-
薰衣草Lavandula angustifolia为唇形科Lamiaceae多年生小灌木,原产于地中海沿岸地区,兼具观赏价值与经济价值,其精油广泛应用于美容、食品、香料及医药保健领域,是重要的天然香料植物[1−2]。中国新疆伊犁地区自20世纪中期起引种栽培薰衣草,现已成为仅次于法国普罗旺斯的全球第二大薰衣草产区,被农业农村部授予“中国薰衣草之乡”称号[3]。薰衣草品种繁多,花色以蓝紫色为主,兼具蓝色、白色、粉色等色系[4]。解析花色形成相关结构基因的功能,对阐明植物花色形成机制、促进物种进化研究及花色遗传改良具有重要意义。
查尔酮合成酶(chalcone synthase,CHS)属于植物聚酮合酶超家族成员(polyketide synthase,PKS)。根据蛋白结构可将PKS分为Ⅰ、Ⅱ和Ⅲ型,PKS-Ⅰ和PKS-Ⅱ型仅存在于微生物,而PKS-Ⅲ型(CHS超家族)主要分布于植物中[5]。CHS作为类黄酮和花青素生物合成的关键限速酶,可为黄酮类化合物的合成提供碳骨架,其主要功能是在植物次生代谢中催化丙二酰CoA和4-香豆酰CoA生成查尔酮。CHS基因与植物颜色变化之间存在着显著的相关性,其催化产物直接影响植物花色、抗逆性及药用活性成分积累[6−8]。CHS在基因及蛋白结构上高度保守,其底物特异性较强,催化机制依赖于高度保守的Cys164-His303-Asn336三联体活性位点,催化生成的一系列结构各异、生理活性不同的植物次生代谢产物参与植物生长发育[9−10]。在掌叶覆盆子Rubus chingii的CHS基因家族成员中,15个RcCHS基因具有组织特异性表达,并在果实不同发育时期差异表达,且RcCHS8和RcCHS11基因的表达与果实中类黄酮的积累呈正相关[11]。棘葡萄Vitis davidii的VdCHS2基因过表达显著增加了其总花色苷的积累,促进花青素成分的合成和稳定积累,CHS2基因的过表达影响了CHI、F3Hb、F3′5′H、DFR4和LDOX等关键基因的差异表达[12]。高丛蓝莓Vaccinium corymbosum的VcCHSs基因过表达可显著增强蓝莓叶片和苹果Malus pumila 果实中花青素的积累,并上调类黄酮生物合成结构基因的表达[13]。
目前,对薰衣草的研究多聚焦于萜类物质合成的代谢途径[14−17],对类黄酮合成机制尚待深入研究。本研究首次克隆了位于GroupⅠ亚家族的薰衣草类黄酮生物合成关键酶基因LaCHS,并对该基因进行生物信息学、组织特异性表达、异源过表达及病毒诱导基因沉默分析,旨在揭示其对花色发育与类黄酮合成的调控功能,为完善薰衣草次生代谢分子机制研究提供理论依据。
-
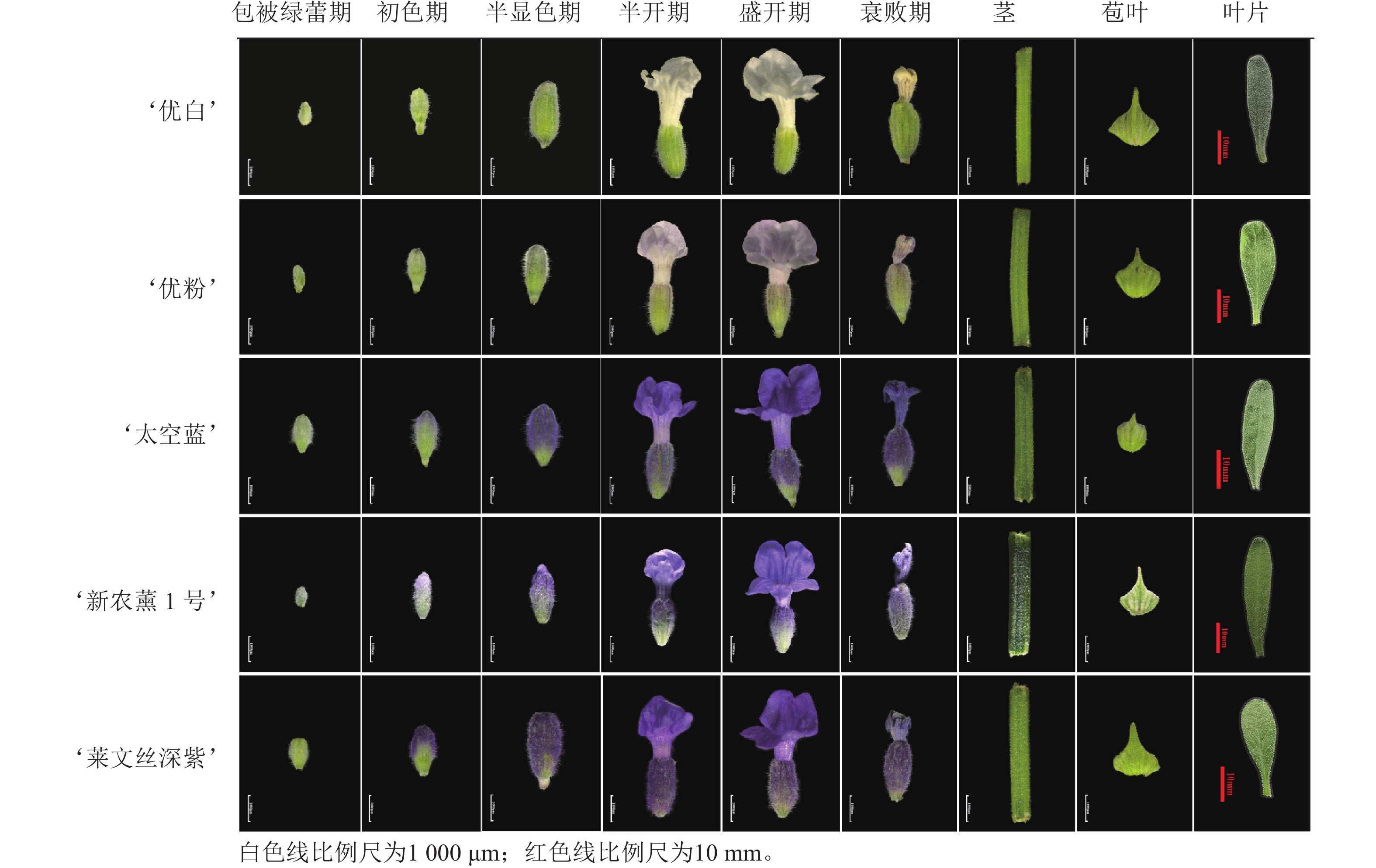
所有材料均来自新疆农业大学薰衣草研究所种质资源圃,对‘莱文丝深紫’薰衣草L. angustifolia ‘Levens Deep Purple’的幼嫩叶片进行基因克隆,以‘优白’ ‘Superior White’、‘优粉’ ‘Premium Pink’、‘太空蓝’ ‘Space Blue’、‘新农薰1号’ ‘Xinnongxun No.1’和‘莱文丝深紫’薰衣草的不同器官为表达模式分析材料。将花器官分为6个发育时期:包被绿蕾期(S1,花瓣完全被花萼包裹,绿色)、初色期(S2,花瓣完全被花萼包裹,花瓣逐渐开始上色)、半显色期(S3,花瓣完全被花萼包裹,花色开始变化为一半绿色一半紫色或白色或粉色)、半开期(S4,花瓣为半开,完全着色)、盛开期(S5,花瓣完全开放,全部着色)和衰败期(S6,花瓣衰败),以及营养器官为盛开期(S5)的茎、苞叶和叶片(图1)。基因克隆用新鲜样品,其余样品于−80 ℃超低温冰箱保存备用。
-
使用Total RNA Extractor(Trizol)提取试剂盒,提取‘莱文丝深紫’幼嫩叶片、其他参试薰衣草品种花器官6个发育时期的花蕾及盛花期(S5)的茎、苞叶和叶片的总RNA。使用分光光度计检测各组织总RNA浓度。使用生工生物工程(上海)股份有限公司的MightyScript Plus 第1链 cDNA 合成 Master Mix (去基因组DNA)试剂盒处理cDNA。总RNA和cDNA分别放入−80和−20 ℃冰箱保存备用。
-
使用美国国家生物技术信息中心(NCBI)网站Conserved Domains Search软件对LaCHS蛋白保守结构域进行预测比对。使用Expasy ProtParam Tool软件预测LaCHS蛋白的分子量、等电点等基本理化性质。通过Plant-mPLoc软件预测LaCHS蛋白的亚细胞定位情况。使用Expasy Protscale软件分析LaCHS蛋白的亲疏水性。使用SOPMA软件预测LaCHS蛋白的二级结构,利用Swiss-Model软件预测LaCHS蛋白的三级结构。从NCBI数据库的Protein Blast中下载与LaCHS蛋白有较高同源性的CHS蛋白序列,使用DNAMAN软件进行多序列比对;使用MEGA11软件将紫苏Perilla frutescens、圣罗勒Ocimum tenuiflorum等其他科属植物的CHS蛋白序列采用邻接法(neighbor-joining)构建系统发育树(bootstrap),值设置为1 000,其余参数默认,并利用evolview作图。
-
基于课题组完成的白色花‘优白’与深紫色花‘莱文丝深紫’盛开期花蕾比较转录组数据筛选获得LaCHS基因的参考序列,设计LaCHS基因扩增特异引物(表1),以‘莱文丝深紫’叶片cDNA为材料进行PCR扩增,将回收到的片段连接到pEASY®-T5载体上,通过热激法将连接产物转化大肠埃希菌Escherichia coli DH5α,经菌落PCR初筛,将阳性菌株送生工生物工程(上海)股份有限公司进行测序分析。
引物名称 正向(反向)引物序列(5′→3′) 引物用途 LaCHS-F ATGGTGACCGTTGAGGAGATCC 序列扩增 LaCHS-R TTAATTAATCGCCACGCTGTGCAG RT-LaCHS-F AACACCACAACCCCCACTTT 荧光定量 RT-LaCHS-R TGCGGAAGTAGTAGTCGGGA Actin-F TGTGGATTGCCAAGGCAGAGT 薰衣草内参 Actin-R AATGAGCAGGCAGCAACAGCA pCAMBIA3301-LaCHS-MF atgaccatgattacgaattcATGGTGACCGTTGAGGAGATC 过表达载体构建 pCAMBIA3301-LaCHS-MR aggtcgactctagaggatccTTAATTAATCGCCACGCTGTGCA pTRV2-LaCHS-MF TGAGTAAGGTTACCGAATTCATGGTGACCGTTGAGGAGATC VIGS载体构建 pTRV2-LaCHS-MR GTGAGCTCGGTACCGGATCCCACCACCACGATGTCTTGC Nb-Actin-F AGTCCTCTTCCAGCCATCCA 本氏烟草内参 Nb-Actin-R TAGGAGCCAAAGCCGTGATT Nb-RT-LaCHS-F ATGGTGACCGTTGAGGAGATCC 目的基因LaCHS Nb-RT-LaCHS-R TTGATGCTCGCTGTTGGTGATG Nb-RT-NbF3′H-F TAAGGCTTCATCCATCCACC 下游基因NbF3′H Nb-RT-NbF3′H-R CAAAGTCATTTCCTCGCACA Nb-RT-NbDFR-F TTGTTGGTCCATTCCTCACG 下游基因NbDFR Nb-RT-NbDFR-R CCACGGGCAAGTCCTTATCG Table 1. Primers used in this study
-
以不同品种薰衣草花器官6个发育时期花蕾及盛开期(S5)茎、苞叶和叶片的cDNA为材料,设计LaCHS基因的实时荧光定量PCR(RT-qPCR)引物(表1)。按照Sangon Biotech的SG Excel Fast SYBR Mixture试剂盒说明书进行RT-qPCR扩增,使用7500 FAST荧光定量PCR仪(Applied Biosystems)完成。结果以薰衣草Actin为内参基因,采用2–∆∆Ct法计算LaCHS基因相对表达量[18−19]。
-
以不同品种薰衣草花器官6个发育时期花蕾及盛开期(S5)茎、苞叶和叶片为材料,每个样品设3个生物学重复。分别称取0.3 g鲜样于适量液氮中研磨,后转移至有3 mL花青素提取液(体积分数为1%盐酸-甲醇溶液)的10 mL离心管中,4 ℃静置12~14 h,4 000 r·min−1离心10 min,利用酶标仪分别测定上清液在530和657 nm处的吸光度D(530)、D(657),根据公式[D(530)-0.25×D(657)]/WF计算样品中花青素的相对含量,其中WF为样品鲜质量[20]。
-
提取测序正确的pEASY®-T5-LaCHS阳性克隆大肠埃希菌的质粒,并进行EcoRⅠ和BamH Ⅰ双酶切线性化,回收LaCHS基因片段。以LaCHS基因回收片段为模板,pCAMBIA3301-LaCHS F/R为特异性引物(表1)进行PCR扩增,将纯化后的LaCHS基因重组到EcoR Ⅰ和BamHⅠ线性化后的pCAMBIA3301过表达载体上,通过热激法转化农杆菌Agrobacterium tumefaciens GV3101感受态细胞,获得pCAMBIA3301-LaCHS表达载体。利用农杆菌介导的叶盘转化法转化本氏烟草[21]。
-
以上述LaCHS基因回收片段为模板,pTRV2-LaCHS F/R为特异性引物(表1)进行PCR扩增,将纯化后的LaCHS基因重组到EcoR Ⅰ和BamH Ⅰ线性化pTRV2载体上,获得pTRV2-LaCHS基因沉默载体。利用热激法转化农杆菌感受态细胞GV3101。以pEASY®-T5-LaCHS载体为模板构建基因沉默载体,热激法转化农杆菌,并使用重悬液调整菌体D(600)至1.3~1.5。将携带pTRV1的菌悬液分别与pTRV2重组质粒菌悬液按体积比为1∶1混合,混合后置于室温避光条件下静置4~5 h以备后续侵染试验使用。选在人工培养室培养5~7周生长健壮且无杂乱分支的‘莱文丝深紫’薰衣草植株幼苗中部4~6片较大的叶片进行注射。注射后避光培养24 h,期间断水3 d。以注射15 d后新长出的嫩芽提取的RNA反转录的cDNA为模板,选择Actin为内参基因进行RT-qPCR分析[4, 22]。
-
组织特异性表达分析以各品种花器官不同发育时期和盛花期不同营养器官中的最低表达量为对照,异源过表达及基因沉默试验以野生型(WT)为对照,采用2-∆∆Ct法计算LaCHS基因的相对表达量[4, 18−22]。花青素相对含量计算方法见1.2.5。数据使用Excel 2021处理,使用SPSS 26.0进行单因素显著性分析,使用GraphPad Prism 10.1.2分析作图。
-
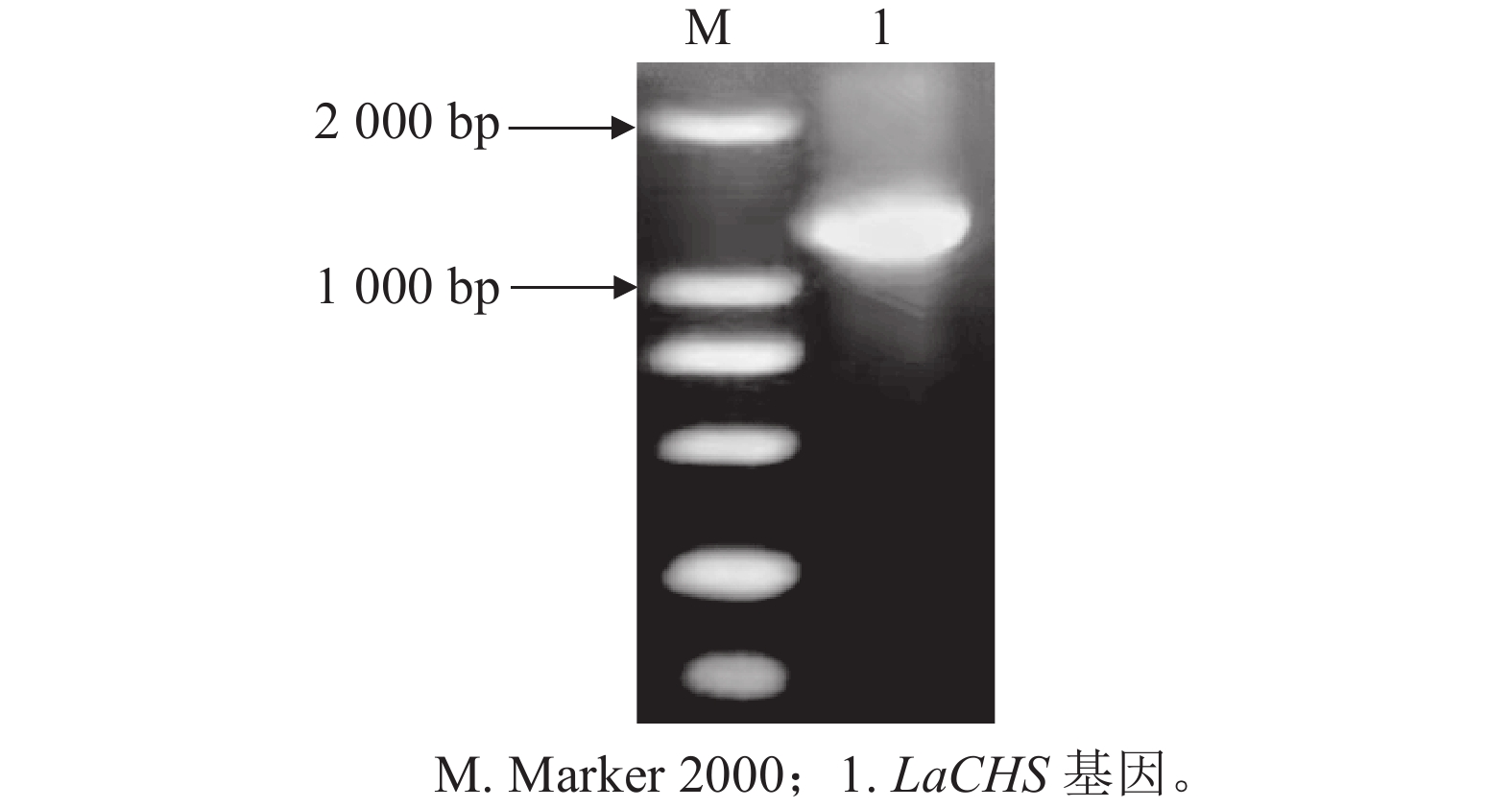
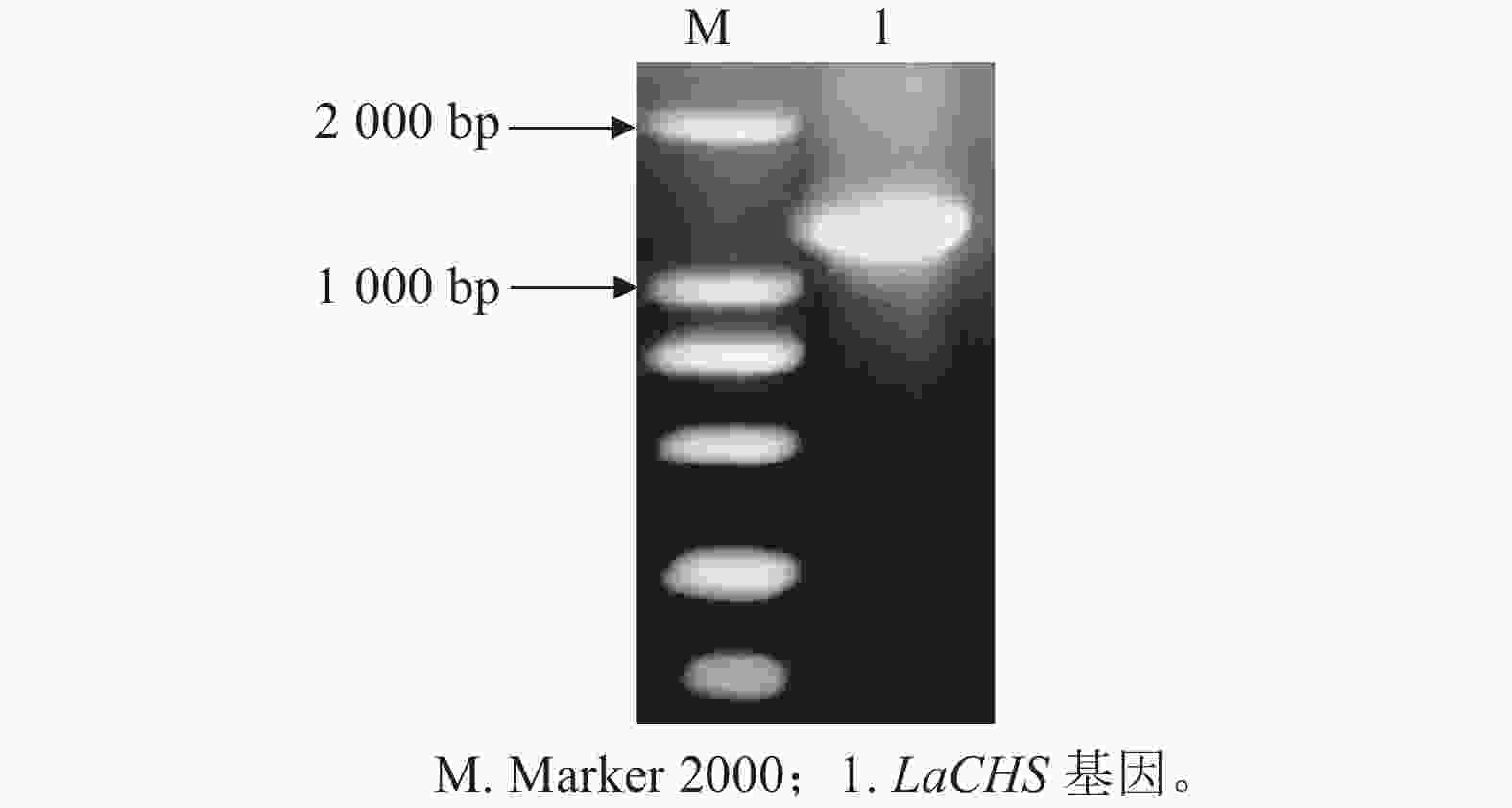
以‘莱文丝深紫’叶片cDNA为模板,通过特异引物LaCHS-R/F进行PCR扩增,获得1条大于1 000 bp大小的条带,与目标条带大小相似(图2)。将回收纯化PCR产物连接到pEASY®-T5载体上,对阳性单克隆菌株进行测序分析。目的片段长度为1 173 bp,编码390个氨基酸,命名为LaCHS基因。
-
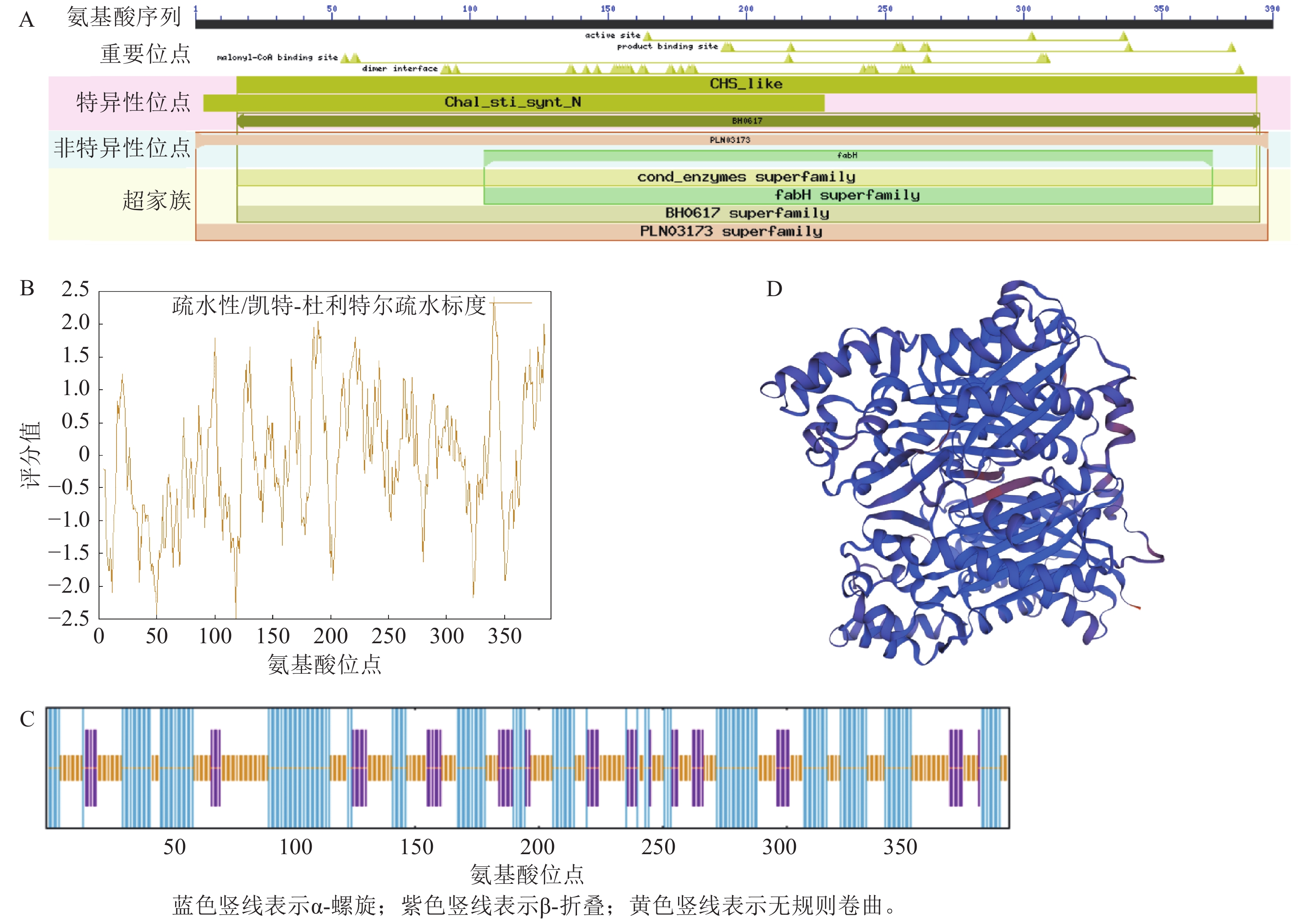
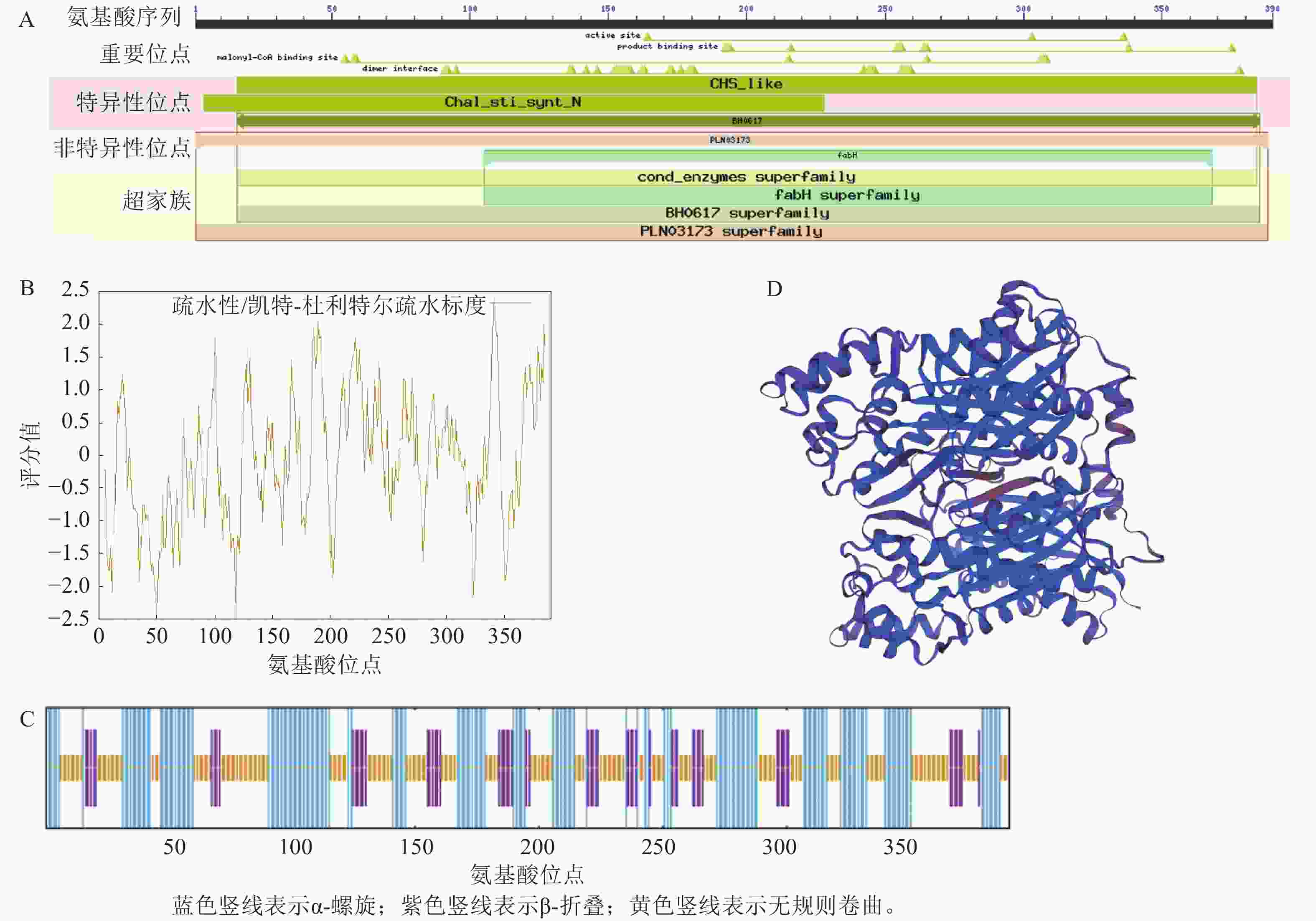
LaCHS蛋白拥有CHS_like区域以及PLN03173和fabH等3个保守结构域,属于查尔酮合酶超级家族(图3A)。LaCHS蛋白分子式为C1886H
3013 N511O563S19,相对分子量为42.46 kDa,理论等电点为6.09,脂肪系数为90.51,不稳定系数为39.27,属于稳定蛋白。LaCHS蛋白平均亲水指数为−0.058,最大疏水分值(−2.489)位于肽链第118位氨基酸,最大亲水分值(2.422)位于肽链第341位氨基酸,属于亲水性蛋白(图3B)。对LaCHS蛋白的二级结构进行预测:发现该蛋白存在α-螺旋、β-折叠、延伸链和无规则卷曲4种结构,其中以α-螺旋和无规则卷曲占比最大,分别占40.51%和44.10%,而延伸链占15.38%(图3C)。以水稻Oryza sativa蛋白(登录号:4yjy.1.A)为模板,对LaCHS蛋白的三级结构进行建模,建模的三级结构覆盖率为92.00%,序列一致性为83.80%。该结果与二级结构预测结果一致(图3D)。 -
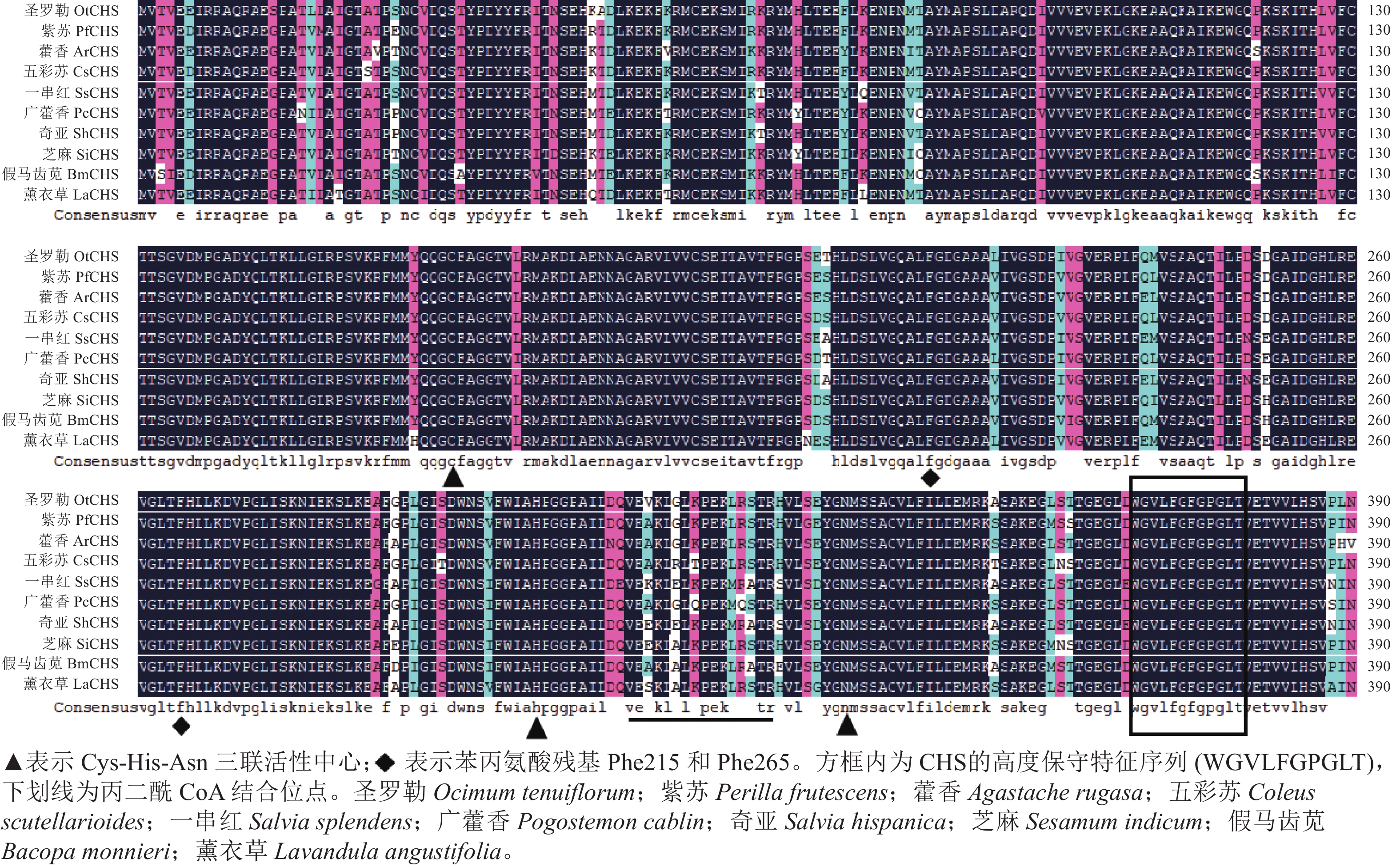
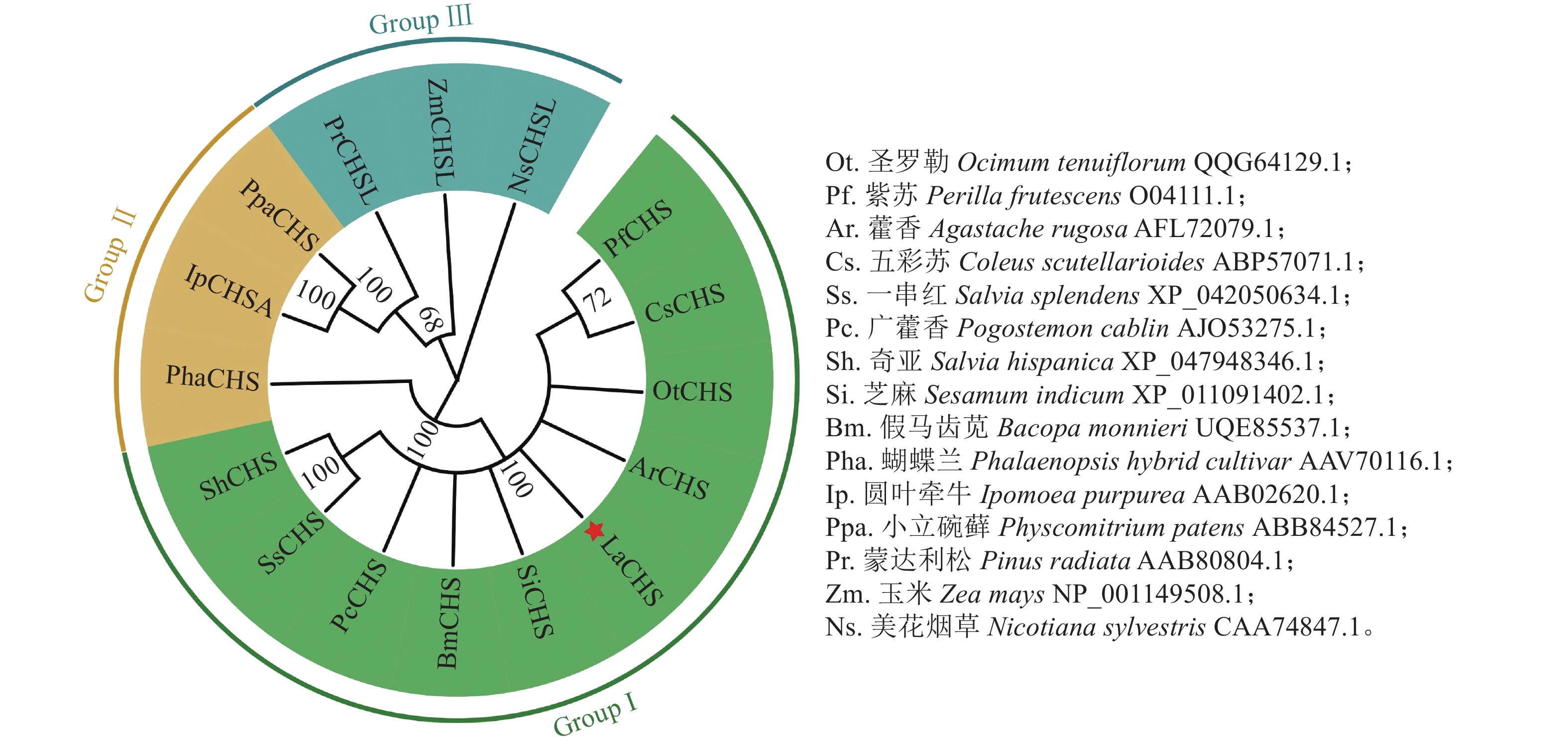
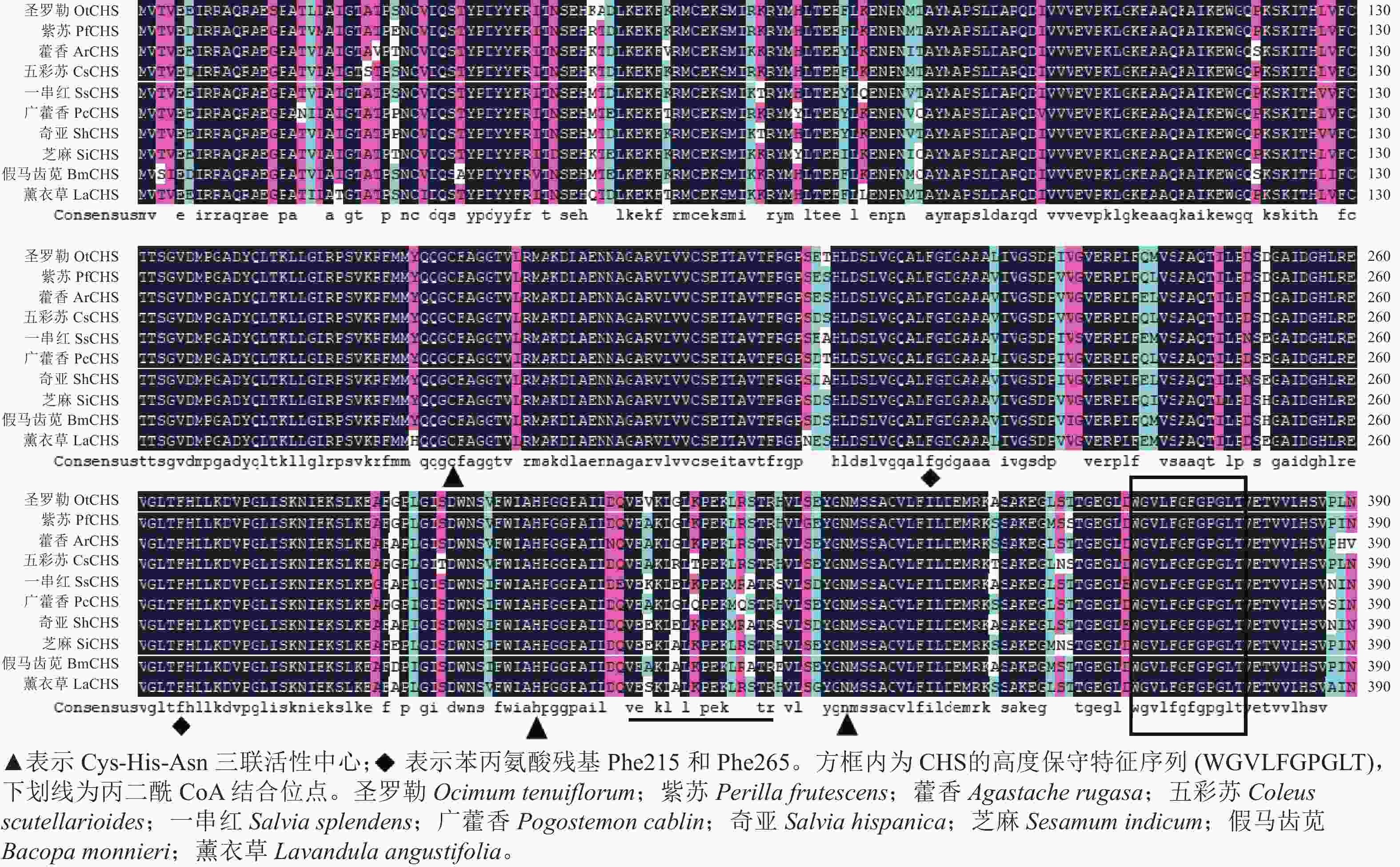
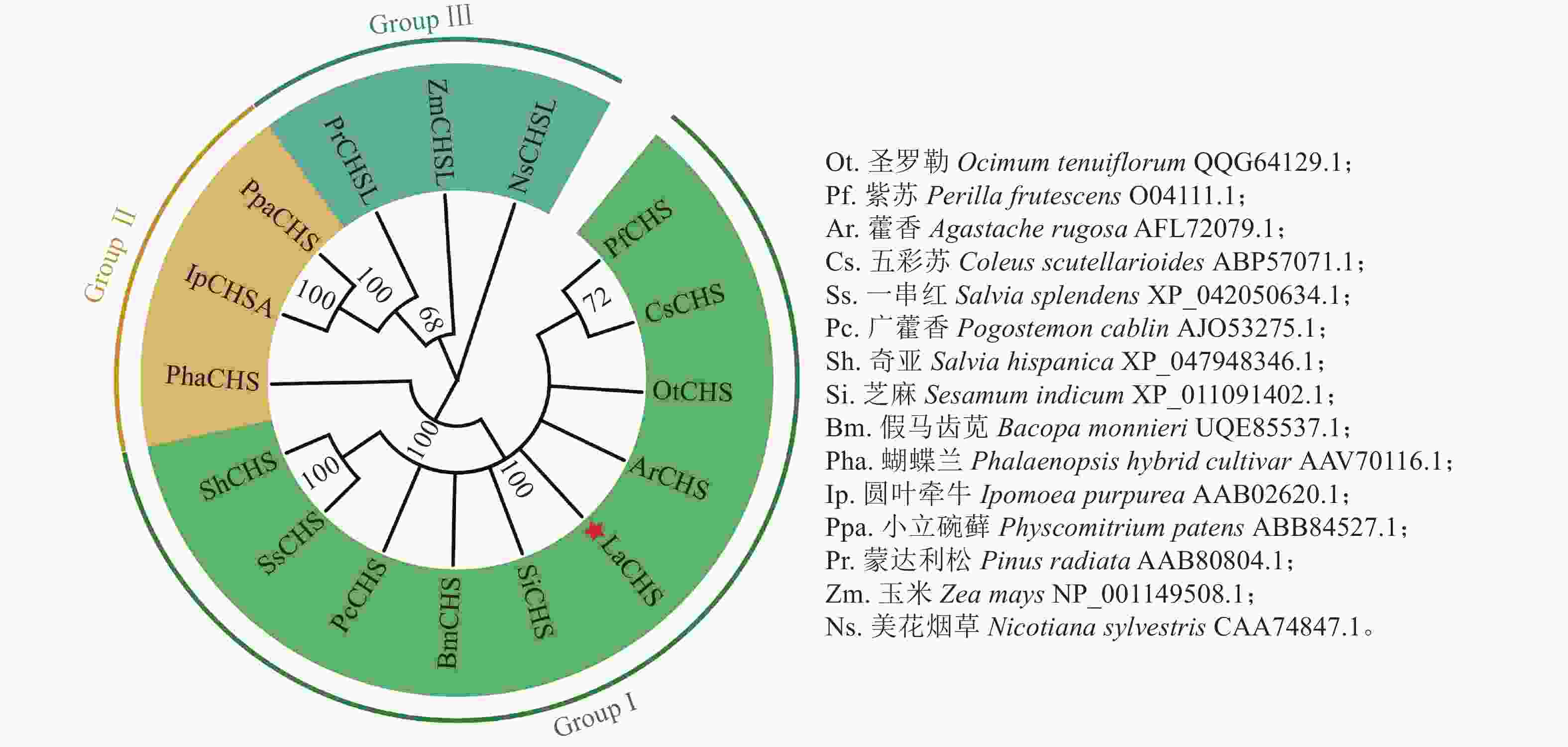
氨基酸多序列比对分析发现:薰衣草LaCHS蛋白具有典型的关键催化三联体(Cys-His-Asn)以及高度保守的CHS特征序列(WGVLFGPGLT)。LaCHS蛋白与圣罗勒、紫苏和藿香Agastache rugosa、五彩苏Coleus scutellarioides的CHS蛋白相似性超过93%(图4)。系统进化树分析结果表明:LaCHS蛋白与圣罗勒(登录号:QQG64129.1)亲缘关系较近,都属于Group Ⅰ型蛋白(图5),该类型CHS蛋白在植物次生代谢催化丙二酰CoA和4-香豆酰CoA生成查尔酮中起重要作用,同科属的CHS蛋白处于同一分支,说明薰衣草LaCHS蛋白在生物进化过程中较为保守。
-
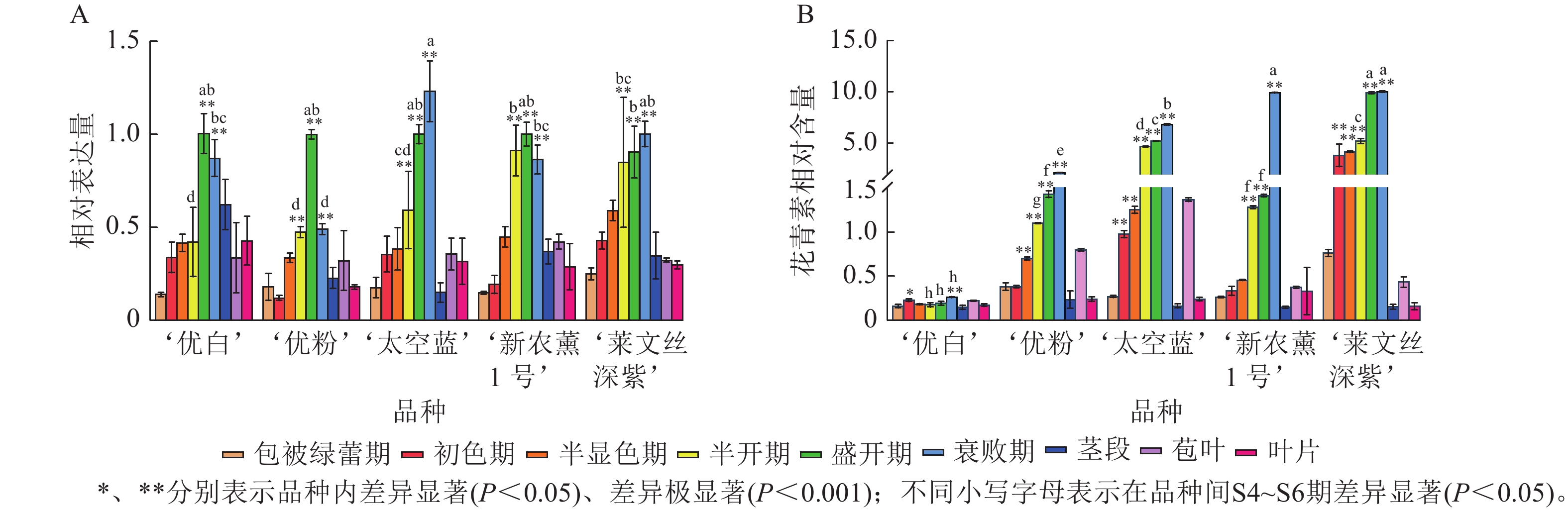
利用RT-qPCR分析薰衣草LaCHS基因在不同品种、花器官不同发育时期及盛开期(S5)不同营养器官中的表达水平。结果表明:LaCHS基因在薰衣草不同品种、不同营养器官间均有表达,且品种内盛开期(S5)的表达水平显著高于包被绿蕾期(S1);品种间,在营养器官中,茎、苞叶和叶片的表达水平与花器官存在显著差异(P<0.05),具有明显的组织特异性。LaCHS基因的相对表达量与花青素相对含量基本随花发育时期而逐渐增加,其中LaCHS基因的表达水平在‘优白’‘优粉’和‘新农薰1号’盛开期(S5)达到最高(图6A),在‘太空蓝’和‘莱文丝深紫’衰败期(S6)达到最高,而5个品种的花青素相对含量均于衰败期(S6)达到峰值(图6B);在盛开期(S5)不同营养器官中,‘优粉’‘新农薰1号’‘太空蓝’在苞叶中的LaCHS基因相对表达量最高,而‘优白’‘莱文丝深紫’在茎中的表达量最高,而5个品种的花青素相对含量在苞叶中均为最高,其次是叶和茎(图6B)。除‘优白’外,其余品种中LaCHS基因的表达变化水平与花青素积累规律呈极显著正相关(P<0.01,表2)。
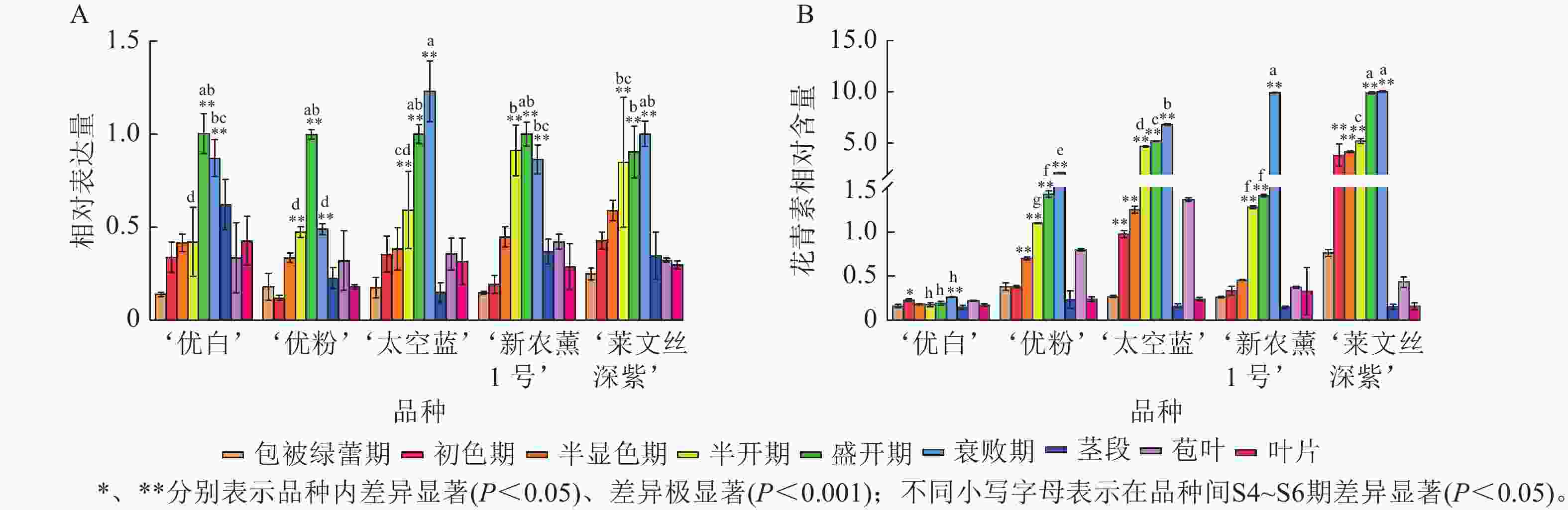
Figure 6. Spatiotemporal expression of the LaCHS gene and relative anthocyanin content analysis in L. angustifolia
项目 ‘优白’ ‘优粉’ ‘太空蓝’ ‘新农薰1号’ ‘莱文丝深紫’ LaCHS 花青素 LaCHS 花青素 LaCHS 花青素 LaCHS 花青素 LaCHS 花青素 LaCHS 1 0.201 1 0.690** 1 0.924** 1 0.511** 1 0.873** 花青素 0.201 1 0.690** 1 0.924** 1 0.511** 1 0.873** 1 说明:** 表示极显著相关(P<0.01)。 Table 2. Correlation of LaCHS expression dynamics and anthocyanin accumulation patterns in L. angustifolia
-
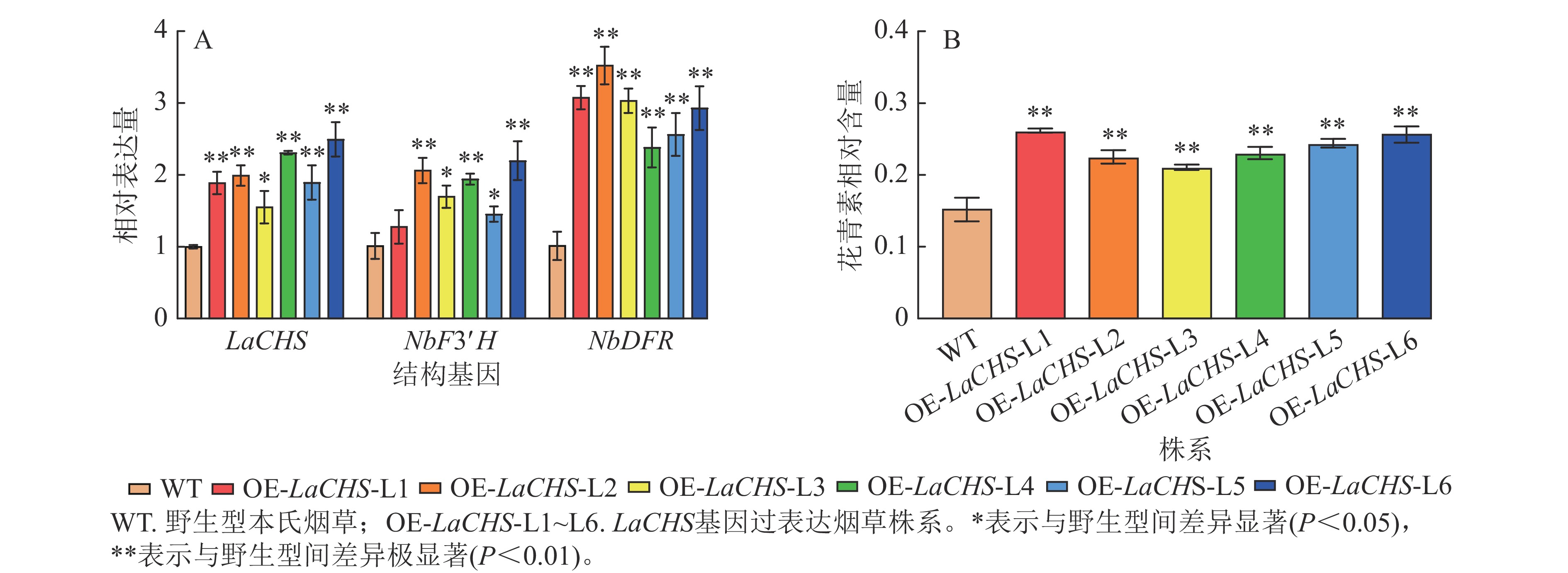
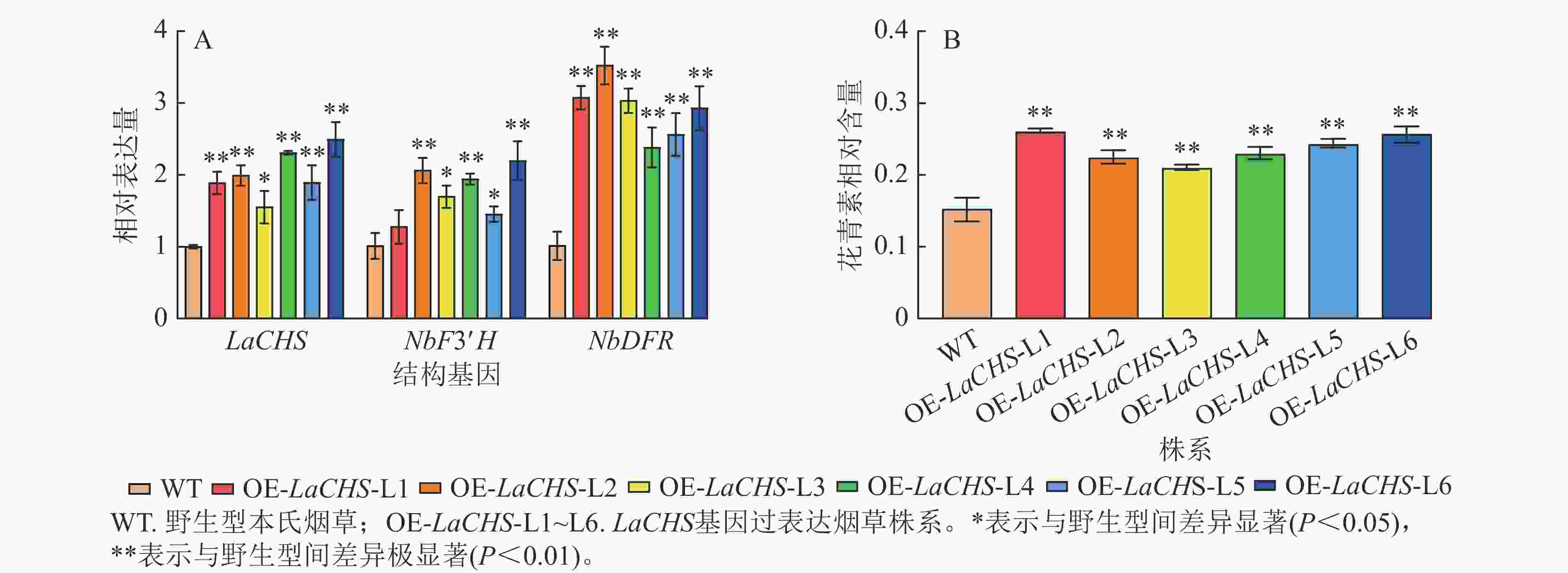
利用农杆菌介导的叶盘法转化本氏烟草共获得22株抗性植株,提取转基因烟草植株叶片DNA并进行目的基因PCR检测,获得6株阳性植株(L1~L6),阳性率为27%。以烟草叶片cDNA为模板,检测烟草内参基因、LaCHS基因及烟草中与花青素生物合成关键结构基因NbF3′H和NbDFR的表达水平。相较于野生型,转基因各株系中LaCHS基因的表达水平上调了0.55~1.49倍,结构基因NbF3′H和NbDFR的表达水平分别上调了0.27~1.19和1.38~2.52倍(图7A)。此外,转基因株系L1~L6叶片中花青素相对含量分别极显著提高了0.72、0.48、0.39、0.52、0.61和0.69倍(图7B,P<0.01)。说明LaCHS能促进花青素合成关键结构基因NbF3′H与NbDFR的表达,可能与花青素合成通路中的其他基因共同调控花青素的合成。
-
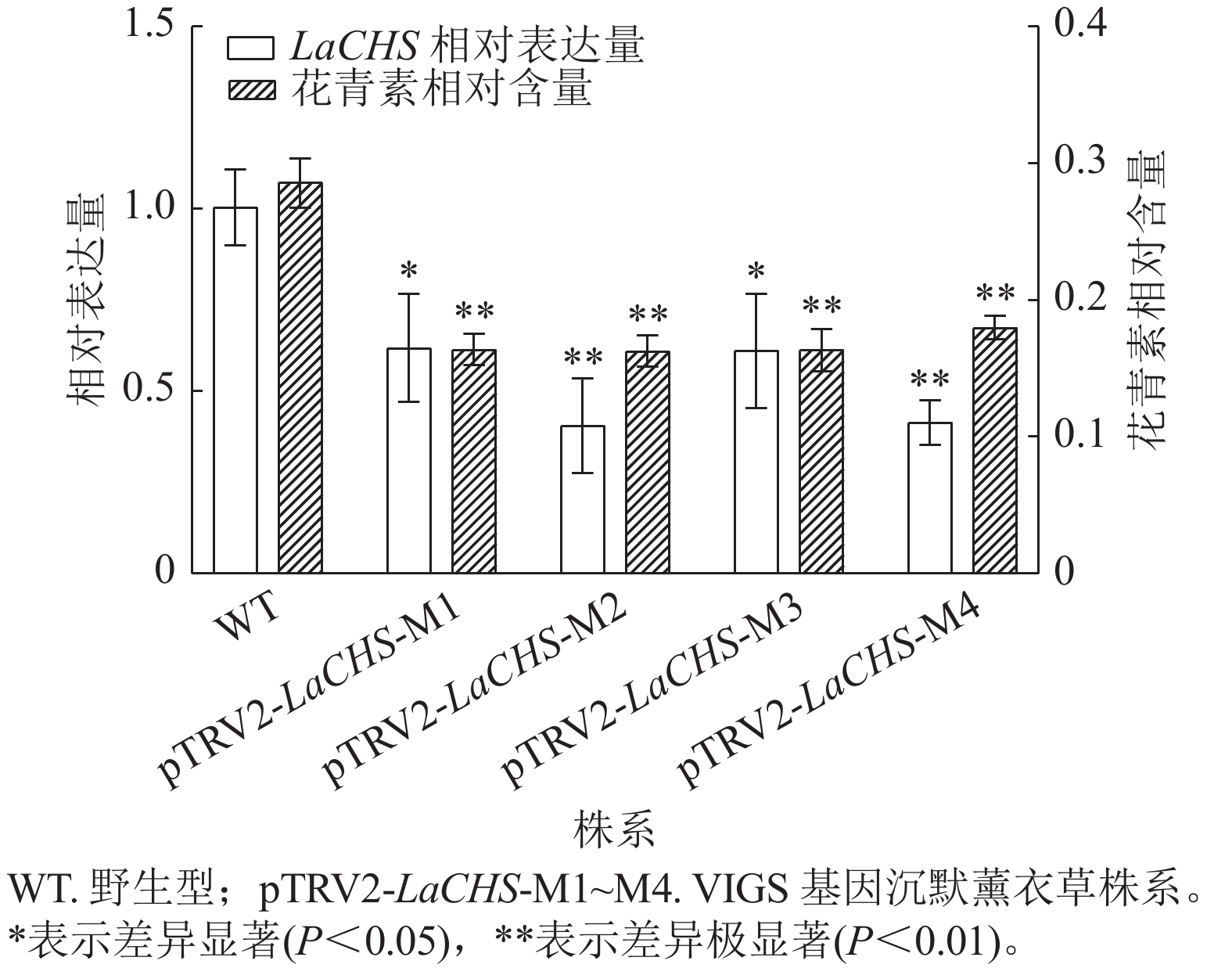
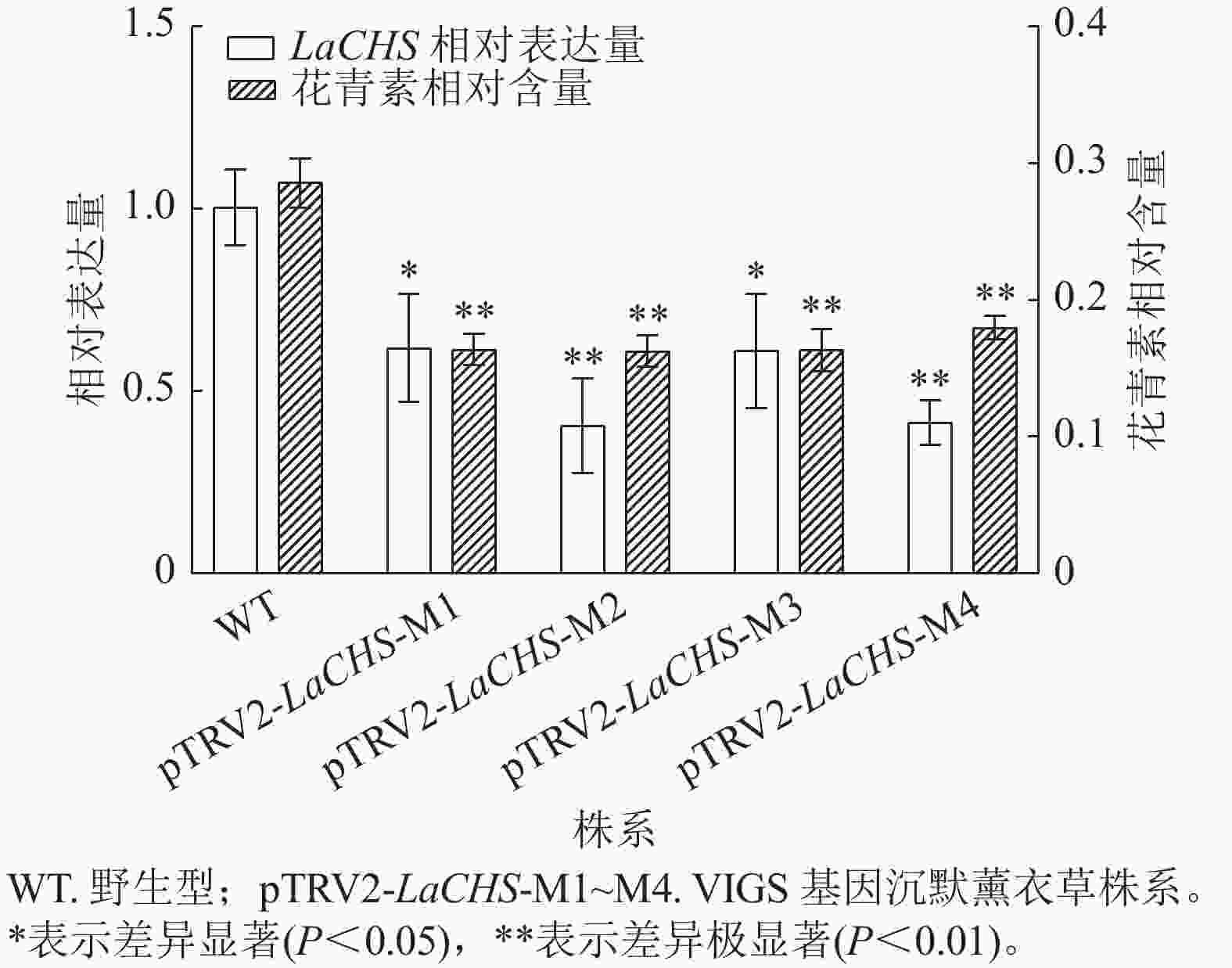
利用pTRV2-LaCHS F/R特异性引物,通过RT-qPCR检测经基因沉默处理后薰衣草新长出的嫩芽中LaCHS基因的表达水平。相较于野生型,薰衣草LaCHS基因沉默株系(M1~M4) 中,LaCHS基因的表达水平显著(P<0.05)或极显著(P<0.01)下降,分别下降了0.38、0.60、0.39和0.59倍(图8),说明所有基因沉默株系中目的基因均被有效沉默。为进一步验证LaCHS基因对花青素合成的调控作用,分别测定薰衣草LaCHS基因沉默各株系中花青素的相对含量。结果显示:相较于野生型,基因沉默各株系中花青素相对含量均极显著下降(P<0.01),分别降低了0.43、0.43、0.43和0.37倍(图8)。结果表明:沉默LaCHS基因可降低薰衣草花青素相对含量,证明LaCHS基因可调节花青素的合成。
-
花青素与植物花色形成密切相关。类黄酮合成途径中的关键酶基因CHS的表达水平变化可影响植物花色呈现[8−10, 23]。研究表明:广藿香Pogostemon cablin的PcCHS蛋白可催化丙二酰CoA和4-香豆酰CoA生成查尔酮[24]。位于Group Ⅰ亚家族的薰衣草LaCHS蛋白拥有CHS_like区域以及PLN03173和fabH等3个保守结构域,属于查尔酮合酶超级家族,且具有WGVLFGPGLT和关键催化三联体(Cys-His-Asn)高度保守的CHS特征序列。LaCHS蛋白与同科圣罗勒、广藿香、紫苏等9种物种的CHS蛋白高度同源且属于Group Ⅰ亚族,因此,推测LaCHS蛋白也可通过催化丙二酰CoA和4-香豆酰CoA而生成查尔酮。
花青素生物合成的调控机制是一个多因子协同作用的复杂网络,主要涉及转录因子(MYB、bHLH等)调控、代谢途径结构基因互作、植物激素(赤霉素、乙烯等)信号转导及环境(光照、温度及机械损伤等)胁迫响应等4个层面[25]。据报道,紫色马铃薯Solanum tuberosum块茎中StCHS4基因与StCHS5基因表达水平显著高于白色和黄色马铃薯,且与马铃薯块茎中花青素的积累呈正相关[26];在丁香Syringa oblata中,花和其他组织中SoCHS1基因的表达水平与花发育过程中花青素相对含量显著正相关[27];在藤茶Ampelopsis grossedentata中,AgCHS1基因的表达水平呈先升后降的趋势,总黄酮含量变化趋势与AgCHS1基因的表达水平变化趋势基本一致[28];在杜鹃花Rhododendron simsii 中,花青素在开花期总体呈增加趋势且在盛花期达到高峰,与RhCHS基因表达水平的变化规律一致[29]。在本研究中,随着花的发育,‘优粉’‘太空蓝’‘新农薰1号’及‘莱文丝深紫’等品种的LaCHS基因表达量与花青素相对含量呈正相关,表现为先增加后降低的趋势,在盛开期或衰败期达到最高。这与上述研究的结果相同。但本研究发现:在白花‘优白’中,LaCHS基因表达量随着花的发展进程逐渐上调,但花青素相对含量始终保持在同一水平。这可能是LaCHS基因与其他调节因子及结构基因互作共同调控,最终生成白色的类黄酮物质并积累,进而使花色呈现为白色[30]。
过表达黄牡丹Paeonia delavayi var. lutea PdCHS转基因烟草品系中大部分花青素成分呈现出不同程度上调[31]。紫苏中,CHS基因的高表达则促进其叶片中黄酮类化合物的积累[32]。对石竹Dianthus chinensis DchCHSs基因、茄子Solanum melongena SmCHS基因进行沉默后,其沉默株系中花青素相对含量、DchCHSs基因表达量及SmCHS基因表达量均显著降低[33−34]。同理,对大白菜Brassica rapa var. glabra BrCHS4基因进行沉默后,抑制了其BrCHS4基因的表达,同时降低了叶片中类黄酮和花青素的相对含量[35]。在本研究中,烟草过表达LaCHS基因株系中,LaCHS基因和与烟草花青素合成相关的NbF3′H与NbDFR结构基因的表达水平均显著上调,花青素的相对含量也同步增加。此外,薰衣草基因沉默株系中,LaCHS基因的表达水平与花青素相对含量均显著降低。推测薰衣草LaCHS基因作为其花青素合成的正向调节基因,与其他结构基因共同调控花青素的合成。这一结果可为LaCHS基因调控薰衣草花青素物质代谢合成分子机制的阐释提供理论参考。
-
本研究在薰衣草中克隆了LaCHS基因。该基因具有典型的关键催化三联体(Cys-His-Asn)和高度保守的CHS特征序列(WGVLFGPGLT),与圣罗勒亲缘关系较近。LaCHS基因具有明显的组织特异性,并随着花的发育呈逐渐升高的趋势,且与花青素的积累规律基本一致。LaCHS基因能促进结构基因NbF3′H和NbDFR的表达,进而促进花青素的积累。本研究探究了薰衣草LaCHS基因的功能,为进一步解析该基因在薰衣草花青素生物合成中的分子机制提供参考,并为薰衣草新品种选育提供理论基础。
Cloning and functional analysis of LaCHS gene related to anthocyanin biosynthesis in Lavandula angustifolia
doi: 10.11833/j.issn.2095-0756.20250283
- Received Date: 2025-05-09
- Accepted Date: 2025-10-02
- Rev Recd Date: 2025-09-26
- Publish Date: 2025-10-20
-
Key words:
- Lavandula angustifolia /
- chalcone synthase /
- gene cloning /
- functional verification /
- anthocyanins
Abstract:
| Citation: | LI Pengfei, WANG Aifan, HUANG Jiahui, et al. Cloning and functional analysis of LaCHS gene related to anthocyanin biosynthesis in Lavandula angustifolia[J]. Journal of Zhejiang A&F University, 2025, 42(5): 1048−1058 doi: 10.11833/j.issn.2095-0756.20250283 |









 DownLoad:
DownLoad: