-
桂花Osmanthus fragrans是中国十大传统名花之一,深受人们喜爱。不同学者已经从桂花的花芽分化[1-3]、开花机制[4-5]、花色呈现[6-7]、花香释放[8-9]等不同层面展开了研究。随着分子水平研究的不断深入,利用实时荧光定量聚合酶链式反应(qRT-PCR)分析基因的表达已成为研究这些过程机理的重要手段,而筛选得到适合不同研究的内参基因格外重要。研究认为[10-11],内参基因并不存在通用性,不同处理、不同组织中内参基因的表达水平都会发生改变[12];选用一种内参基因无法准确反映不同植物组织、同一组织不同处理下的基因表达水平[13-14]。因此,根据具体的实验材料和处理条件筛选合适的内参基因十分必要[15]。光周期和温度处理能对桂花的基因表达水平、花芽分化进程等产生影响。研究表明[4, 16]:长日照和相对低温都能够加快桂花花芽分化进程,而短日照条件下桂花的花芽分化相对较慢,低温或高温都会抑制桂花的花芽分化。本研究在不同光周期和温度条件下对桂花常用的7个候选内参基因进行比较分析,筛选最佳内参基因,为后期研究光周期和温度影响桂花开花的机理奠定基础。
-
实验材料为多年生‘佛顶珠’O. fragrans‘Foding Zhu’盆栽植株,种植于浙江农林大学桂花资源圃。设置4个处理:长日照高温(光照/黑暗=14 h/10 h,26 ℃),长日照低温(光照/黑暗=14 h/10 h,19 ℃),短日照高温(光照/黑暗=10 h/14 h,26 ℃)和短日照低温(光照/黑暗=10 h /14 h,19 ℃),光照强度为100%,相对湿度为60%~80%。实验材料在上述环境中培养40 d。处理结束后从8:00起,每隔4 h取当年生枝条的第2片或第3片嫩叶,液氮速冻,放于-80 ℃保存。
-
每样品取50~100 mg,充分研磨后按照RNAprep pure Plant Kit(天根,北京)说明书步骤提取各样品RNA。通过紫外分光光度计和质量浓度为1%的琼脂糖凝胶电泳检测所提RNA浓度和品质。以所提样品RNA为模板,按照Reverse Transcriptase M-MLV(Takara,大连)说明书合成cDNA第1条链,合成后于-20 ℃储存备用。
-
从桂花转录组数据库中选择肌动蛋白基因(OfACT)、延伸因子1α蛋白基因(OfEF1α)、NADP-异柠檬酸脱氢酶基因(OfIDH)、GTP结合蛋白RNA1基因(OfRNA1)、β微管蛋白基因(OfTUB)、泛素结合酶E2基因(OfUBC2)和18S核糖体RNA基因(Of18S)[12, 17]等7个使用较多的内参基因作为候选内参基因。采用SYBR green Ⅱ荧光染料嵌合法进行内参基因的筛选,设计候选内参基因的引物,通过Bioxman和Primer Premier 5软件对引物进行校验,并利用Primer-BLAST进行检测以确认引物序列的特异性。选用的引物序列由上海捷瑞生物工程有限公司合成(表 1)。
表 1 qRT-PCR相关内参基因引物信息
Table 1. List of primers for qRT-PCR
基因简称 引物序列(5′→3′) 退火温度/ ℃ OfACT CCCAAGGCAAACAGAGAAAAAAT 55.0 ACCCCATCACCAGAATCAAGAA OfEF1α CGTTTGCCACTTCAGGATGTCTA 58.0 GTACCAGGTTTCAGGACTCCAGTTT OfIDH CTTGAAGCAGATGTGGAAGAGTC 57.5 CTTTGTCCATCCTGGGACCAGTC OfRAN1 AGAACCGACAGGTGAAGGCAA 57.5 TGGCAAGGTACAGAAAGGGCT OfTUB AGAAGGGATGGATGGAATGGA 56.0 GTCTTCTTCGTCCTCGGCAGT OfUBC2 TGTTGACAAAACCGATGGAAGGA 57.5 GTGGAGTGTGGAGGATAAGGGTG Of18S AGCCTGAGAAACGGCTACCAC 55.0 ATACGCTATTGGAGCTGGAA -
qRT-PCR实验方法参考SYBR® Premix Ex TaqTMⅡ(Tli RNaseH Plus)(Takara,大连)试剂盒。采用20.0 μL的反应体系:其中双蒸水6.4 μL,上下游引物各0.8 μL,cDNA模板2.0 μL,SYBRⅡ荧光染料10.0 μL。反应在ABI 7300实时PCR仪中进行,反应程序为:95 ℃下预变性30 s;95 ℃下反应5 s,60 ℃反应31 s,共40个循环;95 ℃下反应15 s,60 ℃下反应1 min,95 ℃下反应30 s,60 ℃下反应15 s[17]。3次生物学重复。
-
用geNorm,NormFinder和BestKeeper软件分别对qRT-PCR中所得到样品的反应循环数(Ct值)进行分析,比较7个候选内参基因表达的稳定性,并选择最佳内参基因。geNorm软件以各候选内参基因在样品中的表达水平为依据,计算各候选基因的表达稳定性(M),M值越小越稳定;M<1.5时认为是理想内参[18-19]。geNorm软件还可以根据候选内参基因标准化因子的配对差异性分析(Vn /Vn+1)得出最佳内参基因的个数;其中V表示配对差异值,n表示内参基因个数。软件默认的(Vn /Vn+1)值为0.15,当Vn /Vn+1<0.15时,表明引入的n个内参基因已经稳定,不需要引入(n+1)内参基因,当Vn /Vn+1>0.15时,则需要引入(n+1)个内参基因[17, 20]。因geNorm软件最终挑选出2个或2个以上的内参基因组合来校正数据,所以n≥2。为了了解引用1个内参基因的稳定性,需要通过NormFinder和BestKeeper软件做进一步分析。NormFinder软件对内参基因的筛选是基于方差分析的结果,利用ΔCt法计算候选内参基因表达的稳定值(S),从而评估候选内参基因的表达稳定性;S越小,基因越稳定[21]。BestKeeper软件通过计算标准偏差(SD)对候选内参基因的表达稳定性进行评价和排序,SD越小,则该基因的稳定性越好;若SD>1,则认为该基因不稳定[20-21]。最后通过昼夜节律基因GI的相对表达水平,对选择的最佳内参基因进行验证。
-
提取不同处理桂花样品的总RNA,经紫外分光光度计检测,所有样品总RNA的吸光度值[D(260)/D(280)和D(260)/D(230)]均为1.8~2.1。将所有样品的cDNA等量混合后作为模板,进行普通PCR和定量PCR检测。为检测引物的扩增效率,设置6个cDNA浓度梯度(原液,5-1,5-2,5-3,5-4,5-5原液),根据qRT-PCR结果,利用公式E=(10-1/s-1)×100%得出基因的扩增效率(表 2),其中:E(%)表示扩增效率,s表示曲线斜率。经计算,扩增效率为95%~110%,相关系数为0.993~0.998,符合qRT-PCR的基本要求。
表 2 7个候选内参基因扩增参数
Table 2. Amplification parameters of seven candidate reference genes
基因 Ct 扩增效率/% 曲线斜率 线性相关系数 条带长度/bp OfACT 22.77 ± 0.55 103.66 -3.237 3 0.995 5 143 OfEF1α 20.61 ± 0.42 103.19 -3.247 8 0.995 7 89 OfIDH 23.59 ± 0.39 95.78 -3.453 8 0.997 7 118 OfRNA1 21.68 ± 0.07 95.43 -3.436 5 0.996 2 117 OfTUB 28.44 ± 0.40 107.49 -3.154 6 0.993 2 106 OfUBC2 25.73 ± 0.54 107.15 -3.161 8 0.994 5 75 Of18S 8.99 ± 0.55 108.15 -3.140 9 0.996 8 208 -
根据扩增效率,将各样品的原始cDNA稀释50倍后作为模板进行qRT-PCR。通过比较循环数(Ct)来评估各候选内参基因在所有样品中的转录水平(表 2),候选内参基因平均Ct为8.99~28.44,其中Of18S的Ct最小,说明平均表达水平最高;OfTUB最大,说明平均表达水平最低;其他候选内参基因平均表达水平差异则相对较小。
-
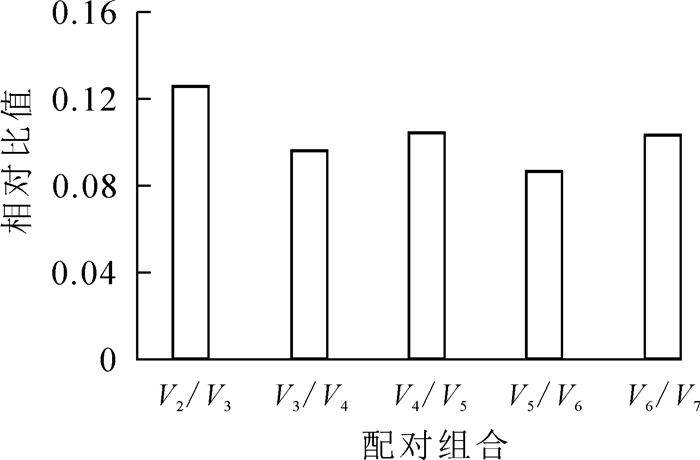
geNorm软件分析发现(表 3),所有候选内参基因的稳定值均小于1.5,符合内参基因筛选的标准;其中OfRAN1和OfIDH的稳定值相等且最小(M=0.347),在不同样品中表达最为稳定,其次是OfACT基因(M=0.393),Of18S稳定值最大(M=0.601),说明Of18S表达稳定性最差。对候选内参基因的配对差异值测算可知(图 1):V2/3=0.126<0.15,表明引入2个内参基因已经稳定,无需引入更多的内参基因。geNorm软件分析结果表明,最佳候选内参基因为OfRAN1和OfIDH。
表 3 不同软件分析7个候选内参基因表达稳定性
Table 3. Gene expression stability of seven reference genes by different software
基因名称 geNorm分析 NormFinder分析 BestKeeper分析 稳定值 排序 稳定值 排序 稳定值 排序 OfACT 0.393 2 0.232 3 0.288 3 OfEF1α 0.524 5 0.390 6 0.497 6 OfIDH 0.347 1 0.206 2 0.133 1 OfRNA1 0.347 1 0.135 1 0.180 2 OfTUB 0.479 4 0.361 5 0.372 5 OfUBC2 0.422 3 0.343 4 0.346 4 Of18S 0.601 6 0.474 7 0.611 7 
图 1 geNorm软件分析候选内参基因的配对差异值
Figure 1. Pairwise variationof candidate reference genes as calculated by geNorm
NormFinder软件分析发现,OfRAN1基因的表达稳定性最高,其次是OfIDH,Of18S基因表达稳定最差,与geNorm软件分析的结果一致(表 3)。BestKeeper软件的分析结果显示(表 3),在7个候选内参基因中,OfIDH基因的表达稳定性最高,其次是OfRAN1,Of18S基因的表达稳定性最差。分析结果尽管与geNorm和NormFinder软件的结果有些差异,但最佳候选内参基因结果一致。
综合3个软件的分析结果可以得出,不同处理条件下,OfRAN1和OfIDH的表达稳定性均为最高,可作为最佳内参基因,而Of18S是表达稳定性最差的基因。
-
GIGANTEA基因(GI)是光周期途径的重要调控基因,其表达呈现明显的昼夜节律。选用桂花昼夜节律相关基因GI对候选内参基因进行验证,可提高内参基因筛选的准确性,保证最佳内参基因对光周期途径基因表达水平的校准作用。选择OfRAN1/OfIDH,OfRAN1,OfIDH以及其他候选内参基因对GI基因的相对表达水平进行校准(图 2),结果显示:相比其他内参基因,不同处理下OfRAN1/OfIDH校准的GI基因的表达模式表现为光照条件下积累,黑暗条件下降低的周期性表达,符合GI基因的昼夜表达规律。说明3个内参基因筛选软件对候选内参基因筛选的结果是正确的,即OfRAN1和OfIDH基因可作为桂花不同光周期和温度处理的最佳内参基因。
-
荧光定量PCR因其众多优点被广泛运用于基因表达的研究中,成为量化基因转录表达,揭示基因表达规律的有效方法[22-23];但引用适当的内参基因对qRT-PCR的结果进行校准是非常必要的[20]。理想内参基因在不同组织、不同发育阶段、不同生长条件下均能稳定表达[17, 24],但事实上并没有哪一个基因能够满足所有条件[25]。因此,根据不同实验材料和处理条件,筛选出合适的、特定的内参基因是确保实验成功的必要条件。
研究发现[26]:选用2个或更多内参基因组合可以提高内参基因校准的精确度,弱化单一内参基因带来的不准确性。RAN(ras-related nuclear protein)是小G蛋白的一类,在细胞生理进化中起到重要的作用,它的很多同源基因是不同光周期、温度条件下常用的内参基因[12, 17]。IDH的同源基因被证实为不同温度条件下桉树Eucalyptus robusta的最佳内参基因[27];ACT和18S同源基因分别可作为不同光周期[28]、温度条件[29]下大豆Glycine max的内参基因;EF1a和TUB基因可作为不同光周期、温度条件下玉米Zea mays的内参基因[30];UBC2的同源基因可作为不同温度条件下欧芹Petroselinum crispum的内参基因[31]。本研究选用了桂花中常用的7个候选内参基因,对其在不同光周期和温度处理下嫩叶中的表达稳定性进行研究;利用geNorm,NormFinder和BestKeeper等3个常用于内参基因筛选的软件进行评估,发现这7个候选内参基因在所有样品中表达稳定性的排序基本相同,均显示OfRAN1和OfIDH基因是最稳定的2个内参基因,将两者进行组合可以准确的校准不同处理下桂花叶片中目的基因的表达水平;而Of18S表达最不稳定,在所有实验材料中都存在较高的表达丰度,因而不适合作为本研究的内参基因。
光周期和温度途径在调控植物成花过程中发挥重要作用。筛选得到的桂花不同光周期和温度条件下的内参基因,能够为研究桂花在不同温度和光周期处理条件下的成花机制提供参考依据,为了解相关成花基因表达模式提供保障。
Screening reference genes of Osmanthus fragrans with differing photoperiod and temperature treatments
-
摘要: 光周期和温度处理能够影响桂花Osmanthus fragrans的基因表达水平和花芽分化过程,通过实时荧光定量聚合酶链式反应(qRT-PCR)分析基因表达已成为研究这些过程机制的重要手段,而筛选出适合光周期和温度处理的内参基因在研究桂花分子机制中尤为重要。为了得到不同光周期和温度处理下桂花中稳定的内参基因,以桂花品种‘佛顶珠’O.fragrans‘Foding Zhu’当年生枝条的第2或第3对幼叶为材料,以OfACT,OfEF1α,OfIDH,OfRAN1,OfTUB,OfUBC2和Of18S等7个基因作为候选内参基因,利用geNorm,NormFinder和BestKeeper 3个软件对候选内参基因的表达稳定性进行比较分析;以昼夜节律相关基因GIGANTEA(GI)对筛选出的最佳内参基因进行验证。结果表明:在不同处理条件下,桂花叶片中OfRAN1和OfIDH为最佳内参基因,Of18S的稳定性最差(geNorm分析结果显示OfRAN1和OfIDH的稳定值为0.347,而Of18S的稳定值为0.601;NormFinder显示OfRAN1和OfIDH的稳定值为0.135和0.206,而Of18S为0.474;BestKeeper显示OfRAN1的稳定值为0.80,OfIDH为0.133,而Of18S为0.474);GI基因的相对表达水平分析表明(昼夜节律基因GI的表达模式为在白天累积并在夜间下降),在7个内参基因及OfRAN1和OfIDH的基因组合中,使用OfRAN1和OfIDH的内参基因组合可获得GI基因更为精确的基因表达结果。综上所述,OfRAN1和OfIDH内参基因组合是桂花在不同光周期和温度处理下最佳内参基因组合。这为桂花分子机制研究奠定基础。Abstract: Photoperiod and temperature treatment can affect the gene expression level and flower bud differentiation process of Osmanthus fragrans. Analysis of gene expression by quantitative real-time polymerase chain reaction (qRT-PCR) has become an important means to study the mechanism of these processes and it is especially important to screen for an internal reference gene suitable for photoperiod and temperature treatment. To screen reference genes in O. fragrans using different photoperiod and temperature treatments, the young leaves of O. fragrans 'Foding Zhu' were used as materials. The O. fragrans treatment conditions were 14 h D (light)/10 h N (dark) +26℃, 14 h D/10 h N +19℃, 10 h D/14 h N +26℃ and 10 h D/14 h N +19℃, light intensity 100%, relative humidity 60%-80%. After treatment for 40 days, the second or third young leaves of the current shoots were taken every 4 h from 8:00 in the morning frozen in the liquid nitrogen and stored at -80℃. The seven genes, including OfACT, OfEF1α, OfIDH, OfRAN1, OfTUB, OfUBC2, and Of18S, were chosen as candidate reference genes. qRT-PCR was used to screen the reference genes, and GeNorm, NormFinder, and BestKeeper were used to analyze the stability of the candidate reference genes and screen out the best ones. Finally, the circadian rhythm gene GIGANTEA (GI) was used to verify the best reference genes. Results showed that OfRAN1 and OfIDH had the highest stability(GeNorm showed that the stable value of OfRAN1 and OfIDH were 0.347, while Of18S was 0.601; NormFinder showed that the stable value of OfRAN1 and OfIDH were 0.135 and 0.206, while Of18S was 0.474; BestKeeper showed that the stable value of OfRAN1 was 0.800 and OfIDH was 0.133, while Of18S was 0.474), which meant they were the best reference genes for the different conditions and Of18S had the worst stability(The greater the stability value, the worse the stability). The relative expression level of GI balanced by seven reference genes and the genetic combinations of OfRAN1 and OfIDH meant more accurate expression results(The expression pattern of circadian rhythm gene GI is accumulated during the day and falls at night). In summary, the genetic combinations of OfRAN1 and OfIDH were the best combination of reference genes used to study the molecular mechanism of O. fragrans under different photoperiod and temperature treatments.
-
Key words:
- botany /
- Osmanthus fragrans /
- reference gene /
- qRT-PCR /
- GI
-
表 1 qRT-PCR相关内参基因引物信息
Table 1. List of primers for qRT-PCR
基因简称 引物序列(5′→3′) 退火温度/ ℃ OfACT CCCAAGGCAAACAGAGAAAAAAT 55.0 ACCCCATCACCAGAATCAAGAA OfEF1α CGTTTGCCACTTCAGGATGTCTA 58.0 GTACCAGGTTTCAGGACTCCAGTTT OfIDH CTTGAAGCAGATGTGGAAGAGTC 57.5 CTTTGTCCATCCTGGGACCAGTC OfRAN1 AGAACCGACAGGTGAAGGCAA 57.5 TGGCAAGGTACAGAAAGGGCT OfTUB AGAAGGGATGGATGGAATGGA 56.0 GTCTTCTTCGTCCTCGGCAGT OfUBC2 TGTTGACAAAACCGATGGAAGGA 57.5 GTGGAGTGTGGAGGATAAGGGTG Of18S AGCCTGAGAAACGGCTACCAC 55.0 ATACGCTATTGGAGCTGGAA 表 2 7个候选内参基因扩增参数
Table 2. Amplification parameters of seven candidate reference genes
基因 Ct 扩增效率/% 曲线斜率 线性相关系数 条带长度/bp OfACT 22.77 ± 0.55 103.66 -3.237 3 0.995 5 143 OfEF1α 20.61 ± 0.42 103.19 -3.247 8 0.995 7 89 OfIDH 23.59 ± 0.39 95.78 -3.453 8 0.997 7 118 OfRNA1 21.68 ± 0.07 95.43 -3.436 5 0.996 2 117 OfTUB 28.44 ± 0.40 107.49 -3.154 6 0.993 2 106 OfUBC2 25.73 ± 0.54 107.15 -3.161 8 0.994 5 75 Of18S 8.99 ± 0.55 108.15 -3.140 9 0.996 8 208 表 3 不同软件分析7个候选内参基因表达稳定性
Table 3. Gene expression stability of seven reference genes by different software
基因名称 geNorm分析 NormFinder分析 BestKeeper分析 稳定值 排序 稳定值 排序 稳定值 排序 OfACT 0.393 2 0.232 3 0.288 3 OfEF1α 0.524 5 0.390 6 0.497 6 OfIDH 0.347 1 0.206 2 0.133 1 OfRNA1 0.347 1 0.135 1 0.180 2 OfTUB 0.479 4 0.361 5 0.372 5 OfUBC2 0.422 3 0.343 4 0.346 4 Of18S 0.601 6 0.474 7 0.611 7 -
[1] 李瑾.桂花花芽分化的研究[D].开封: 河南大学, 2007. LI Jin. Study on The Flower Bud Differentiation of Osmanthus fragrans Lour.[D]. Kaifeng: Henan University, 2007. [2] 邱芳, 张波, 史玉敏, 等.不同龄期桂花花芽分化期间矿质元素和可溶性糖含量的变化[J].安徽农业科学, 2012, 41(24):10024-10026. QIU Fang, ZHANG Bo, SHI Yumin, et al. Changes of mineral elements and the soluble sugar content at flower bud differentiation of Osmanthus fragrans (Thunb.) Lour. on different ages[J]. J Anhui Agric Sci, 2013, 41(24):10024-10026. [3] 王彩云, 高丽萍, 鲁涤非, 等. '厚瓣金桂'桂花花芽形态分化的研究[J].园艺学报, 2002, 29(1):52-56. WANG Caiyun, GAO Liping, LU Difei, et al. A study on morphological differentiation of flower bud of Osmanthus fragrans 'Houban Jingui'[J]. Acta Hortic Sin, 2002, 29(1):52-56. [4] 王英, 张超, 付建新, 等.桂花花芽分化和花开放研究进展[J].浙江农林大学学报, 2016, 33(2):340-347. WANG Ying, ZHANG Chao, FU Jianxin, et al. Progresses on flower bud differentiation and flower opening in Osmanthus fragrans[J]. J Zhejiang A&F Univ, 2016, 33(2):340-347. [5] 曹际云.桂花不同栽培变种的成花条件及花器官发育的形态解剖学研究[D].济南: 山东大学, 2008. CAO Jiyun. The Study on the Qualification of Floral Formation and Development of Floral Orgrans of Osmanthus fragrans[D]. Ji'nan: Shandong University, 2008. [6] 韩远记.桂花花色变异的机理和不同花色品种花瓣的cDNa-AFLP差异分析[D].开封: 河南大学, 2014. HAN Yuanji. Mechanism of Flower Color Variation and cDNA-AFLP Analysis of 2 Clutivar with Different Flower Color in Osmanthus fragrans[D]. Kaifeng: Henan University, 2014. [7] 母洪娜, 孙陶泽, 杨秀莲, 等.两个桂花品种花色色素相关基因的差异表达[J].南京林业大学学报(自然科学版), 2015, 39(3):183-186. MU Hongna, SUN Taoze, YANG Xiulian, et al. Differential expression of flower color related genes of Osmanthus fragrans Lour. 'Chenghongdangui' and 'Zaoyingui'[J]. J Nanjing For Univ Nat Sci Ed, 2015, 39(3):183-186. [8] LI Fafang, HUANG Qizhi. Analysis of fragrance composition in three cultivars of Osmanthus fragrans Albus group flower by gas chromatography-mass spectrometry[J]. Wuhan Univ J Nat Sci, 2011, 16(4):342-348. [9] 侯丹, 付建新, 张超, 等.桂花品种'堰虹桂''玉玲珑'和'杭州黄'的香气成分及释放节律[J].浙江农林大学学报, 2015, 32(2):208-220. HOU Dan, FU Jianxin, ZHANG Chao, et al. Flower scent composition of Osmanthus fragrans 'Yanhong Gui' 'Yu Linglong' and 'Hangzhou Huang', and their emission patterns[J]. J Zhejiang A&F Univ, 2015, 32(2):208-220. [10] de OLIVEIRA L A, BRETON M C, BASTOLLA F M, et al. Reference genes for the normalization of gene expression in Eucalyptus species[J]. Plant Cell Physiol, 2012, 53(2):405-422. [11] PINTO F, PACHECO C C, FERREIRA D, et al. Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria[J]. PLoS One, 2012, 7(4):e34983. doi:10.1371/journal.pone.0034983. [12] ZHANG Chao, FU Jianxin, WANG Yiguang, et al. Identification of suitable reference genes for gene expression normalization in the quantitative real-time PCR analysis of sweet osmanthus (Osmanthus fragrans Lour.)[J]. PLoS One, 2015, 10(8):e0136355. doi:10.1371/journal.pone.0136355. [13] ARTICO S, NARDELI S, BRILHANTE O, et al. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data[J]. BMC Plant Biol, 2010, 10(1):49-60. [14] JAIN M, NIJHAWAN A, TYAGI A K, et al. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR[J]. Biochem Biophysic Res Commun, 2006, 345(2):646-651. [15] 胡瑞波, 范成明, 傅永福.植物实时荧光定量PCR内参基因的选择[J].中国农业科技导报, 2009, 11(6):30-36. HU Ruibo, FAN Chengming, FU Yongfu. Reference gene selection in plant real-time quantitative reserve transcription PCR (qRT-PCR)[J]. J Agric Sci Technol, 2009, 11(6):30-36. [16] 陆婷, 王艺光, 骆义波, 等.'四季桂'不同季节的花芽分化与发育比较[J].园艺学报, 2017, 44(6):1145-1156. LU Ting, WANG Yiguang, LUO Yibo, et al. Comparison on flower bud differentiation and development of different seasons in Osmanthus fragrans 'Sijigui'[J]. Acta Hortic Sin, 2017, 44(6):1145-1156. [17] 付建新, 张超, 王艺光, 等.桂花组织基因表达中荧光定量PCR内参基因的筛选[J].浙江农林大学学报, 2016, 33(5):727-733. FU Jianxin, ZHANG Chao, WANG Yiguang, et al. Reference gene selection for quantitative real-time polymerase chain reaction (qRT-PCR) normalization in the gene expression of sweet osmanthus tissue[J]. J Zhejiang A&F Univ, 2016, 33(5):727-733. [18] WEI Libin, MIAO Hongmei, ZHAO Ruihong, et al. Identification and testing of reference genes for sesame gene expression analysis by quantitative real-time PCR[J]. Planta, 2013, 237(3):873-889. [19] DIE J V, ROMÁ N B, NADAL S, et al. Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions[J]. Planta, 2010, 232(1):145-153. [20] 涂礼莉, 张献龙, 刘迪秋, 等.棉花纤维发育和体细胞胚发生过程中实时定量PCR内对照基因的筛选[J].科学通报, 2007, 52(20):2379-2385. TU Lili, ZHANG Xianlong, LIU Diqiu, et al. Suitable internal control genes in qRT-PCR normalization in cotton fiber development and somatic embryogenesis[J]. Chin Sci Bull, 2007, 52(20):2379-2385. [21] LI Haiyan, DONG Yuanyuan, YANG Jing, et al. De novo transcriptome of safflower and the identification of putative genes for oleosin and the biosynthesis of flavonoids[J]. PLoS One, 2012, 7(2):e30987. doi:10.1371/journal.pone. 0030987. [22] GUTIERREZ L, MAURIAT M, GUÉNIN S, et al. The lack of a systematic validation of reference genes:a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants[J]. Plant Biotechnol J, 2008, 6(6):609-618. [23] BUSTIN S A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR):trends and problems[J]. J Mol Endocrinol, 2002, 29(1):23-39. [24] 尹静, 任春林, 詹亚光, 等.可用于实时荧光定量PCR标准化的白桦内参基因[J].植物生理学通讯, 2010, 46(10):1061-1066. YIN Jing, REN Chunlin, ZHAN Yaguang, et al. Selection of internal control genes for real-time RT-PCR normalization in white birch (Betula platyphylla Suk.)[J]. Plant Physiol Commun, 2010, 46(10):1061-1066. [25] van GUILDER H D, VRANA K, FREEMAN W A. Twenty-five years of quantitative PCR for gene expression analysis[J]. BioTechniques, 2008, 44(5):619-626. [26] 杨晶, 卢玉彬, 迟孟涵, 等.红花种子不同发育时期内参基因表达稳定性分析[J].中草药, 2017, 48(9):1845-1850. YANG Jing, LU Yubin, CHI Menghan, et al. Analysis on stability of reference genes in different developmental stages of seeds from Carthamus tinctorius[J]. Chin Tradit Herbal Med, 2017, 48(9):1845-1850. [27] MOURA J C, ARAÚJO P, BRITO M S, et al. Validation of reference genes from Eucalyptus spp. under different stress conditions[J]. BMC Res Notes, 2012, 5:634. [28] 胡瑞波.大豆FT/TFL1基因克隆、表达模式及功能分析[D].北京: 中国农业科学院, 2009. HU Ruibo. Molecular Cloning, Expression Profiles and Functional Analysis of FT/TFL1 Genes in Soybean (Glycine max)[D]. Beijing: Chinese Academy of Agricultural Sciences, 2009. [29] 李琼琼.大豆NCED1基因及其启动子的克隆、表达载体的构建及植物转化[D].长春: 吉林农业大学, 2012. LI Qiongqiong. Cloning, Construction of Plant Expression Vector and Transformation of GmNCED1 Gene and Promoter from Glycine max[D]. Changchun: Jilin Agricultural University, 2012. [30] 吝月爱.玉米在非生物胁迫和激素处理条件下实时荧光定量PCR内参基因的选择[D].成都: 四川农业大学, 2012. LIN Yueai. Reference Gene Selection for Quantitative Real-time PCR in Maize Treated with Abiotic Stresses and Hormones[D]. Chengdu: Sichuan Agricultural University, 2012. [31] 宋雄.欧芹不同逆境条件下适宜内参基因的筛选[D].南京: 南京农业大学, 2016. SONG Xiong. Screening Suitable Reference Genes under Different Stresss Conditions in Parsley[D]. Nanjing: Nanjing Agricultural University, 2016. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.2019.05.011






 下载:
下载: