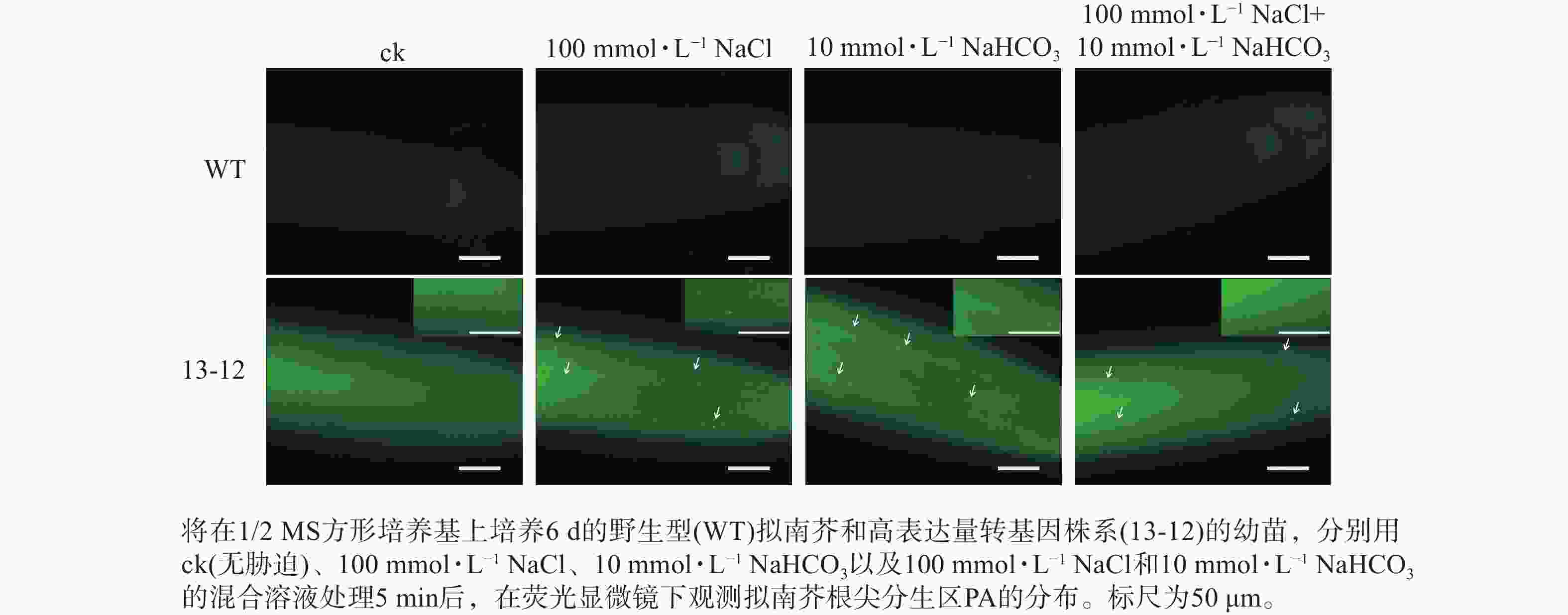
为了进一步验证 PA荧光探针的实用性,利用荧光探针表达量较高的转基因株系(13-12)检测盐碱胁迫条件下根尖细胞中PA含量的变化。结果显示:盐、碱单独或混合处理转基因株系,5 min后即可在根尖分生区细胞中观察到PA的点状分布(图7)。因此推断,本研究构建的PA荧光探针可用于监测逆境条件下植物细胞中PA含量的变化。另外,盐碱胁迫可快速诱导PA积累这一现象表明在植物早期逆境响应中PA发挥某种重要作用。
-
已知PA在植物生长发育与逆境响应过程中发挥重要作用,但由于缺乏PA的可视化分析工具,对细胞中PA含量动态变化的了解十分有限。这一方面限制了PA作用机制的研究,另一方面给评估基因工程手段操控植物细胞PA含量变化所产生的实际应用效果带来困难。因此,本研究将酵母蛋白Spo20p中对PA高度专一的结合域与荧光蛋白融合,成功构建了可对细胞内PA进行可视化检测的荧光探针。
为了使荧光探针适用于监测整个生长周期不同组织细胞中的PA水平,选择组成型表达启动子UBQ10驱动GFP-PABD融合基因的表达。研究发现:植物细胞中PA的检测灵敏度与PA荧光探针的表达量密切相关,表达量较高的荧光探针可有效检测到外源PA处理带来的胞内PA水平的变化,而表达量较低的探针检测效果则相反。如13-12株系中PA荧光探针的表达量高于其他株系,该株系经2 μmol·L−1外源PA处理10 min,其根尖细胞中PA的点状分布即可被观察到,这也说明外源PA能够被拟南芥根尖快速吸收,继而与PA荧光探针结合。鉴于已有研究中常用浓度高于10 μmol·L−1的外源PA检测PA探针的灵敏度[35],本研究中构建的荧光探针可有效监测到2 μmol·L−1外源PA处理所带来的细胞内PA含量的变化,推测基于13-12转基因株系的PA荧光探针具有较高的PA检测灵敏度。
需要指出的是,PA荧光探针使用过程中,样品处理时间的延长和观察光源的增强均会使植物细胞造成某种损伤,导致细胞产生自发荧光,从而降低PA检测的信噪比。因此在实验过程中,需尽量缩短植物样本的制片时间及荧光观察的时间,以提高观察结果的真实性。
盐碱地中盐和碱是2种共存但不同的非生物胁迫,共同影响植物的正常生长发育。利用本研究构建的PA荧光探针发现:盐胁迫处理5 min就会造成PA在细胞中的积累,这与已有的研究结果一致[22, 36]。同时,碱胁迫和盐碱混合胁迫也会快速诱导PA的积累。因此,为了全面了解植物早期逆境响应机制,有必要监测早期细胞中PA含量的动态变化。本研究开发的PA荧光探针为这些方面的研究提供了重要技术支撑。
-
本研究成功构建了一种基于酵母蛋白Spo20p中PA结合域的荧光探针,可有效监测植物细胞中PA含量变化。应用该探针发现盐碱胁迫可快速诱导拟南芥根尖细胞质膜上和胞内PA的积累,这一结果表明PA在植物对盐碱胁迫早期应答过程中发挥重要作用。
Generation and application of a fluorescent probe for phosphatidic acid in Arabidopsis thaliana
doi: 10.11833/j.issn.2095-0756.20230355
- Received Date: 2023-06-08
- Accepted Date: 2023-07-28
- Rev Recd Date: 2023-07-27
- Available Online: 2024-01-19
- Publish Date: 2024-02-20
-
Key words:
- phosphatidic acid /
- fluorescent probe /
- transgene /
- saline-alkaline stress /
- Arabidopsis thaliana
Abstract:
| Citation: | MA Shuyan, ZHENG Yueping, ZHENG Zhifu. Generation and application of a fluorescent probe for phosphatidic acid in Arabidopsis thaliana[J]. Journal of Zhejiang A&F University, 2024, 41(1): 104-112. DOI: 10.11833/j.issn.2095-0756.20230355 |












 DownLoad:
DownLoad: