-
长链非编码 RNA (lncRNA)是一类长度大于200个核苷酸的调控RNA,不具有编码蛋白质能力[1−2]。根据 lncRNA 在基因组上的位置,通常被分为长内含子非编码 RNA (intronic lncRNA)、长基因间非编码RNA (lincRNAs) 和长非编码自然反义转录本 (lncNATs)[3−4]。lncRNA 具有顺式调控和反式调控2种调控模式。顺式调控是指 lncRNA 对距离较近的 mRNA 的一种转录激活与表达调控方式。lncRNA 主要以序列互补为原理结合在未解链的染色质大沟中,或者解链的 DNA 链上。反式调控是指 lncRNA 通过与启动子和增强子或与这些位点结合并受影响的其他蛋白质相互作用来调节非连锁基因的表达、染色质状态和 RNA 聚合酶活性[5]。
lncRNA 可以起到支架的作用,组织蛋白复合物,充当亚细胞结构,从而行使功能。lncRNA 以剂量依赖形式在细胞质或者细胞核内调控结合的蛋白或者 RNA 的活性[6]。lncRNA 还可以通过与 RNA 或 DNA 的序列互补性或同源性,在结构上形成大分子复合物组装的分子框架和支架[7]。lncRNA 可作为与其互作分子的招募者、引导者和信号分子等,通过表观遗传调控、转录调控、转录后调控和翻译调控而发挥其功能[8−12]。研究表明:lncRNA 在毛竹Phyllostachys edulis次生壁的合成过程中发挥重要作用[13]。
转座子是生物体的重要组成成分。研究表明:许多 lncRNA 来源于转座子序列,称为转座子来源的lncRNA (TE-lncRNA1)[14−15]。23%的拟南芥Arabidopsis thaliana、50%的水稻Oryza sativa、51%的玉米Zea mays的 TE-lncRNA 主要来源于逆转座子序列[16−17]。它主要参与作物的生长发育、非生物胁迫应答等不同的生物过程[18]。转座子来源的 lncRNA 受胁迫诱导表达,且表达量较高,具有较高的研究价值[19]。
毛竹分布范围广,种植面积大,是中国极为重要的非木材森林资源[20]。吴佳军[21]构建了低温、高温、紫外、高盐等4种胁迫毛竹全转录组数据库。在本研究中,我们利用4种胁迫全转录组数据库筛选并鉴定了在紫外胁迫下差异表达的1个lncRNA,它来源于 LARD 转座子序列,命名为Ph-TElncRNA1。本研究以毛竹Ph-TElncRNA1为研究对象,鉴定其调控的靶基因,分析了其与靶基因的表达协同性,以及Ph-TElncRNA1和靶基因在叶片着色过程中的表达趋势,初步探究了Ph-TElncRNA1的功能。
-
毛竹种子采自广西省桂林市灵川县同一母竹,发芽后放置在室温人工气候室中进行培养。紫外胁迫处理:取五叶一心期实生苗,30 W紫外灯分别处理2、4、6、8 h[22]。对照为正常的日常光。每个处理做3个重复,处理后立即取样,每个样取5张叶片和心。处理好的样品进行液氮速冻,存于−80 ℃冰箱,用于RNA提取。
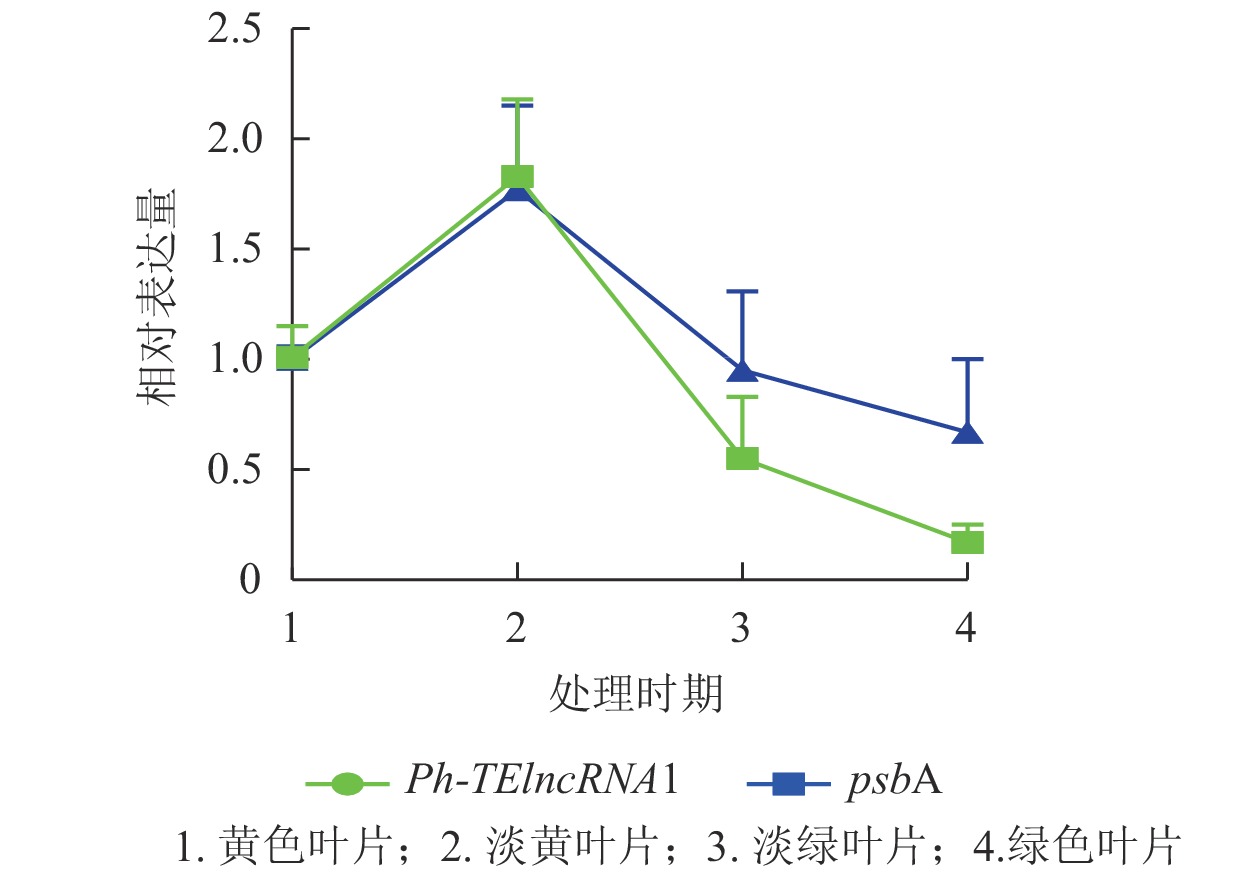
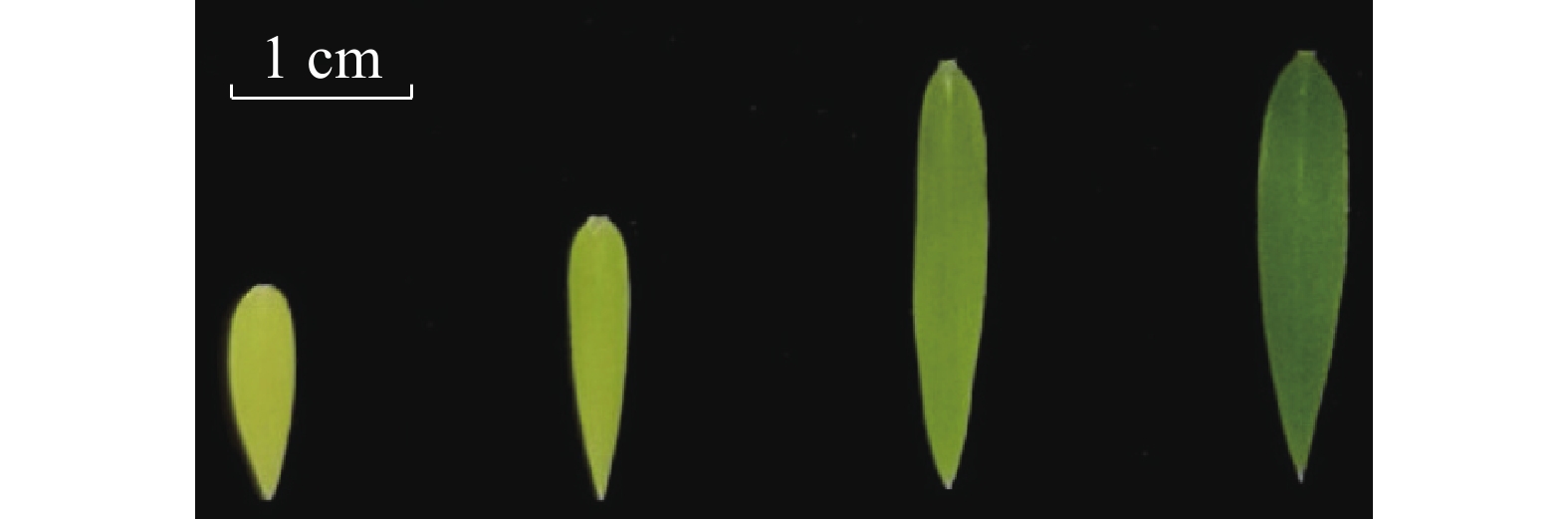
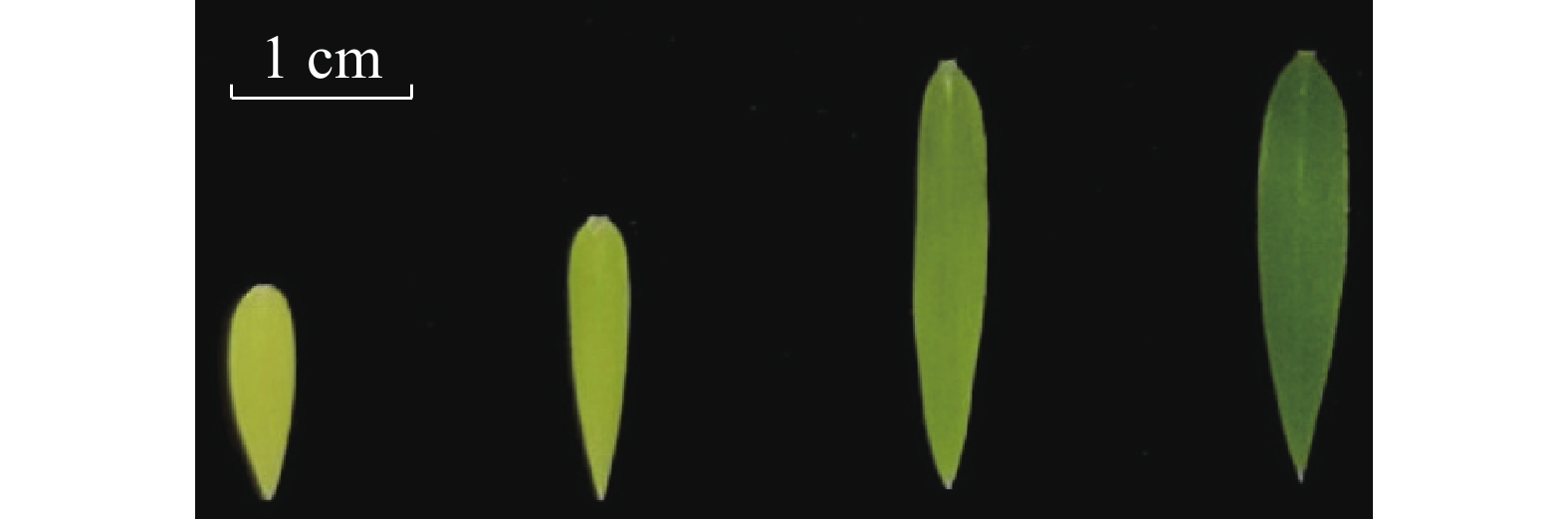
不同着色程度毛竹叶片采集:选取长势良好且一致的毛竹实生幼苗,待其在暗培养箱里长到三叶一心时,取黄色叶片的幼苗,并将其中3盆转移到光照培养箱中;培养第2天取淡黄色叶片;培养第4天,取淡绿色叶片;培养第6天,取绿色的叶片(图1)。每个类型的叶片取3张,进行液氮速冻,存于−80 ℃冰箱,用于RNA提取。
-
以前期转录组测序后的低温、高温、紫外、高盐等4种胁迫全转录组为基础,在吴佳军[21]构建的毛竹 lncRNA 数据库中,筛选到在紫外胁迫处理中差异表达,且靶基因差异表达,缺乏开放阅读框的 lncRNA。使用以下4个的软件:CNCI (CodingNon-CodingIndex)、CPC2 (Coding Potential Calculator)、PLEK (predictor of long non-coding RNAs and messenger RNAs based on an improved k-mer scheme)和Pfam (PfamScan) 判断转录本的编码情况,确定典型的 lncRNA。
根据其在基因组的位置和转座子位置关系,确定其为来源转座子的 lncRNA,命名为Ph-TElncRNA1。
在毛竹基因组中提取Ph-TElncRNA1的序列,进行引物设计(F:AGGGGCGCTTCCGGTTCTG; R:AAGAGATATATTTTACATCCCAGA),PCR 扩增,测序确认。
-
使用2种方法预测lncRNA的靶基因。第1种是顺式 (cis) 靶基因,根据lncRNA与基因的位置关系,将lncRNA附近100 kb范围以内的基因作为其靶基因,通过使用perl脚本查找这类靶基因[23]。第2种是根据lncRNA和mRNA之间的碱基互补配对关系,利用在线软件LncTar计算lncRNA和mRNA配对位点的自由能和标准化自由能,将低于标准化自由能阈值的基因认定为lncRNA的靶基因,称作反式 (trans) 靶基因。通过转录组数据筛选与对应TE-lncRNA变化趋势一致的靶基因,且差异表达的基因作为研究对象。
-
提取经胁迫处理后毛竹实生苗RNA,以反转录后的cDNA作为定量模板,进行实时荧光定量PCR (RT-qPCR)验证,分析Ph-TElncRNA1和靶基因表达趋势的关联性,剔除相关性不强的靶基因。使用IBM SPSS Statistics软件对Ph-TElncRNA1和靶基因的相对表达量进行皮尔逊相关性分析,确定Ph-TElncRNA1和靶基因的相关情况。
-
提取4种颜色(黄色、淡黄、淡绿、绿色)毛竹实生苗叶片(图1)RNA,以反转录后的cDNA作为定量模板,进行RT-qPCR验证,分析Ph-TElncRNA1和靶基因psbA在叶片着色过程中的表达模式。
-
以吴佳军[21]构建的 lncRNA 数据库为基础,筛选到紫外胁迫处理中差异表达,且靶基因差异表达的 lncRNA。通过 CNCI、Pfam、CPC2、PLEK 等4个软件分析lncRNA 的真实性。 CNCI 和Pfam软件均表明筛选的 lncRNA 不具有编码蛋白的能力,CPC2[编码可能性(coding probability)为0.144 099,菲克特分数(Fickettscore)为0.465880)]和 PLEK (软件评分为−2.282730,该软件评分小于 0 为非编码 RNA)也表明筛选的 lncRNA 编码蛋白的可能性较低。由此可以判定筛选的 lncRNA 是一个典型的非编码 RNA。
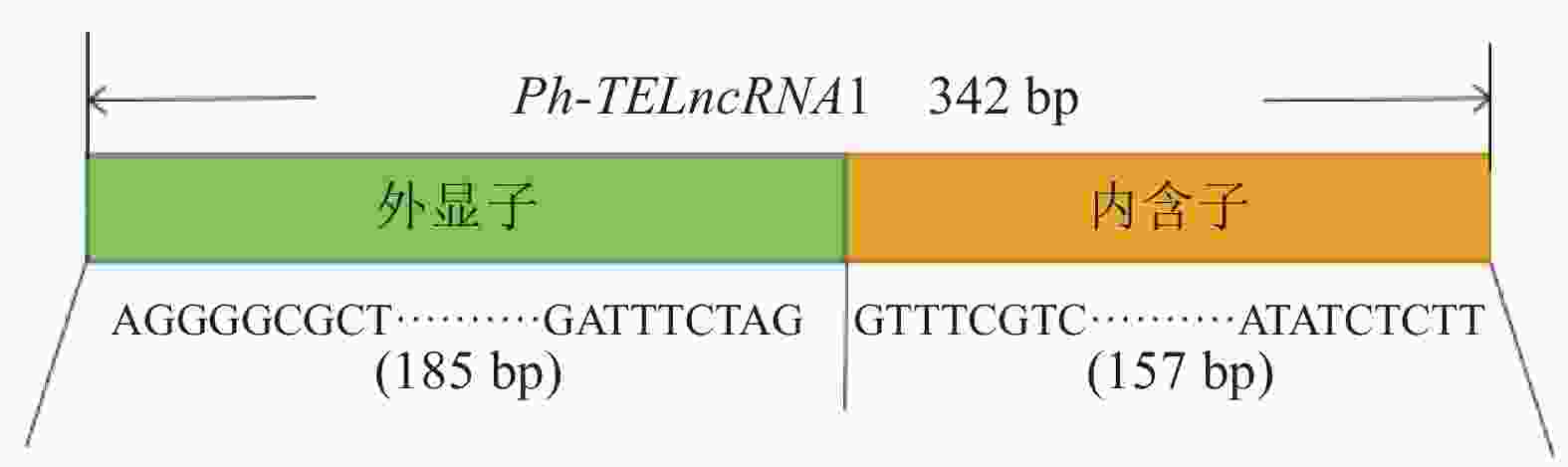
分析表明:该lncRNA 位于一个反转录转座子大片段(large retrotransposons derivatives, LARD)内部(图2),为转座子来源的lncRNA,命名为Ph-TElncRNA1。Ph-TElncRNA1全长为 342 bp,其中 185 个碱基来源于外显子,157 个碱基来源于内含子(图3)。

图 2 转座子LARD和Ph-TElncRNA1位置关系结构图
Figure 2. 2Positional relationship between the LARD transposon and Ph-TElncRNA1
利用在线软件LncTar计算lncRNA和mRNA配对位点的自由能和标准化自由能,低于标准化自由能的靶基因认定为是该 lncRNA 的靶基因。进一步通过转录组数据筛选与对应Ph-TElncRNA1变化趋势一致的靶基因,且差异表达的基因作为研究对象。得到该 lncRNA 的3个靶基因,分别为PH010-02523G0060、PH01002523G0140 和 PH010025-23G0050。
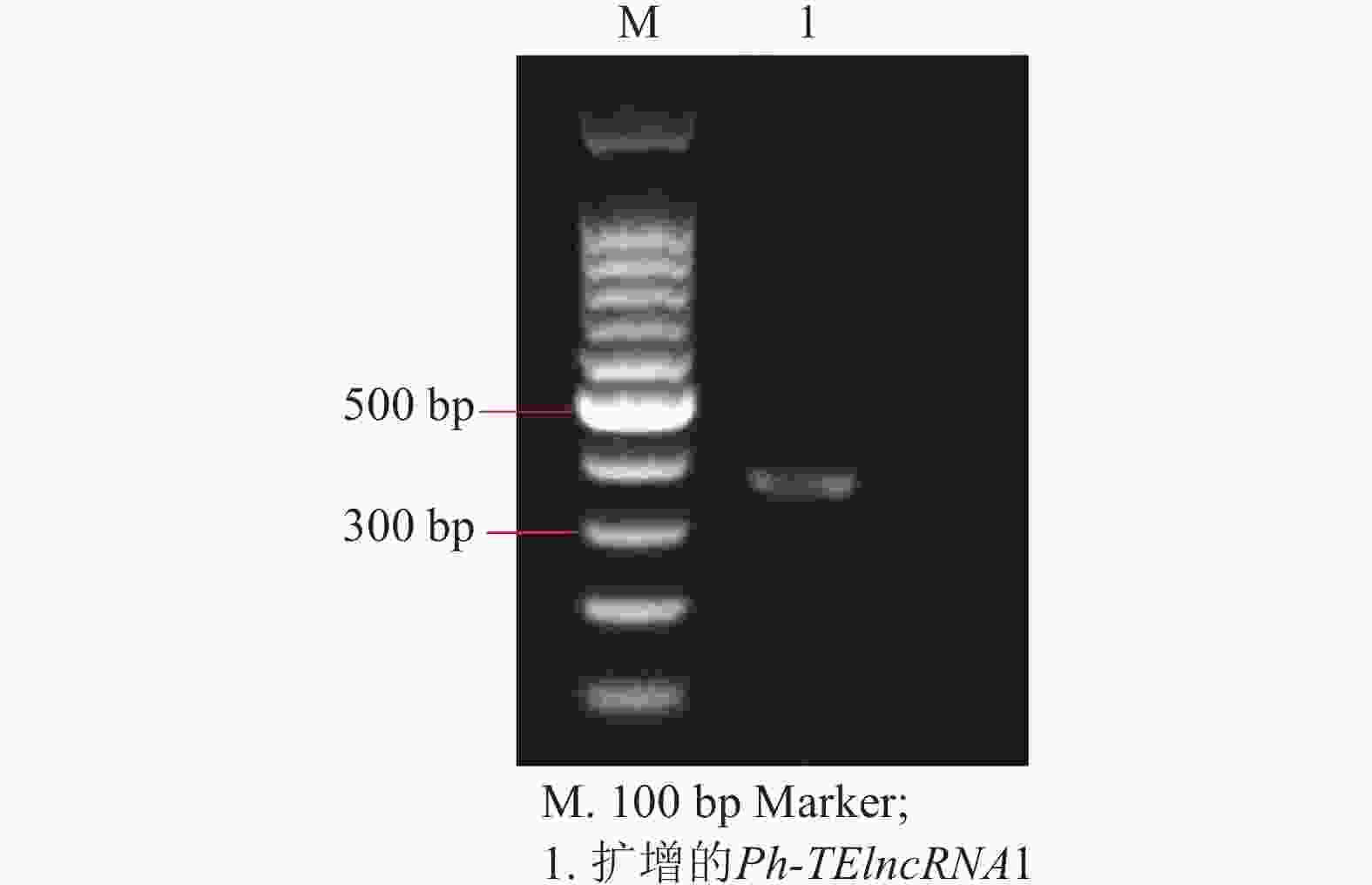
以紫外处理2 h的毛竹叶片 cDNA 为模板,通过 PCR 扩增出相应的片段(图4),测序结果表明成功克隆到Ph-TElncRNA1 (图5)。
-
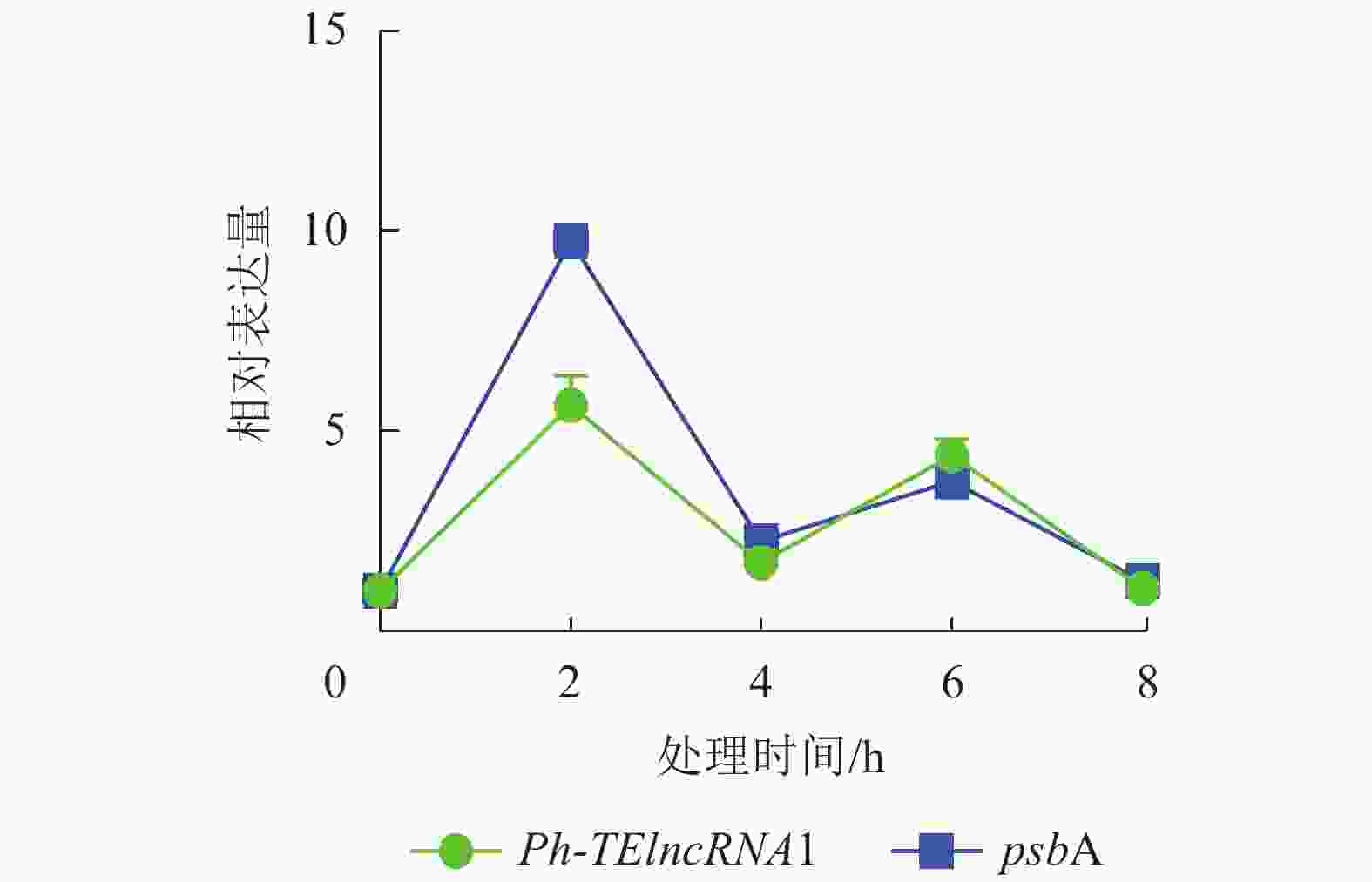
通过实时荧光定量 PCR 验证,发现 PH01002523G0060 和PH01002523G0050 在紫外胁迫处理 2、4、6、8 h 后,相对表达量的趋势杂乱,与Ph-TElncRNA1不一致,说明 PH01002523G0060 和 PH01002523G0050 这2个靶基因与Ph-TElncRNA1的关联性不强,从而剔除掉。最终得到靶基因 PH01002523G0140 与Ph-TElncRNA1关联性较强,且相对表达量呈现完全一致的趋势。靶基因 PH01002523G0140 是psbA同源基因,编码 photosystem Ⅱ protein D1。与对照相比,Ph-TElncRNA1和靶基因psbA的相对表达量在紫外胁迫处理初期表达量上升显著,随着紫外灯照射时间的延长逐渐降低。Ph-TElncRNA1和psbA在紫外胁迫处理 2 h 时,相对表达量达到最高值(图6),Ph-TElncRNA1与psbA呈显著正相关(P=0.000,皮尔逊相关系数为0.898)。
-
Ph-TElncRNA1的靶基因psbA编码光系统Ⅱ (PSⅡ) 反应中心蛋白 D1,是叶绿体的重要组成部分。通过对叶片着色的4个时期样本(黄色、淡黄、淡绿、绿色)进行RT-qPCR发现:Ph-TElncRNA1和靶基因psbA在4个时期的相对表达量呈正相关(图7)。根据图7中的表达趋势可知:在淡黄时期,也就是将毛竹放入培养箱2 d时,表达量最高,在这一时期叶绿体正在形成,而光系统Ⅱ (PSⅡ) 反应中心蛋白 D1作为重要的蛋白亚基,也处于表达高峰时期。Ph-TElncRNA1和靶基因psbA在叶绿体形成过程中表达量最高,在叶绿体形成后呈现逐渐下降的趋势。两者在叶片着色过程中,相对表达量的变化趋势呈显著正相关(P=0.000,皮尔逊相关系数为0.926)。
-
由于lncRNA具有低表达和低序列保守性的特点,曾被认为是RNA聚合酶Ⅱ转录的副产物[24]。随着人们对lncRNA研究的不断深入,逐渐发现其在动植物等真核生物生长发育的过程中扮演着至关重要的作用[25]。目前,对lncRNA的研究大多集中于治疗人体各类疾病的研究上,如胃癌[26]、口腔鳞状细胞癌[27]、变应性鼻炎[28]等。相比于动物中lncRNA的研究,对植物lncRNA的探索仍处于生物信息学分析的初级阶段,研究主要集中在拟南芥[29]、烟草Nicotiana tabacum[30]及水稻[31]等植物中。研究表明:大量非编码RNA来源于转座子序列,如lncRNA、miRNA、siRNA等。转座子来源的lncRNA参与了真核生物生长发育等活动中[16],但大多数转座子来源的lncRNA的功能尚未研究。
从实验室前期转录组数据分析可知:毛竹63.59%的lncRNA是转座子起源的,毛竹中受非生物胁迫诱导的 TE-ncRNA 非常丰富[21]。本研究通过对Ph-TElncRNA1和psbA在紫外胁迫下相对表达量的分析发现:Ph-TElncRNA1与psbA的相对表达量呈完全相同的趋势,在紫外胁迫下这两者是协同表达的。Ph-TElncRNA1与psbA相对表达量显著相关,进一步说明psbA可能是Ph-TElncRNA1靶基因。
psbA基因编码光系统Ⅱ(PSⅡ)反应中心蛋白D1[32]。D1蛋白是PSⅡ反应中心复合物中的一个非常重要的蛋白亚基,D1蛋白在叶绿体中的合成效率决定PSⅡ复合体的修复效率,进而决定植物的光合作用效率。研究表明:光可以诱导D1蛋白的表达,实现精准控制[33]。本研究对毛竹实生苗叶片着色过程Ph-TElncRNA1和psbA相对表达量的分析表明:Ph-TElncRNA1与psbA在叶绿体正在形成时期达到表达峰值,呈正相关,说明lncRNA可以通过靶基因参与调控毛竹叶片的着色过程。本研究为进一步深入分析lncRNA与靶基因的互作机制奠定基础,对于研究lncRNA的生物学功能也有一定的意义。
Identification of Ph-TElncRNA1 in Phyllostachys edulis and its regulation of target genes
-
摘要:
目的 旨在探究毛竹Phyllostachys edulis Ph-TElncRNA1及靶基因的表达模式,初步分析lncRNA的功能。 方法 基于高温、低温、紫外、高盐胁迫处理毛竹实生苗全转录组测序数据,筛选在胁迫下差异表达的lncRNA。通过CNCI、Pfam、CPC2、PLEK等4个软件对lncRNA的编码特性进行分析,用LncRNATar软件鉴定lncRNA的靶基因,通过实时荧光定量PCR分析lncRNA和靶基因在紫外胁迫下和叶片着色过程中的表达模式。 结果 筛选到1个在紫外胁迫下差异表达的lncRNA,此lncRNA来源于LARD转座子序列(large retrotransposons derivatives),命名为Ph-TElncRNA1 (Phyllostachys edulis transposable element derived lncRNA1)。Ph-TElncRNA1是一个典型的非编码RNA,全长为342 bp,其中,185个碱基来源于外显子,157个碱基来源于内含子。Ph-TElncRNA1的靶基因为psbA,编码photosystem Ⅱprotein D1。实时荧光定量结果表明:Ph-TElncRNA1与psbA的相对表达量呈完全相同的趋势,说明在紫外胁迫下Ph-TElncRNA1与psbA呈正相关。通过Ph-TElncRNA1和psbA在不同着色时期的毛竹叶片的相对表达量分析,发现在叶绿体形成期,Ph-TElncRNA1和psbA表达量达到峰值。 结论 Ph-TElncRNA1和psbA在紫外胁迫下协同表达,Ph-TElncRNA1可以通过调控靶基因psbA参与毛竹叶片发育。图7参33 -
关键词:
- Ph-TElncRNA1 /
- 叶片着色 /
- D1蛋白 /
- 毛竹
Abstract:Objective The purpose of this study is to explore the expression patterns of Ph-TElncRNA1 and target genes in Phyllostachys edulis, and to preliminarily analyze the function of lncRNA. Method Based on the whole transcriptome sequencing data of Ph. edulis seedlings under high temperature, low temperature, ultraviolet and high salt stress, lncRNAs differentially expressed under stress were screened. The coding characteristics of lncRNA were analyzed by four kinds of software (CNCI, Pfam, CPC2 and PLEK), and the target genes of lncRNA were identified by LncTar software. The expression patterns of lncRNA and target genes under UV stress and leaf coloration were analyzed by real-time fluorescent quantitative PCR. Result One differentially expressed lncRNA under UV stress was screened, which was derived from the lncRNA of the Large retrotransposons derivatives and named Ph-TElncRNA1 (Ph. edulis transposable element derived lncRNA1). Ph-TElncRNA1 was a typical non-coding RNA with a total length of 342 bp, 185 bases from exons and 157 bases from introns. The target gene of Ph-TElncRNA1 was psbA, encoding photosystem Ⅱ protein D1. Real time fluorescence quantitative results showed that the relative expression of Ph-TElncRNA1 and psbA was the same, indicating that Ph-TElncRNA1 was positively correlated with psbA under UV stress. Through the analysis of the relative expression of Ph-TElncRNA1 and psbA in Ph. edulis leavesat different coloring stages, it was found that the expression of psbA and Ph-TElncRNA1 reached the peak during chloroplast formation stage. Conclusion Ph-TElncRNA1 and psbA are co-expressed under UV stress, and Ph-TElncRNA1 can participate in the leaf development of Ph. edulis by regulating the target gene psbA. [Ch, 7 fig. 33 ref.] -
Key words:
- Ph-TElncRNA1 /
- leaf coloration /
- D1 protein /
- Phyllostachys edulis
-
-
[1] ZHANG Xiaopei, DONG Jie, DENG Fenni, et al. The long non-coding RNA lncRNA973 is involved in cotton response to salt stress [J]. BMC Plant Biology, 2019, 19(1): 443 − 459. [2] HEO J B, LEE Y S, SUNG S. Epigenetic regulation by long noncoding RNAs in plants [J]. Chromosome Research, 2013, 21(6/7): 685 − 693. [3] DERRIEN T, JOHNSON R, BUSSOTTI G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression [J]. Genome Reseach, 2012, 22(9): 1775 − 1789. [4] CHEKANOVA J A. Long non-coding RNAs and their functions in plants [J]. Current Opinion Plant Biology, 2015, 27: 207 − 216. [5] KOPP F, MENDELL J T. Functional classification and experimental dissection of long noncoding RNAs [J]. Cell, 2018, 172(3): 393 − 407. [6] ENGREITZ J M, PANDYA-JONES A, MCDONEL P, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X-chromosome [J/OL]. Science, 2013, 341(6147): 1237973[2022-05-10]. doi: 10.1126/science.1237973. [7] YUAN Tingting, ZHU Chenglei, LI Guangzhu, et al. An integrated regulatory network of mRNAs, microRNAs, and lncRNAs involved in nitrogen metabolism of moso bamboo[J]. Frontiers in Genetics, 2022, 13: 854346. doi:10.3389/fgene.2022.854346. [8] GUIL S, ESTELLER M. Cis-acting noncoding RNAs: friends and foes [J]. Nature Structural &Molecular Biology, 2012, 19(11): 1068 − 1075. [9] ZHANG Yuchan, LIAO Jianyou, LI Zeyuan, et al. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice [J]. Genome Biology, 2014, 15(12): 512 − 528. [10] ARIEL F, JEGU T, LATRASSE D, et al. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop [J]. Molecular Cell, 2014, 55(3): 383 − 396. [11] BERRY S, DEAN C. Environmental perception and epigenetic memory: mechanistic insight through FLC [J]. The Plant Journal, 2015, 83(1): 133 − 148. [12] MOISON M, PACHECO J M, LUCERO L, et al. The lncRNA APOLO interacts with the transcription factor WRKY42 to trigger root hair cell expansion in response to cold [J]. Moleclar Plant, 2021, 14(6): 937 − 948. [13] MATZKE M A, MOSHER R A. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity [J]. Nature Reviews Genetics, 2014, 15(6): 394 − 408. [14] WANG Jiongliang, HOU Yinguang, WANG Yu, et al. Integrative lncRNA landscape reveals lncRNA-coding gene networks in the secondary cell wall biosynthesis pathway of moso bamboo (Phyllostachys edulis)[J/OL]. BMC Genomics, 2021, 22(1): 638[2022-05-10]. doi: 10.1186/s12864-021-07953-z. [15] GOLICZ A A, SINGH M B, BHALLA P L. The long intergenic noncoding RNA (LincRNA) landscape of the soybean genome [J]. Plant Physiology, 2018, 176(3): 2133 − 2147. [16] WANG Dong, QU Zhipeng, YANG Lan, et al. Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants [J]. The Plant Journal, 2017, 90(1): 133 − 146. [17] ZHAO Ting, TAO Xiaoyuan, FENG Shouli, et al. LncRNAs in polyploid cotton interspecific hybrids are derived from transposon neofunctionalization[J/OL]. Genome Biology, 2018, 19(1): 195[2022-05-10]. doi: 10.1186/s13059-018-1574-2. [18] LÜ Yuanda, HU Fengqin, ZHOU Yongfeng, et al. Maize transposable elements contribute to long non-coding RNAs that are regulatory hubs for abiotic stress response [J/OL]. BMC Genomics, 2019, 20(1): 864[2022-05-10]. doi: 10.1186/s12864-019-6245-5. [19] 王益军, 王亚丽, 陈煜东. 转座子来源的植物长链非编码RNA[J]. 植物学报, 2020, 55(6): 768 − 776. WANG Yijun, WANG Yali, CHEN Yudong. Transposon-derived long noncoding RNA in plants [J]. Chinese Bulletin of Botany, 2020, 55(6): 768 − 776. [20] 彭忠明, 徐克凡, 周凯, 等. 毛竹DNA甲基转移酶家族的鉴定与生物信息学分析[J/OL]. 分子植物育种, 2022-04-26[2022-05-10]. https://kns.cnki.net/kcms/detail/46.1068.S.20220426.0814.002.html. PENG Zhongming, XU Kefan, ZHOU Kai, et al. Identification and biological information analysis of Phyllostachys edulis DNMT genefamily [J/OL]. Molecular Plant Breeding, 2022-04-26[2022-05-10]. https://kns.cnki.net/kcms/detail/46.1068.S.20220426.0814.002.html. [21] 吴佳军. 毛竹转座子来源的非编码RNA在非生物胁迫中的响应机制[D]. 杭州: 浙江农林大学, 2019. WU Jiajun. The Response Mechanism of Phyllostachys edulis Transposable Non-coding RNA in Abiotic Stress[D]. Hangzhou: Zhejiang A&F University, 2019. [22] HUNTER K W, AMIN R, DEASY S, et al. Genetic insights into the morass of metastatic heterogeneity [J]. Nature Reviews Cancer, 2018, 18(4): 211 − 223. [23] JIA Hui, OSAK M, BOGU G K, et al. Genome-wide computational identification and manual annotation of human long noncoding RNA genes [J]. RNA, 2010, 16(8): 1478 − 1487. [24] 刘琳营, 苏晓俊, 闵玲. 植物中长链非编码RNA研究进展综述[J]. 江苏农业科学, 2021, 49(12): 12 − 19. LIU Linying, SU Xiaojun, MIN Ling. Research progress of long non-coding RNA in plants: a review [J]. Jiangsu Agricultural Science, 2021, 49(12): 12 − 19. [25] 张玉婵, 陈月琴. 长链非编码RNA在植物生殖发育中的调控作用[J]. 生命科学, 2016, 28(6): 640 − 644. ZHANG Yuchan, CHEN Yueqin. Long non-coding RNAs in plant reproductive growth [J]. Chinese Bulletin of Life Sciences, 2016, 28(6): 640 − 644. [26] 吕秉哲, 马东江, 魏成, 等. LncRNA对胃癌表型的影响及诊疗潜力[J]. 生命的化学, 2022, 42(3): 64 − 73. LÜ Bingzhe, MA Dongjiang, WEI Cheng, et al. Emerging impact of long noncoding RNA and its molecular mechanisms in gastric cancer [J]. Chemistry of Life, 2022, 42(3): 64 − 73. [27] 林诗晗, 邹璐宁, 林雪梅, 等. 基于TCGA/GEO数据库对lncRNA在口腔鳞状细胞癌中甲基化水平及预后相关性的分析[J]. 实用口腔医学杂志, 2022, 38(3): 368 − 372. LIN Shihan, ZOU Ling, LIN Xuemei, et al. Identification of DNA methylationoflnc RNA for predicting prognosis of oral squamous cell carcinoma based on TCGA/GEO database [J]. Journal of Practical Stomatology, 2022, 38(3): 368 − 372. [28] 王益玲, 王留珍, 冯海燕. 生物信息学分析长链非编码RNA在变应性鼻炎中的作用[J]. 中国耳鼻咽喉颅底外科杂志, 2022, 28(1): 51 − 57. WANG Yiling, WANG Liuzhen, FENG Haiyan. Role analysis of long non-coding RNA in allergic rhinitis based on bioinformatics [J]. The Chinese Journal of Otolaryngology-Cranial Bottom Surgery, 2022, 28(1): 51 − 57. [29] 赵雷. 拟南芥长链非编码RNA调控高温胁迫响应的机理研究[D]. 泰安: 山东农业大学, 2017. ZHAO Lei. Functional Analysis of Long Nocoding RNA for Heat Stress Tolerance in Arabidopsis[D]. Tai’an: Shandong Agricultural University, 2017. [30] MA Peng, ZHANG Xiao, LUO Bowen, et al. Transcriptomic and genome-wide association study reveal long noncoding RNAs responding to nitrogen deficiency in maize[J/OL]. BMC Plant Biology, 2021, 21(1): 93[2022-05-10]. doi: 10.1186/s12870-021-02847-4. [31] 张晓佩. 陆地棉长链非编码RNAs-lncRNA354和lncRNA973的功能和作用机制研究[D]. 泰安: 山东农业大学, 2022. ZHANG Xiaopei. Function and Mechanism Studies of Upland Cotton Long Non-coding RNAs-lncRNA354 and lncRNA973[D]. Tai’an: Shandong Agricultural University, 2022. [32] FERREIRA K N, IVERSON T M, MAGHLAOUI K, et al. Architecture of the photosynthetic oxygen-evolving center [J]. Science, 2004, 303(5665): 1831 − 1838. [33] SIROHIWAL A, NEESE F, PANTAZIS D A. Protein matrix control of reaction center excitation in photosystem Ⅱ [J]. Journal of American Chemical Society, 2020, 142(42): 18174 − 18190. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.20220396






 下载:
下载: