-
类黄酮物质参与植物多种生理活动,不仅能为植物生长发育提供帮助,同时还在植物抵抗逆境胁迫方面发挥作用[1]。黄烷酮3-羟化酶(F3H)作为类黄酮代谢途径上游的关键酶之一,负责催化形成二氢黄酮醇,之后经过花青素和黄酮醇合成途径形成各种类黄酮衍生物[2]。F3H在基因及蛋白结构上高度保守,其底物特异性较强,需要在2-氧化戊二酸盐、分子氧、亚铁离子(Fe2+)和抗坏血酸盐的辅助下发挥作用,所以在分类上也被归为2-酮戊二酸依赖性双加氧酶(2-ODD)家族[3−4]。目前,在草莓Fragaria vesca、红花Carthamus tinctorius、拟南芥Arabidopsis thaliana、茶Camellia sinensis、黄花红砂Reaumuria trigyna、独行菜Lepidium apetalum和铁皮石斛Dendrobium officinale等植物中相继报道了关于类黄酮合成关键酶基因F3H的调控研究[5−11]。在功能上,F3H基因对植物的颜色着色有重要作用。苹果 Malus domestica MdLUX和MdPCL-like通过其启动子区域DNA低甲基化以及激活MdF3H,促进果皮中的花青素合成[12]。在草莓中,RNAi介导的F3H基因沉默导致花青素及黄酮醇显著降低,产生无色的草莓果实[5]。此外,F3H基因表达受到多种植物激素的正调控,其转录水平的增加也能提高植物对非生物和生物胁迫的耐受能力[13]。番茄 Solanum lycopersicum SlF3HL能在低温胁迫中刺激转基因烟草 Nicotiana tabacum合成类黄酮物质,使烟草耐冷性提高[14]。CtF3H在不同表型和化学型的红花中作用不同,在茉莉酸甲酯(MeJA)刺激下,CtF3H在花橙黄色型红花中高表达,并与醌式查尔酮和黄酮醇的积累有关,但在花白色型红花中低表达且不影响黄酮醇的积累[6]。
掌叶覆盆子Rubus chingii是中国特色的药食同源植物,也是浙江省新“浙八味”之一,具有补肝益肾、固精缩尿等功效[15]。相关研究表明掌叶覆盆子富含20多种黄酮类活性成分,具有抗菌抗炎等多种药理活性[16−17]。然而掌叶覆盆子类黄酮生物合成及转录调控机制尚不清晰。本研究克隆得到1个掌叶覆盆子类黄酮生物合成途径关键酶基因RcF3H的全长序列,并对其进行生物信息学分析,同时利用转录组和实时荧光定量PCR (RT-qPCR)技术分析RcF3H基因在不同组织、果实发育期以及MeJA刺激下的表达特征,为后期解析RcF3H对掌叶覆盆子类黄酮代谢的调控机制提供参考。
-
掌叶覆盆子植物材料来自杭州市富阳环山掌叶覆盆子种植基地(29.93°N,119.95°E)。4个发育阶段(青果、青转黄果、黄果、红果)的掌叶覆盆子果实于2022年4—5月陆续采收。
激素MeJA处理:于2022年4月,选择生长状态良好的1年生掌叶覆盆子植株,使用100 μmol·L−1的MeJA溶液喷施叶片的正、反两面做胁迫处理(MJ),以清水为对照组(ck)。每2 d喷施1次,处理7 d后,采集每组植株从上往下的第2张叶片作为样本。各3个重复,液氮速冻处理后放于−80 ℃低温冰箱保存。
-
使用FastPure® Universal Plant Total RNA Isolation Kit提取试剂盒(RC411),提取掌叶覆盆子植物样本总RNA。使用分光光度计(NanoDrop 2000)检测提取掌叶覆盆子总RNA浓度。使用Evo M-MLV反转录试剂盒Ⅱ(AG11711)反转录合成掌叶覆盆子cDNA。总RNA和cDNA分别放入−80和−20 ℃低温冰箱保存备用。
-
根据掌叶覆盆子基因组文件[18]得到RcF3H基因的参考序列。使用Primer Premier 6.0设计RcF3H基因的扩增引物(表1)。引物合成由浙江尚亚生物技术有限公司完成。PCR扩增体系:ddH2O 9.5 μL,上下游引物各1 μL (10 μmol·L−1),掌叶覆盆子cDNA模板1 μL,12.5 μL PrimeSTAR Max premix(R045A)。PCR扩增程序:94 ℃ 3 min;98 ℃ 10 s,58 ℃ 15 s,72 ℃ 70 s,35个循环;72 ℃ 5 min;4 ℃ 保温。反应结束后将PCR扩增产物使用琼脂糖凝胶电泳检测并使用产物纯化试剂盒(DC301)回收。最后将RcF3H基因片段连接到pMD19-T载体上(D102A),热激法转化大肠埃希菌Escherichia coli DH5α感受态,挑取阳性单克隆进行菌液PCR鉴定,由浙江尚亚生物技术有限公司测序分析。
引物名称 引物序列(5′→3′) 引物用途 β-actin-F ATCCACGAGACTACATACAACTCC 内参基因 β-actin-R CTGTCTGCAATACCAGGGAAC RcF3H-F ATGGCTCCTACACCTACTAC 序列扩增 RcF3H-R AGCAAAAATACCATCCACTT RcF3H-qPCR-F CAAAGTGGCCTACAACCAATTC 荧光定量 RcF3H-qPCR-R CCTCGACAATCTTCTTGCAAATC Table 1. Primer sequence for this study
-
使用Gene Structure Display Server网站分析RcF3H基因结构,并使用IBS 2.0作图。使用Expasy ProtParam tool网站预测掌叶覆盆子RcF3H蛋白的分子量、等电点等基本理化性质;通过Plant-mPLoc网站预测RcF3H蛋白的亚细胞定位情况。使用Expasy Protscale网站分析掌叶覆盆子RcF3H蛋白亲疏水性;使用美国国家生物技术信息中心(NCBI)中的CD-Search分析RcF3H蛋白的保守结构域。
使用SOPMA网站预测RcF3H蛋白的二级结构,利用Swiss-Model网站预测RcF3H蛋白的三维结构。从NCBI数据库的Protein Blast中下载与RcF3H有较高同源性的蔷薇科Rosaceae植物的F3H蛋白序列,使用DNAMAN软件进行多序列比对;使用MEGA 7软件将拟南芥、水稻Oryza sativa等单、双子叶其他科属植物的F3H蛋白序列采用邻接法(neighbor-joining)构建系统发育树(bootstrap值设置为1 000,其余参数默认),并利用ChiPlot作图。使用PlantCARE网站获得RcF3H基因前2 000 bp的启动子序列,检索得到顺式作用元件[19−20]。
-
参考掌叶覆盆子的组织转录组数据[18],分析不同组织下RcF3H基因相对表达量,并使用TBtools 软件绘制热图。
-
以反转录合成的掌叶覆盆子cDNA为材料,使用Primer Premier 6.0设计RcF3H基因的RT-qPCR引物(表1)。引物由浙江尚亚生物技术有限公司合成。RT-qPCR反应方法参照文献[21],使用iTaq™ universal SYBR® Green supermix (1725121)以及荧光定量PCR仪(ABI 7500)完成。RT-qPCR反应体系:iTaq™ universal SYBR® Green supermix 5 μL,上、下游引物各0.5 μL (10 mmol·L−1),cDNA模板1 μL (50 mg·L−1),ddH2O 3 μL。RT-qPCR反应程序:95 ℃ 30 s;95 ℃ 10 s,60 ℃ 30 s ,40个循环。结果以掌叶覆盆子β-actin为内参基因,采用2–∆∆Ct法[22]计算RcF3H基因相对表达量。
-
数据使用Excel 2021进行统计分析。使用SPSS 26进行单向方差分析以及t检验,使用GraphPad Prism 8作图。
-
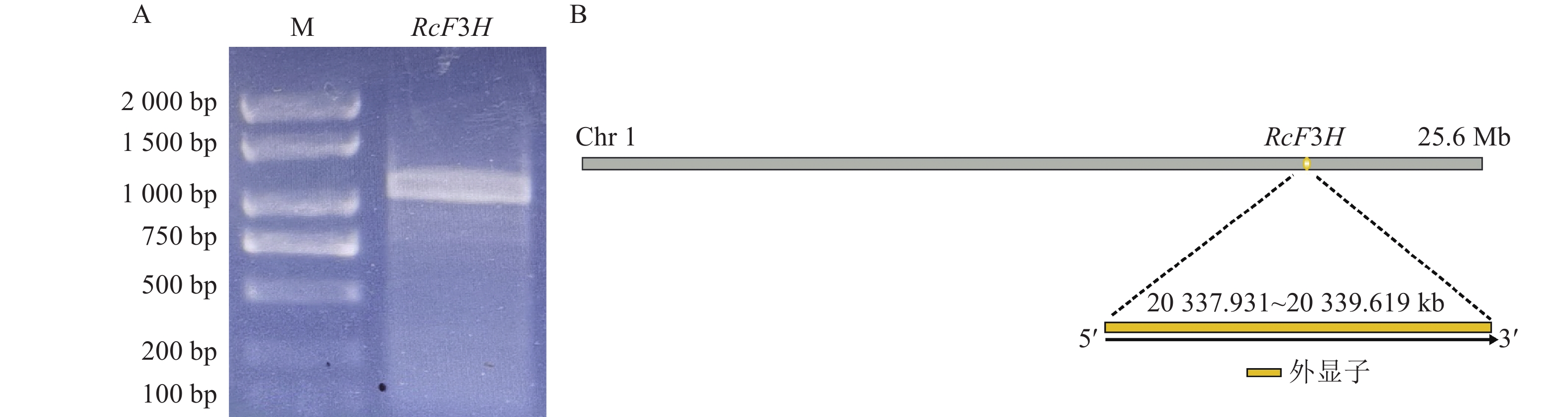
以反转录合成的掌叶覆盆子cDNA为模板,成功扩增得到目的基因片段(图1A)。对基因片段进行切胶回收,连接转化至DH5α感受态,12 h后对阳性单克隆菌液进行PCR验证并测序分析。结果显示:目的片段长度为1 098 bp,将其命名为RcF3H。RcF3H基因定位于第1条染色体内,由1个外显子构成,共编码了365个氨基酸(图2)。
-
RcF3H蛋白分子式为C1831H2900N498O553S13,相对分子量为65.16 kDa;理论等电点为5.64;脂肪系数为83.32;不稳定系数为38.22,属于稳定蛋白。亚细胞定位预测表明:该蛋白定位在细胞质上。
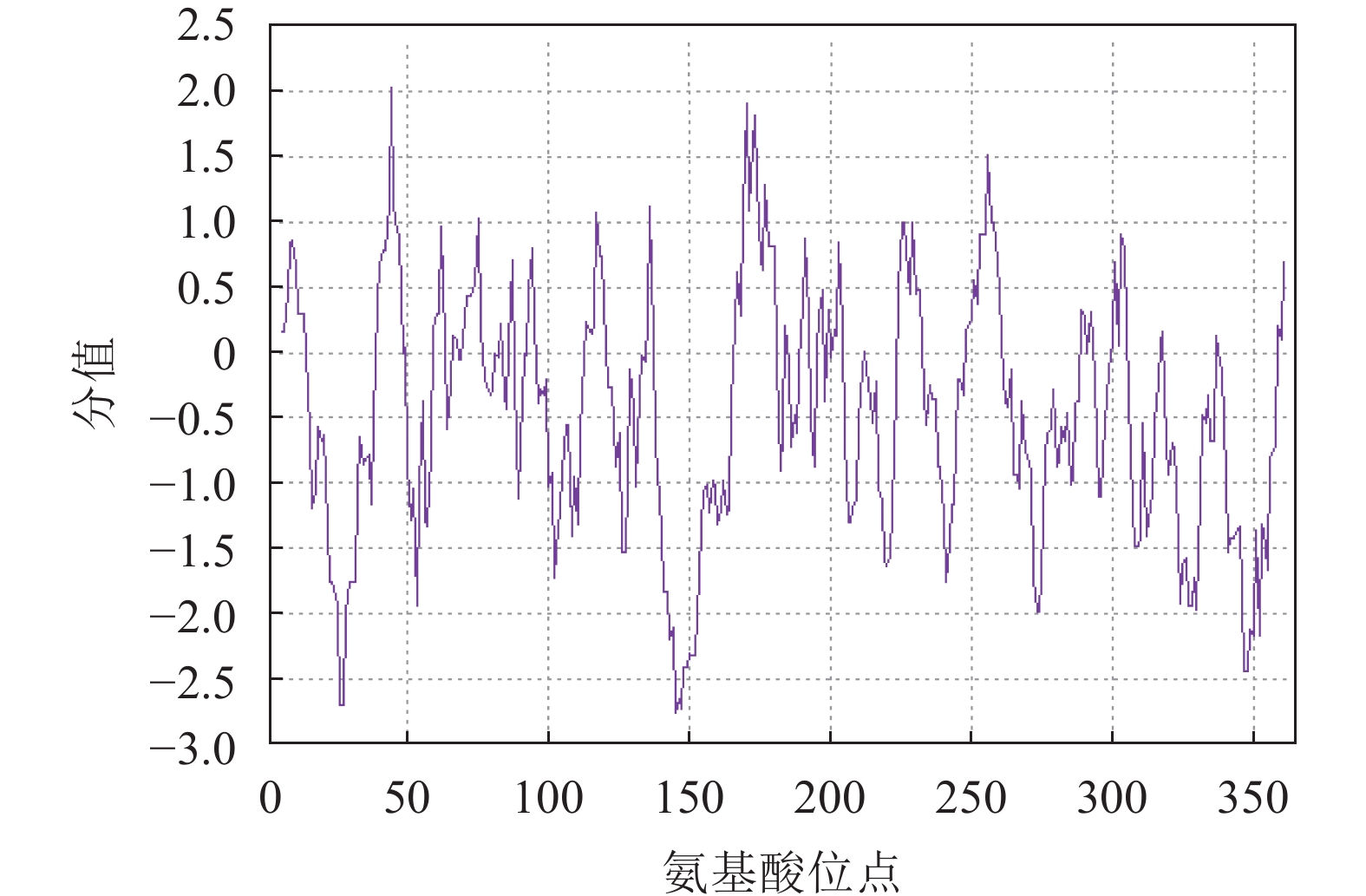
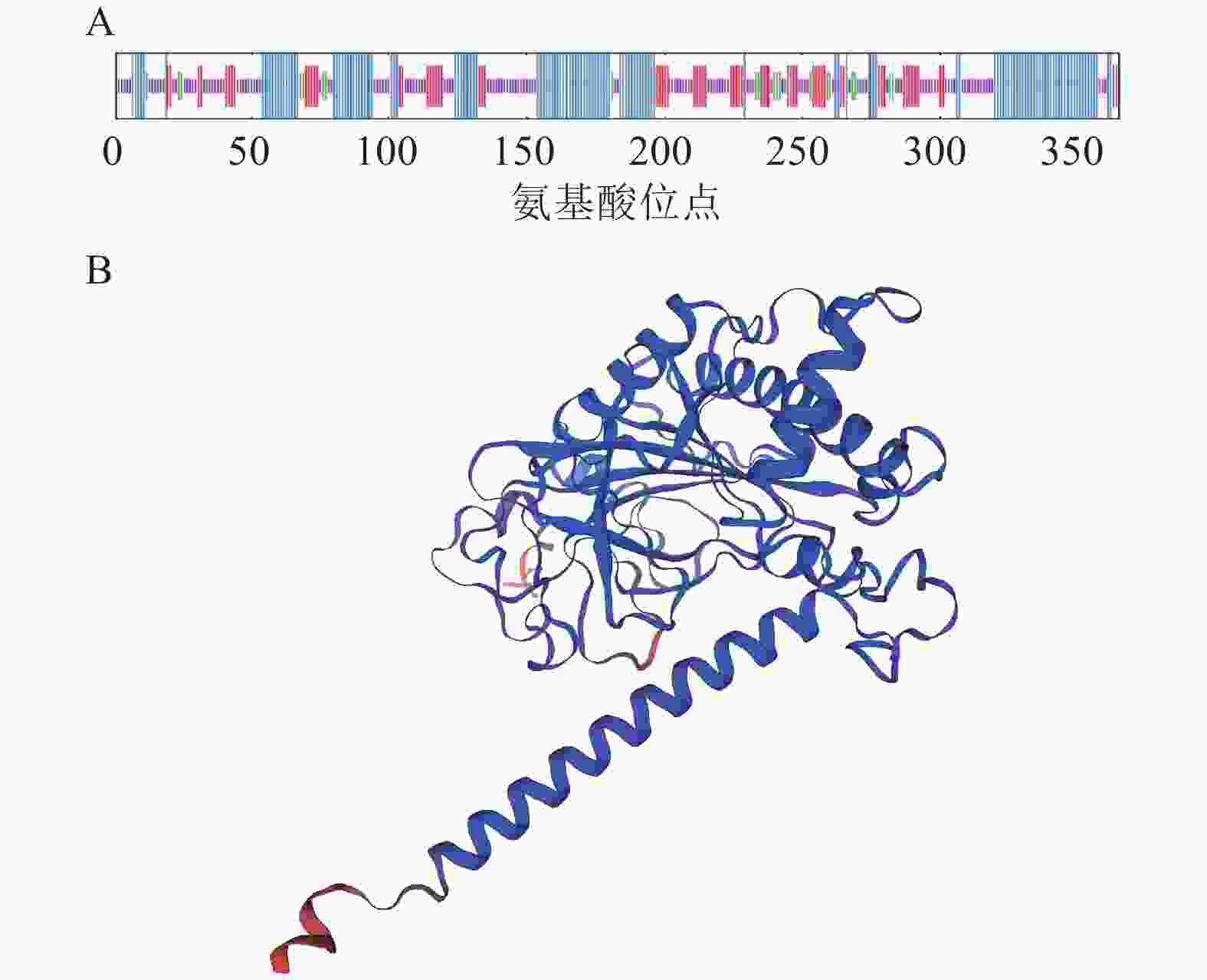
蛋白亲疏水性分析表明:RcF3H蛋白属于亲水性蛋白;蛋白平均亲水指数为−0.448,大部分区域表现出亲水性,最大疏水分值(−2.756)位于肽链第145位氨基酸,最大亲水分值(2.022)位于肽链第44位氨基酸(图3)。
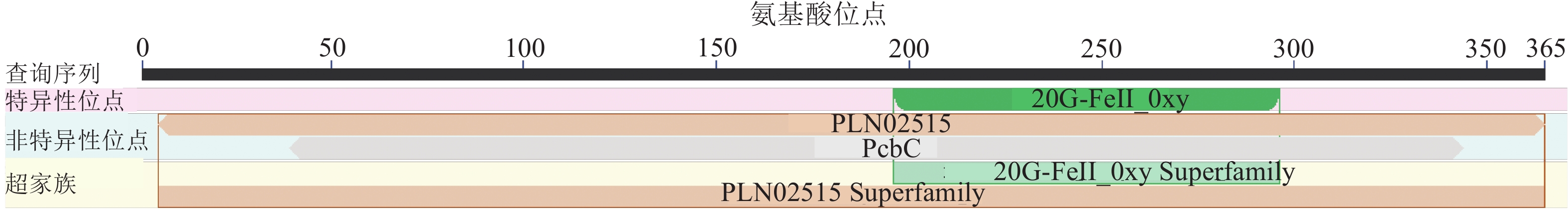
蛋白结构域分析发现:RcF3H蛋白属于2-酮戊二酸依赖性双加氧酶家族,拥有2OG-FeⅡ_0xy区域以及PLN02515和Pcb C等2个保守结构域(图4)。
-
RcF3H蛋白的二级结构预测发现:该蛋白存在α-螺旋、β-折叠、直链延伸和无规则卷曲4种结构,其中以α-螺旋和无规则卷曲占比最大,分别占37.53%和38.63%,而直链延伸和β-折叠分别占18.36%和5.48% (图5)。此外,以石榴Punica granatum蛋白(编号:A0A218XVH5.1)为模板,对RcF3H蛋白的三级结构进行建模。建模的三级结构覆盖率为92.31%,序列一致性为85.75%。该结果与二级结构预测结果一致。
-
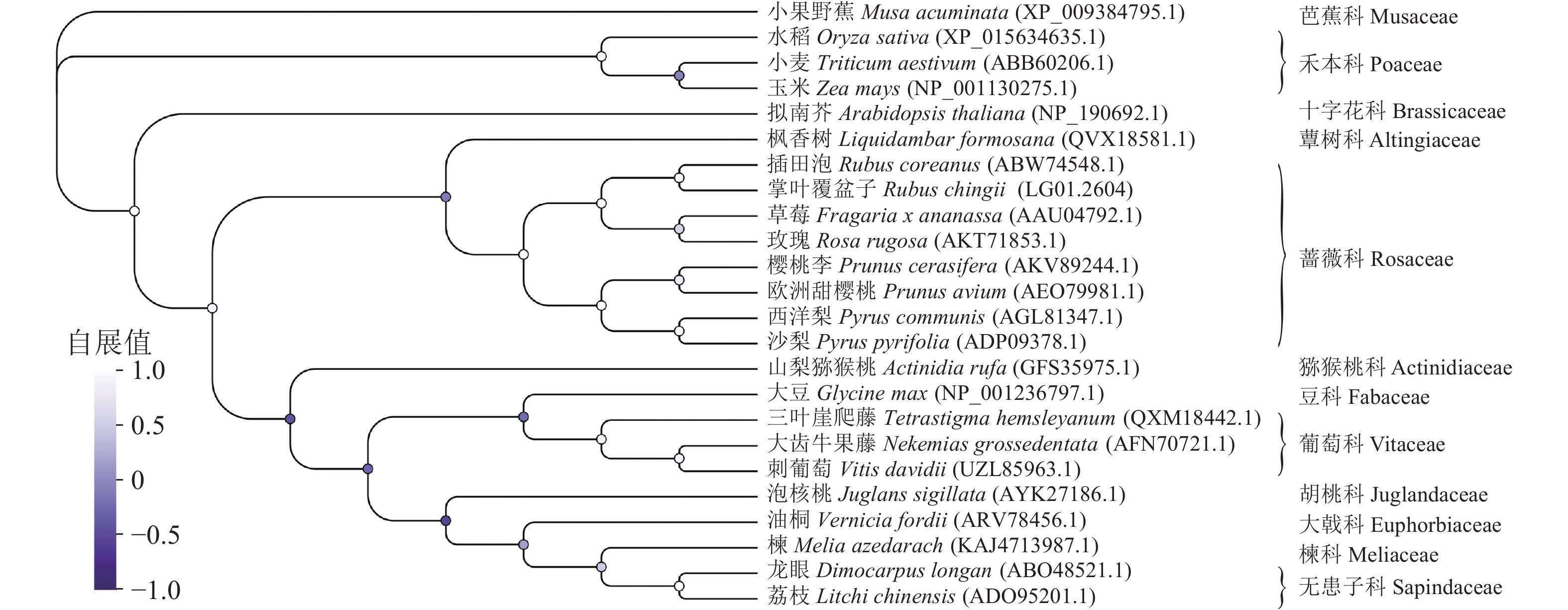
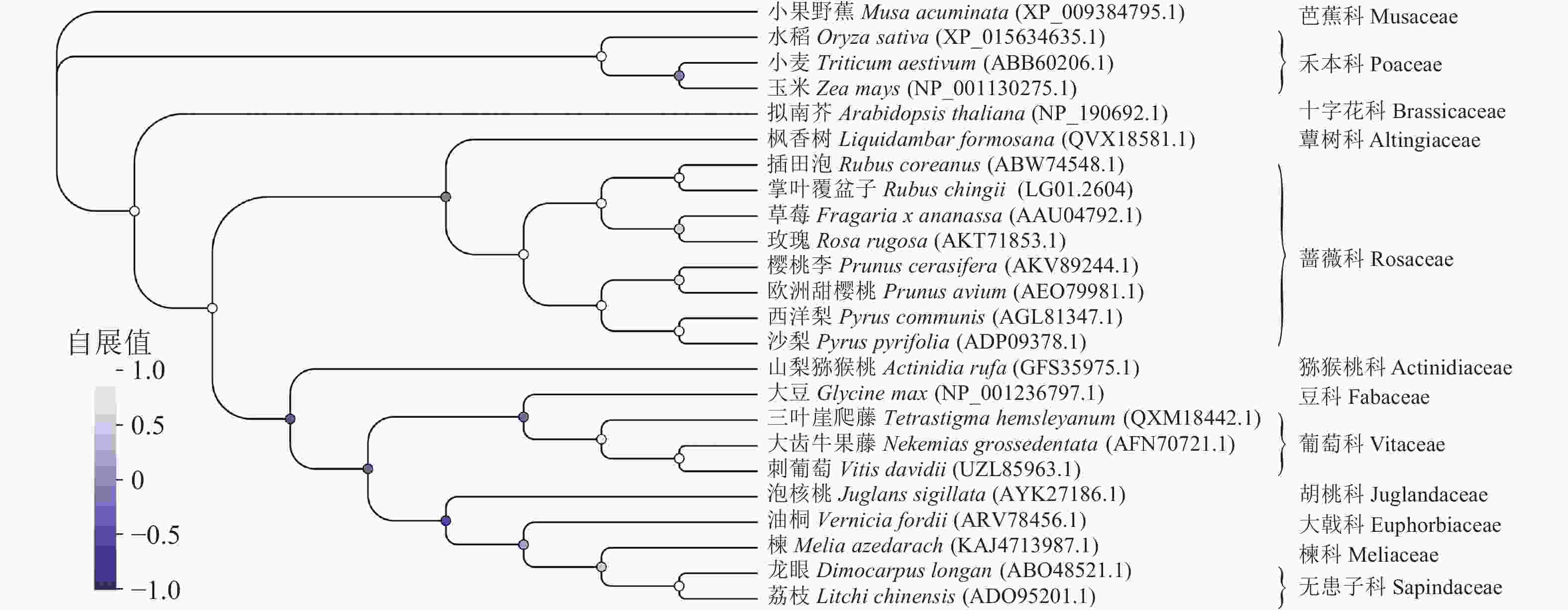
多序列比对分析发现:蔷薇科F3H蛋白序列具有很高的相似度,其中RcF3H与插田泡Rubus coreanus、草莓和玫瑰Rosa rugosa的相似性超过95%,与樱桃李Prunus cerasifera、欧洲甜樱桃Prunus avium和西洋梨Pyrus communis相似性接近90% (图6)。系统进化树分析发现:RcF3H与蔷薇科插田泡蛋白(编号:ABW74548.1)的亲缘关系最密切。同科属的F3H蛋白在系统进化上处于同一分支。与单子叶植物相比,掌叶覆盆子RcF3H与双子叶植物关系更密切,属于同一亚组(图7)。以上结果说明掌叶覆盆子RcF3H蛋白在生物进化过程中较为保守。
-
RcF3H基因启动子中具有大量相对保守的CAAT-box和TATA-box。根据顺式作用元件功能的不同,可以将其分为植物生长发育、激素响应以及胁迫响应3个类别(表2)。RcF3H基因启动子顺式作用元件有2个植物生长发育的顺式调控元件;在激素响应中茉莉酸响应元件数量最多,赤霉素和生长素次之。此外,RcF3H基因在胁迫响应上包含有多数响应光照元件以及少量应对厌氧、缺氧、防御和应激、低温以及干旱的顺式作用元件。以上结果表明:RcF3H基因在辅助掌叶覆盆子响应激素和应对胁迫上发挥了重要作用,且可能受外界刺激后调控合成类黄酮物质。
类别 元件名称 数量 功能 植物生长发育 Circadian 1 参与昼夜节律控制的顺式调控元件 O2-site 2 玉米蛋白代谢调控的顺式调控元件 激素响应 AuxRE 1 生长素反应元件的一部分 CGTCA-motif 1 参与茉莉酸反应性的顺式作用元件 TGACG-motif 1 参与茉莉酸反应性的顺式作用元件 MYC 6 激素响应元件 P-box 1 赤霉素响应元件 Myb 1 MYB识别位点 MYB 1 MYB识别位点 MYB-like sequence 1 MYB识别位点 胁迫响应 ARE 3 对厌氧诱导必要的顺式调节元件 GC-motif 2 参与缺氧特异性诱导性的增强子元件 LTR 1 参与低温反应的顺式元件 MBS 1 MYB结合位点参与干旱诱导 STRE 1 胁迫响应元件 as-1 1 胁迫响应元件 TC-rich repeats 1 参与防御和应激反应的顺式作用元件 Box 4 2 参与光反应的保守DNA模块 GATA-motif 1 光响应元件的一部分 GT1-motif 3 光响应元件 Sp1 1 光响应元件 W box 1 诱导子响应元件 Table 2. Analysis of cis-regulatory elements of RcF3H promoter in R. chingii
-
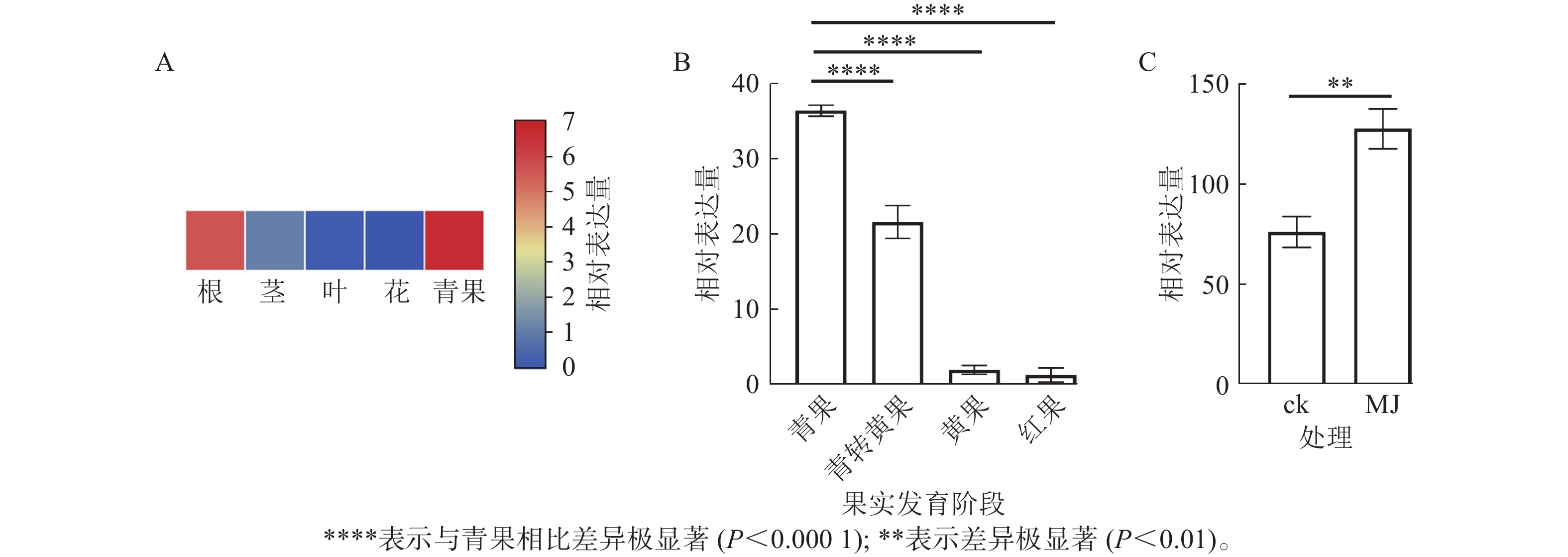
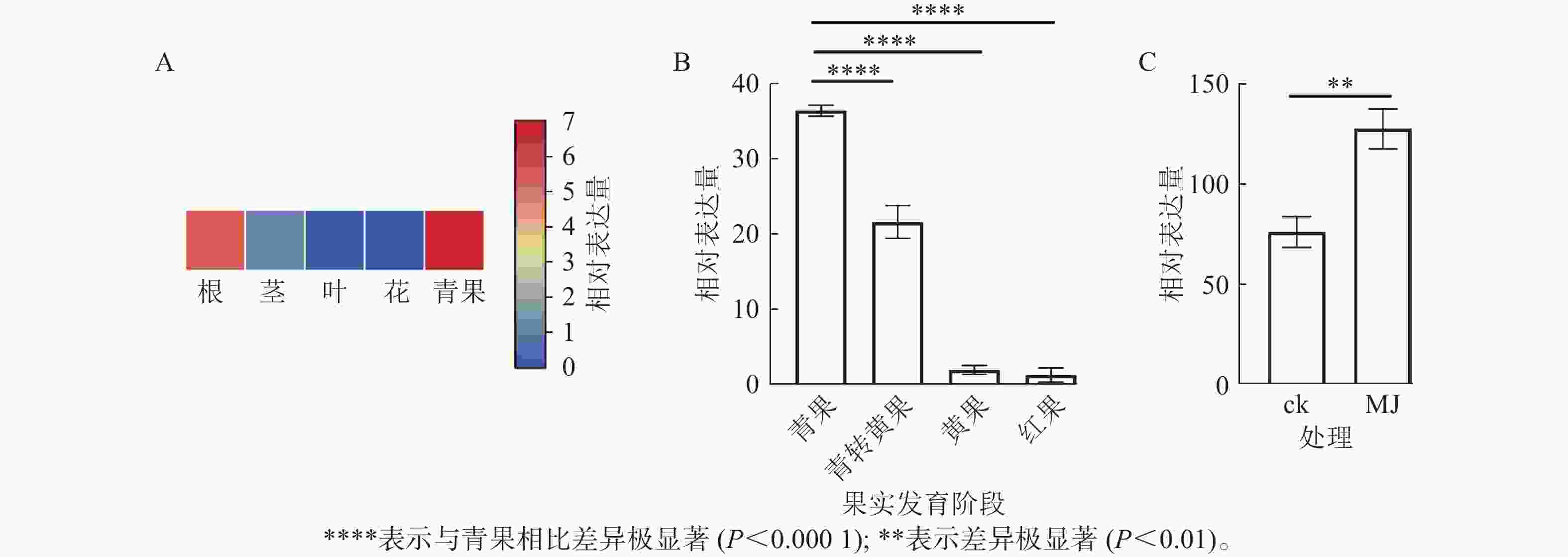
由图8A可知:RcF3H基因在掌叶覆盆子的根、茎、叶和果实中表达;其中青果中的相对表达量最高,分别是根的1.18倍,茎的5.05倍,叶片的46.00倍。表明RcF3H可能与掌叶覆盆子果实的生长发育有关。进一步分析在果实发育成熟过程中RcF3H基因的相对表达量。由图8B可知:RcF3H基因表达在青果期表达量最高,并在发育成熟过程表达量逐渐下降,该趋势与类黄酮质量分数变化一致[23]。启动子顺式作用元件分析发现:掌叶覆盆子RcF3H基因前2 000 bp启动子上存在响应MeJA的顺式作用元件(表2)。结合RT-qPCR技术发现:掌叶覆盆子RcF3H基因经过MeJA诱导处理后基因表达明显上升(图8C)。
-
本研究从掌叶覆盆子cDNA中克隆得到了完整的RcF3H基因序列,该基因由1条外显子构成,全长1 098 bp,编码了365个氨基酸。蛋白序列比对后发现:掌叶覆盆子RcF3H蛋白与蔷薇科插田泡等6种植物F3H的同源关系较近。蛋白质是细胞的功能分子,基因功能的主要执行者。掌叶覆盆子RcF3H拥有 2OG-Fe(Ⅱ)-Oxy结构以及2个典型的PLN02515和Pcb C保守结构域,亚细胞定位预测该蛋白位于细胞质中。一些蛋白质需要转移至特定的位置后才能发挥作用,信号肽能引导新合成的蛋白质向分泌通路转移[24]。RcF3H蛋白为稳定的亲水性蛋白,蛋白二级和三级结构分析发现:RcF3H主要以α-螺旋和无规则卷曲缠绕形成。系统进化树分析发现:RcF3H蛋白与单子叶植物亲缘关系较远,与双子叶植物亲缘关系较近且处于同一亚组,说明在进化过程中RcF3H基因较保守。
转录调控对基因表达起着很大的作用,主要通过基因的启动子及其相关的顺式作用元件控制[25]。对RcF3H启动子分析发现:其拥有茉莉酸、赤霉素和生长素等响应元件。RcF3H基因在MeJA处理7 d后表达量上调了1.68倍。在拟南芥中,茉莉酸和脱落酸能够调控类黄酮生物合成中AtF3H的表达[26]。MeJA处理绿熟期的樱桃番茄S. lycopersicum ‘Xin Taiyang’果实后,促进了类黄酮的积累,其类黄酮代谢途径中PAL1、C4H以及F3H等基因表达水平上调[27]。掌叶覆盆子RcF3H启动子还存在逆境胁迫响应有关的作用元件。黄花红砂在受到UV-B和干旱胁迫后,其RsF3H酶活性增加,类黄酮生物合成产物积累增多[28]。说明RcF3H基因可能在激素和逆境胁迫上发挥作用,调节类黄酮物质的合成来提高对环境适应性。
进一步探究掌叶覆盆子RcF3H表达与类黄酮的关系。RcF3H在掌叶覆盆子的根、茎、叶和果实中表达且具有组织特异性。RcF3H在青果中表达量最高,并在果实成熟过程中表达量逐渐下降,推测RcF3H可能与果实发育有关。随着掌叶覆盆子果实发育成熟,椴树苷和山奈酚-3-O-芸香糖苷等各种黄酮化合物会逐渐减少[23],这与RcF3H表达变化趋势一致,推测RcF3H可能促进类黄酮的积累。一些研究报道了MeJA能促进类黄酮的生物合成[26−27]。掌叶覆盆子RcF3H能响应外源MeJA的刺激,其表达量相较于对照组上升了1.68倍,而MeJA促进掌叶覆盆子类黄酮代谢未见报道。综上所述,掌叶覆盆子RcF3H在多个组织器官中发挥生物学功能,并能影响类黄酮的积累。此外,具体的生物学功能验证以及调控机制解析也有待进一步研究。
-
本研究在掌叶覆盆子中鉴定克隆了2-酮戊二酸依赖性双加氧酶家族基因RcF3H,该基因蛋白定位于细胞质,与双子叶植物同源性较高。该基因启动子上存在大量与激素和逆境响应相关的元件。RcF3H在多个器官中均有表达且具有组织特异性,并随果实成熟表达量逐渐下降。外源MeJA的刺激处理可提高RcF3H基因的表达。本研究初步解析了RcF3H在掌叶覆盆子类黄酮物质积累中的作用,为进一步研究其分子机制提供参考。
Cloning and expression analysis of RcF3H in Rubus chingii
doi: 10.11833/j.issn.2095-0756.20240326
- Received Date: 2024-04-29
- Accepted Date: 2024-07-28
- Rev Recd Date: 2024-07-25
- Available Online: 2024-11-20
- Publish Date: 2024-11-20
-
Key words:
- Rubus chingii /
- flavanone 3-hydroxylase /
- gene cloning /
- expression analysis /
- flavonoids
Abstract:
| Citation: | YING Yuxin, CHEN Junyu, YAO Lingtiao, et al. Cloning and expression analysis of RcF3H in Rubus chingii[J]. Journal of Zhejiang A&F University, 2024, 41(6): 1180-1188. DOI: 10.11833/j.issn.2095-0756.20240326 |














 DownLoad:
DownLoad: