-
陆地棉Gossypium hirsutum是主要的经济作物之一,在经济发展过程中占有重要地位。而今因优质耕地面积减少导致的粮棉争地问题日益突出,中国陆地棉产区又整体呈现西北内陆棉区面积不断扩大,黄河、长江棉区面积持续减少的趋势[1]。在西北内陆地区栽培早熟棉能充分发挥其可晚播的特点,减少因早春干燥、降温,以及晚霜等原因造成的育苗病虫害,降低杀虫剂施用量[2],增加霜前开花率并改善陆地棉品质[3]。因此,筛选早熟棉对提高耕地利用效率具有重要意义[4]。
植物从生理生长转向生殖生长的过程为开花[5],受环境激素影响[6],目前较为广泛的调控开花途径是光周期途径、春化途径、自主途径及年龄途径等[7]。自主及春化途径主要通过开花抑制基因FLC位点进行[8],FLC调控FT和SOC1抑制开花[9],受光周期途径正向调控[10],可被FLD等通路抑制[11]。光周期靠CO/FT表达改变模式[12]。CO是光周期的核心基因[13],其蛋白有2个锌指结构域正向调控FT[14−15],N端蛋白控制光稳定,C端CCT区域用于核定位[16]。对拟南芥Arabidopsis thaliana研究表明:CCA1/LHY在TOC1上游调控光形态建成抑制其节律[17−18]。激活CCA1/LHY和TOC1翻译组蛋白可调控昼夜节律[19]。RVE8/LCL5也可通过结合TOC1启动子调节昼夜节律[20],节律核心基因限制TOC1的降解[21]。TOC1和CCA1的mRNA转录水平受Hesp调控[22]。
PRR亚家族成员是生物钟重要组分。中心环CCA1和LHY通过结合启动子负调控TOC1(APRR1)[23]。CCA1和LHY是PRR9、PRR7的正调控因子[24],也可能是PRR5的正调控因子。3个PRR基因通过结合启动子负调控CCA1和LHY[25]。PRR5促进TOC1积累使其稳定[26],PRR3和PRR5阻断TOC1与ZTL互作使其稳定[24]。对玉米Zea mays研究表明:PRR家族成员参与包括光响应在内的多种信号传导[27]。对大豆Glycine max研究表明:PRR家族CCT结构缺失与无意义突变影响开花时间[28]。对大白菜Brassica pekinensis[29]、大豆突变体[30]研究也表明:TOC1可控制早花,参与非生物胁迫应答[31]。
陆地棉中开花相关基因大多数属于光周期和生物钟相关途径[32]。前人对陆地棉中光周期通路CO[33]、FT[34],赤霉素途径FPF1[35]、SPL3[36]和部分MADS-box[37−38]家族基因进行研究,说明研究开花通路相关基因具有重要意义。结合生物信息学分析的基因功能研究有助于更好地理解基因功能[39−40]。本研究将从陆地棉群体高密度遗传图谱[41]及数量性状基因座(QTL)定位[42]中发掘陆地棉中拟南芥TOC1(APRR1)的同源基因GhPRR9进行家族分析和功能验证,预测GhPRR9的结构及可能行使的功能,并对GhPRR9功能加以验证,为培育早熟棉提供一定的理论参考。
-
陆地棉全基因组数据下载于Cottongen[43],拟南芥全基因组数据下载于TAIR[44],水稻Oryza sativa、草棉Gherbaceum、可可Theobroma caca、玉米、大豆、毛果杨Populus trichocarpa基因组数据下载于Phytozome[45]。
根据拟南芥PRR亚家族的定义,在Pfam上获得CCT (PF06203)和REC (PF00072)结构域隐马模型,用HMMER扫描整个陆地棉基因组取交集,利用在线工具[46]鉴别所筛选出的基因是否同时包含CCT和REC结构域,最终得到GhPRR家族基因成员。使用ExPASY网站[47]分析工具和WoLF对家族成员进行蛋白理化性质分析。
用MEGA[48]对8个物种的PRR亚家族蛋白进行多序列比对,邻接法JJT模型构建系统进化树,校验重复100次。用DNAMAN进行保守序列比对和绘制。
利用TBtool[49]软件制作染色体定位图和domain结构;使用MEME[50]网站分析家族成员所含Motif并进行可视化。使用Plant Care[51]分析GhPRR亚基因家族上游2 000 bp顺式启动子元件,使用TBtools进行可视化。在美国国家生物技术信息中心(NCBI)数据库中下载陆地棉相关的表达数据(序列号:PRJNA490626,编号:490626),用TBtools绘制热图。
-
提取陆地棉标准系‘TM-1’花蕾RNA,并用试剂盒(CAT#037A)反转录得到底物。使用Primer 5设计引物并扩增目标片段。使用TaKaRa纯化试剂盒(9761)纯化片段,pMD18-T Vector Cloning Kit (CAT# 6011)连接T载。热激法转化DH5α感受态菌株,活化涂板后挑单菌落进行菌液PCR分析,选取合理条带单克隆测序。
以T载为模板克隆片段并连接至过表达载体,热激转化农杆菌Agrobacterium tumefaciens GV3101,筛选阳性单克隆后取带花序的健康拟南芥提前剪下角果。浸入活化农杆菌液侵染1 min,沥干后黑暗1 d正常培养,收集种子为T0代。消毒播种T0代种子至相应抗性培养基上,其中,正常生长幼苗转入正常条件培养。筛选并验证拟南芥的阳性植株,成熟后收取T1代种子,如此培养至T3代。
-
从陆地棉基因组中提取GhPRR9起始密码子上游2 000 bp片段并预测顺式启动子元件。从陆地棉标准系‘TM-1’叶片DNA中分别克隆以起始编码为原点,长500、1 000、1 500和2 000 bp的片段,XcmⅠ酶切链接载体pCXGUS-P,热激法转入大肠埃希菌Escherichia coli,测序无误后将质粒转入农杆菌中侵染拟南芥得到种子。在卡那霉素培养基上播种筛选阳性植株培养至开花,取相关组织染色并观察。
-
将完成转化的表达载体以及绿色荧光蛋白(GFP)空载体通过热激法转入农杆菌菌株GV3101。培养后离心收集菌体重新悬浮,注射幼嫩烟草下表皮。注射后的烟草黑暗培养1 d后恢复正常光照周期。取下表皮制成临时切片,在激光共聚焦显微镜(LSM880)下观察记录影像。
-
相对定量使用2−ΔΔCt法,内参基因为GhHistone3 (陆地棉)和AtUBQ5 (拟南芥)。扩增程序为95 ℃ 30 s,95 ℃ 15 s,60 ℃ 30 s,共40个循环。
陆地棉时空表达分析取样:选取4个品种陆地棉材料的不同器官组织,每个品种20株随机取样,混合研磨。日周期节律分析:取三叶期的陆地棉标准系‘TM-1’植株,在人工气候室中培养1周后,隔4 h取1次顶芽,重复3株混样研磨。
-
用SGN VIGS Tool设计最佳VIGS片段。以测序正确的T载为模板进行扩增,SpeI和AscI酶切位点链接到pCLCrVA载体并转化至农杆菌LBA4404中。pCLCrVA-GhPRR9、pCLCrVA、pCLCrVA-PDS重悬液分别与pCLCrVB的重悬液按体积比1∶1混合均匀。
选取子叶完全展平,第1片真叶尚未完全显形的健康植株进行注射。侵染后的陆地棉设置辅助对照、沉默株、烟草花叶病毒株和空白对照植株,避光培养1 d后转入正常光照培养至开花,记录现蕾开花时间。
-
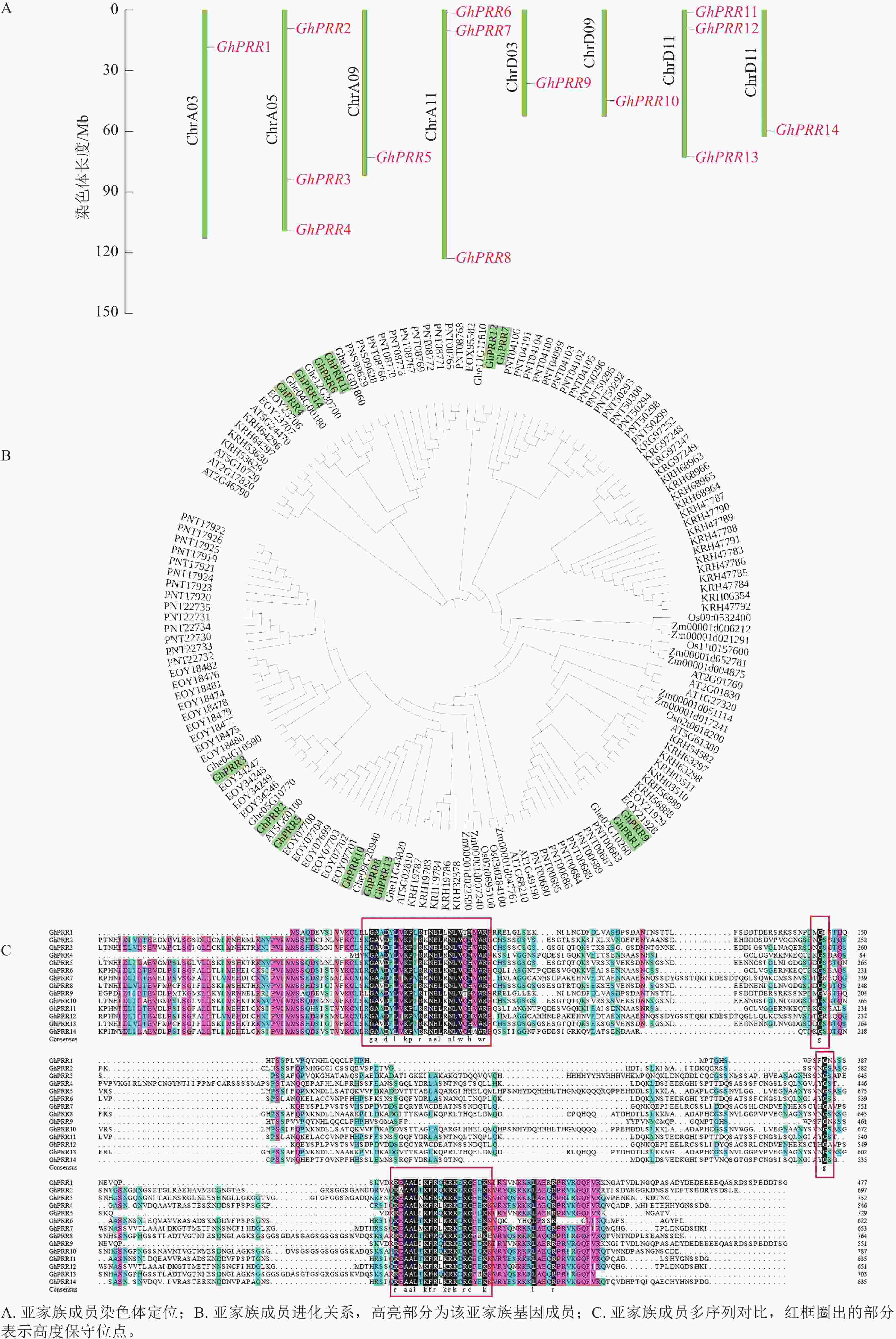
在陆地棉全基因组中共鉴定到14个PRR亚基因家族成员,分别命名为GhPRR1~GhPRR14 (表1)。理化性质分析显示:PRR亚家族成员蛋白有552~775个氨基酸,相对分子量为60.76~85.30 kDa,平均等电点为6.77,酸性蛋白8个,碱性蛋白6个。亚细胞定位结果显示:有11个蛋白定位于细胞核中,2个定位于叶绿体,1个定位于内质网。
表 1 GhPRR亚家族蛋白理化性质
Table 1. Physicochemical properties of protein in GhPRR subfamily
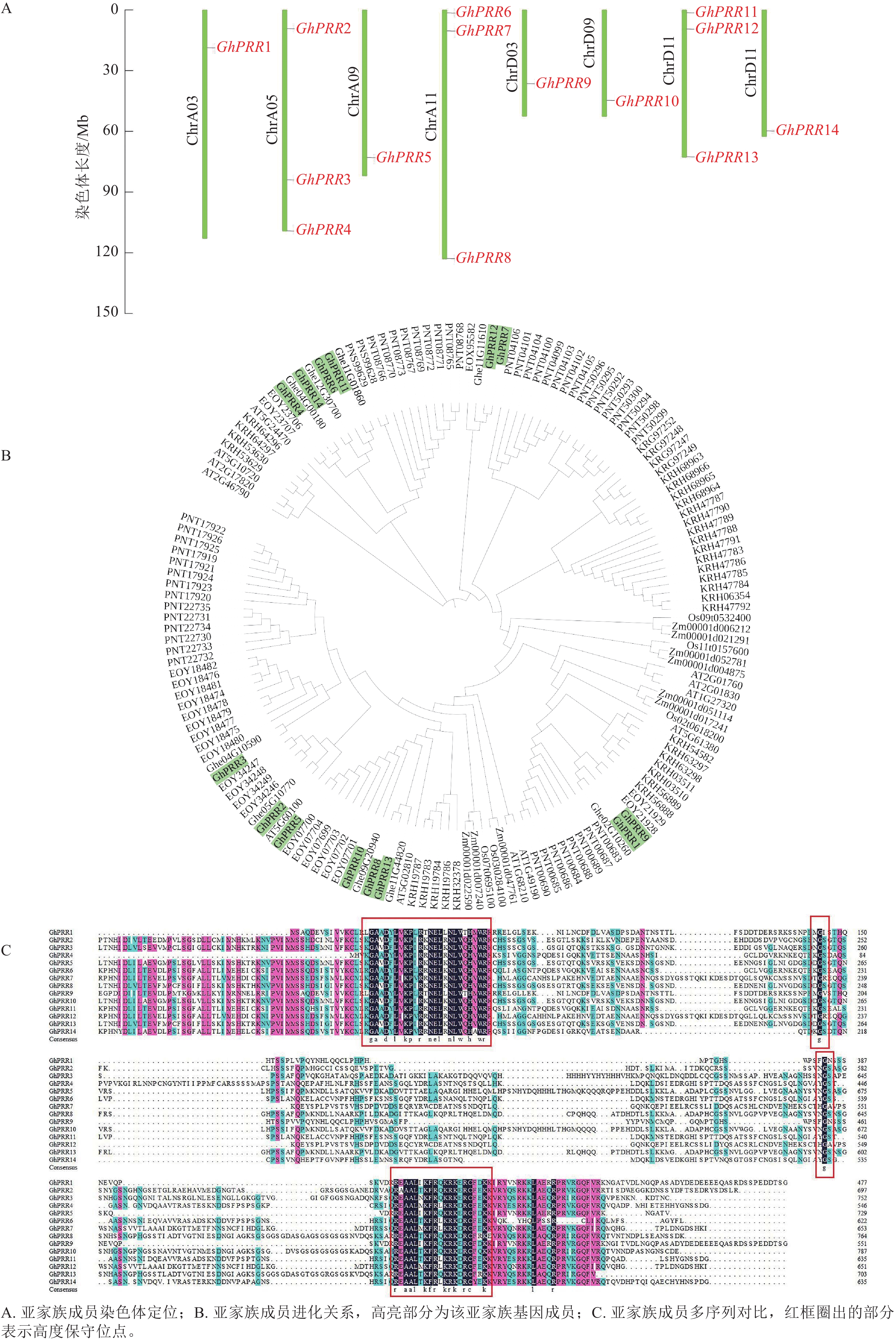
蛋白名称 染色体位置 等电点 分子量/kDa 氨基酸/个 亚细胞定位 亲水性 GhPRR1 ChrA03 5.49 53.53 487 细胞核 −0.876 GhPRR2 ChrA05 7.32 76.62 696 细胞核 −0.725 GhPRR3 ChrA05 8.07 81.80 743 叶绿体 −0.738 GhPRR4 ChrA05 8.55 60.76 552 细胞核 −0.834 GhPRR5 ChrA09 6.33 73.66 669 内质网 −0.592 GhPRR6 ChrA11 6.53 68.72 625 细胞核 −0.565 GhPRR7 ChrA11 5.16 73.11 665 细胞核 −0.688 GhPRR8 ChrA11 6.84 82.54 750 细胞核 −0.689 GhPRR9 ChrD03 5.66 61.96 563 细胞核 −0.743 GhPRR10 ChrD09 7.11 85.30 775 叶绿体 −0.677 GhPRR11 ChrD11 7.56 70.20 638 细胞核 −0.692 GhPRR12 ChrD11 5.62 72.77 661 细胞核 −0.622 GhPRR13 ChrD11 7.91 76.08 691 细胞核 −0.680 GhPRR14 ChrD12 6.67 70.92 645 细胞核 −0.742 用TBtools绘制出染色体定位图(图1A),可观察到GhPRR家族基因保守分布在染色体两端,14个成员分布在8条染色体上,A亚族8个,D亚族6个,其中Chr A05、Chr A11、Chr D11染色体上分别拥有3个该家族的基因,其余染色体均为1个,表明GhPRR亚家族在陆地棉AD亚基因组上呈现不完全均匀分布。
-
从水稻、拟南芥、玉米、草棉、可可、大豆、毛果杨中分别鉴定出5、6、9、9、24、35、49个PRR亚家族成员,与陆地棉GhPRR亚家族成员蛋白构建系统进化树(图1B)。聚类结果显示:陆地棉GhPRR亚家族进化关系最接近的物种是草棉和可可。不同物种中该基因家族成员的数量差异较为明显,也体现出PRR家族成员在不同物种中的多样性。
多重序列比对(图1C)显示:陆地棉GhPRR亚家族蛋白共有4处位点保守性较强,其中有2个高度保守的g位点,说明该家族拥有2段特征结构域(REC与CCT)。
-
蛋白结构分析显示:14个蛋白均含有CCT结构域(图2A)。GhPRR6、GhPRR1、GhPRR2、GhPRR4、GhPRR8和GhPRR13含有REC超家族结构域(cl19078),其余成员含有psREC_PRR结构域(cd17852,属cl19078超家族),亚家族成员有一定的保守性,REC结构域主要功能为核酸识别,CCT结构域主要标志转录因子,以上2个结构的保守性显示了该家族成员的功能。
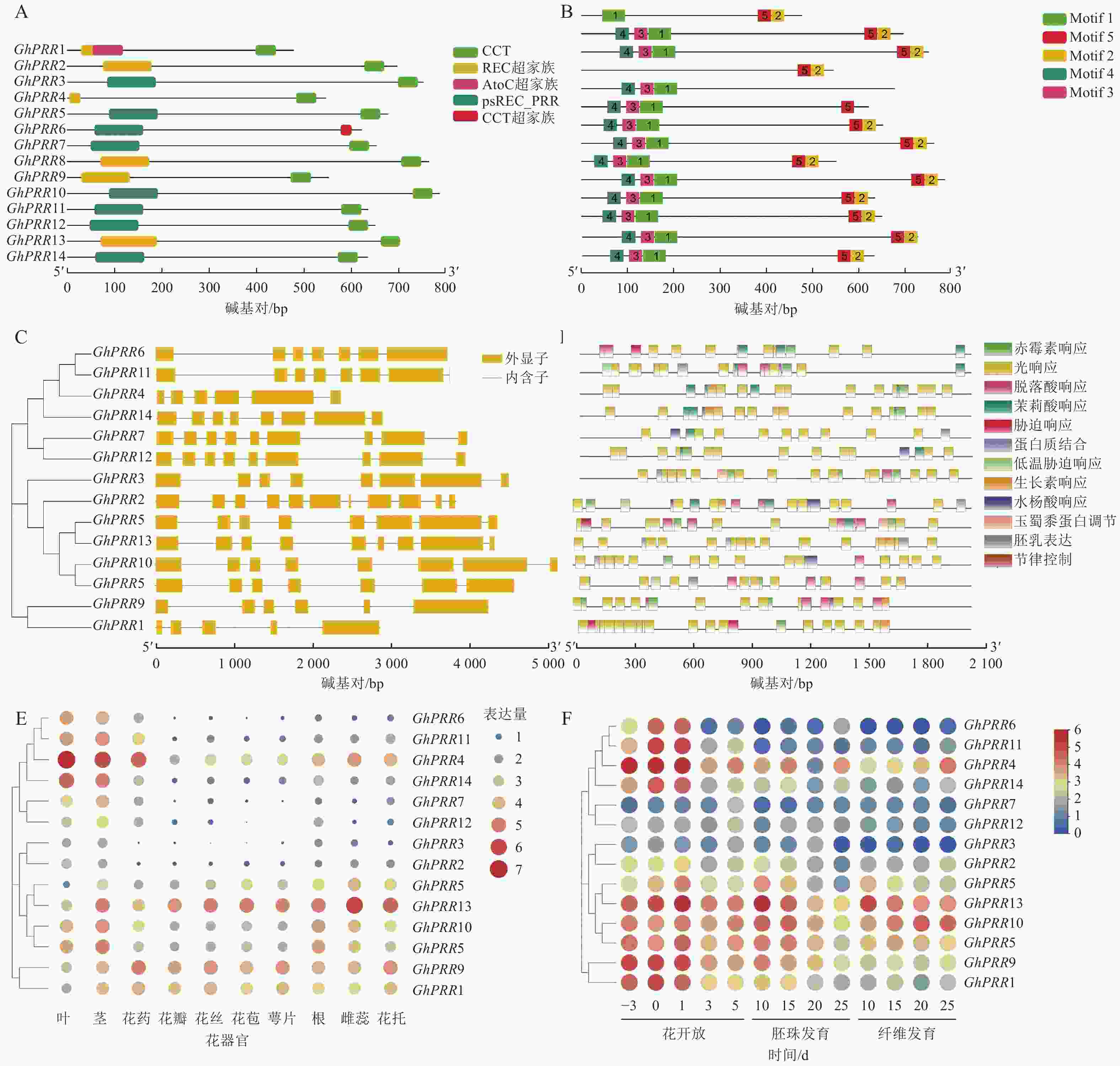
图 2 陆地棉GhPRR亚家族成员结构(A~D)及时空表达量(E~F)分析
Figure 2. Structure (A-D) and spatiotemporal expression (E-F) of GhPRR subfamily members in cotton
MEME分析共得到5个保守基序(图2B)。除GhPRR5外其余成员均含有Motif 2和Motif 5。GhPRR1仅有Motif 1,没有Motif 3和Motif 4,GhPRR4没有Motif 1、Motif 3和Motif 4,其余成员都拥有Motif 1、Motif 3和Motif 4。
基因结构(图2C)显示:该亚家族成员GhPRR1外显子最少(5个),GhPRR2最多(11个),其中,5个成员有8个外显子,3个成员有9个外显子,2个成员有7个外显子,2个成员有6个外显子。最长外显子在3′端较为保守,结构相似度和进化关系基本一致。
-
使用Plant Care对陆地棉GhPRR亚家族成员上游2 000 bp顺式启动子元件进行分析(图2D)发现:主要存在三类顺式元件,一是生长发育响应元件,如光响应元件、生物钟控件;二是激素响应元件,如赤霉素、脱落酸等响应元件;三是非生物胁迫元件,如逆境、盐胁迫等响应元件。其中光响应元件最多(184个),其次为赤霉素响应元件(28个)。说明该亚家族成员主要参与光响应和赤霉素通路。
-
利用公开的转录组数据对陆地棉GhPRR亚家族成员进行组织表达分析(图2E)发现:不同成员组织表达水平差异较大。其中茎叶和花药中表达量最高的是GhPRR4,最少的分别是GhPRR8、GhPRR3和GhPRR2。GhPRR13和GhPRR9在花丝、花苞、花萼中表达量较高,GhPRR2和GhPRR6最少。根中GhPRR13表达量最多,GhPRR7最少,雌蕊花托中GhPRR13表达量最高,GhPRR12和GhPRR2最少。GhPRR4、GhPRR11和GhPRR6可能主要作用于维管组织,GhPRR13和GhPRR9可能作用于花器官组织。
时间表达模式分析(图2F)显示:除GhPRR7、GhPRR12、GhPRR2、GhPRR3和GhPRR10外,GhPRR亚家族其他成员表达量均呈现开花前3 d至开花后1 d逐渐增加,开花后3~5 d逐渐降低的趋势,说明其可能集中在开花前和开花时发挥作用。GhPRR7主要作用在开花后5 d及之后,GhPRR12和GhPRR2可能较少参与开花过程。胚珠中GhPRR13表达量在第10天达到顶峰,说明它在胚珠发育前期可能发挥着一定的作用,GhPRR10、GhPRR5、GhPRR9、GhPRR18、GhPRR3、GhPRR4和GhPRR14也呈现相似的趋势,说明这些基因可能拥有类似的作用模式。纤维发育期间,GhPRR13、GhPRR10和GhPRR4表达量较高,说明这些基因可能参与纤维发育调控。时间模式上,GhPRR13在10~25 d纤维中表达量持续下降,而GhPRR10和GhPRR4则表现出持续上升的趋势,可知GhPRR10和GhPRR4可能参与纤维发育的后期调控,而GhPRR13则参与早期的纤维发育调控。丰富的时空表达说明陆地棉GhPRR家族成员广泛参与到开花前后、胚珠和纤维的发育过程中。
-
使用Plant Care在线工具对该基因上游2 000 bp进行启动子顺式元件分析(表2),发现拟南芥AtTOC1的同源基因GhPRR9上存在着大量的光响应元件,说明光对该基因的转录有着重要的调控作用。除此之外,在GhPRR9基因的启动子区域还存在茉莉酸等激素响应元件,说明该基因可能参与激素相关通路的调节。
表 2 GhPRR9启动子顺式元件预测
Table 2. Cis-acting element prediction of GhPRR9 promoter
名称 起始位置/bp 所在链 功能 名称 起始位置/bp 所在链 功能 ARE 43 − 厌氧胁迫响应 TATA-box 635 − 核心元件 P-box 1 121 − 赤霉素响应 TATA-box 636 − 核心元件 G-box 167 + 光响应 Sp1 1 057 − 光响应 G-box 1 070 + 光响应 G-Box 1 009 − 光响应 A-box 882 − 顺式调节 ABRE 168 + 脱落酸响应 TCCC-motif 871 + 光响应 TGACG-motif 878 + 茉莉酸响应 CAAT-box 249 + 增强区域 TGACG-motif 1 991 − 茉莉酸响应 CAAT-box 354 + 增强区域 Box Ⅱ 1 007 − 光响应 AE-box 535 − 光响应 Box 4 419 + 光响应 GATA-motif 710 + 光响应 MRE 1 513 − 光响应MYB结合 ATCT-motif 1 343 − 光响应 CGTCA-motif 878 − 茉莉酸响应 TATA-box 634 − 核心元件 对陆地棉标准系‘TM-1’进行荧光定量分析(图3A)表明:GhPRR9在花丝、萼片、花托中表达量较高,叶片最低,表明GhPRR9可能更多参与陆地棉的生殖生长。
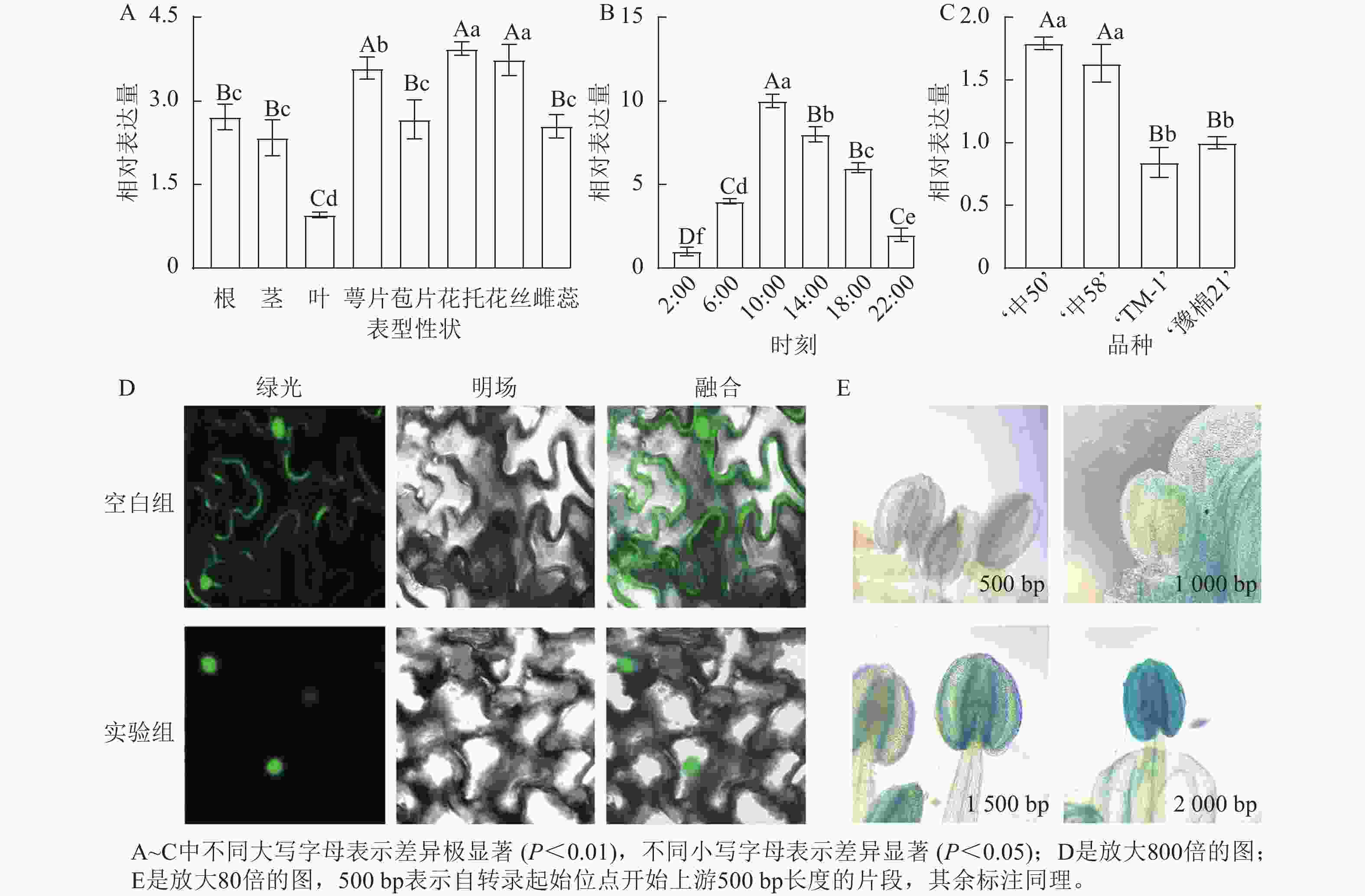
图 3 基因GhPRR9的时空表达量、亚细胞定位和启动子染色
Figure 3. Spatial and temporal expression of GhPRR9 gene, subcellular localization and promoter staining
对在人工光照条件下陆地棉三叶期标准系‘TM-1’顶芽隔4 h取样并进行荧光定量分析(图3B)表明:GhPRR9在光照开始后逐渐积累,并在中午达到顶峰,之后慢慢下降,在光周期内的表达呈现出一定的周期性。
进一步对GhPRR9早熟品种‘中50’‘ZHONG 50’、‘中58’‘ZHONG 58’和晚熟品种‘TM-1’、‘豫棉21号’‘YM21’的表达量分析发现:GhPRR9在早熟品种中表达量显著高于晚熟品种(图3C),说明GhPRR9和早熟性状呈正向相关。
将未转化的GFP质粒和35S::GhPRR9-GFP质粒分别转入农杆菌GV3101,并侵染烟草叶片组织,制作表皮切片置于激光共聚焦显微镜下发现:对照组分布于整个细胞中,而GFP融合蛋白荧光仅分布于细胞核(图3D)。
截取GhPRR9不同长度的启动子与携带GUS报告基因的质粒进行重组,分别转入农杆菌GV3101后通过沾花法侵染拟南芥,获得纯合转基因株系染色观察,结果显示上游500 bp启动子几乎没有表达(图3E),而上游2 000 bp的启动子着色程度最深,说明GhPRR9启动子上游500~2 000 bp内可能存在关键调控元件诱导基因的表达。
-
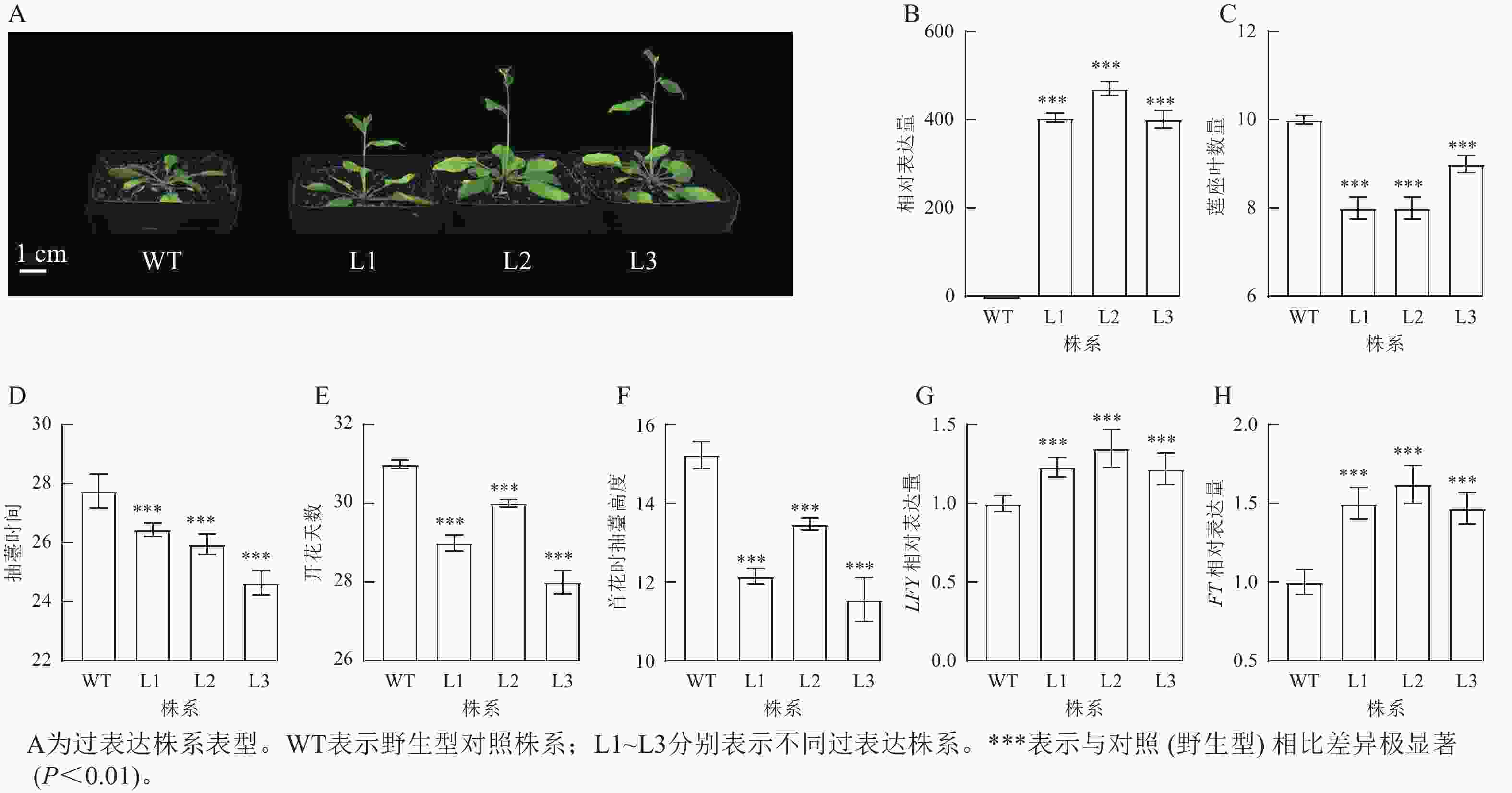
将拟南芥GhPRR9过表达株系培养至抽薹,并观察表型性状(图4A)发现:过表达株系GhPRR9表达量比野生型明显提高(图4B),且转基因过表达株系连座叶数量明显减少(图4C),抽薹时间和开花提前(图4D和图4E),首花抽薹高度极显著矮于野生型(图4F,P<0.01),说明GhPRR9正向调控植物的早花性状。过表达GhPRR9能促进开花关键基因LFY与FT表达(图4G和图4H),表明GhPRR9也可能通过影响关键基因表达调控通路进而影响开花时间。
-
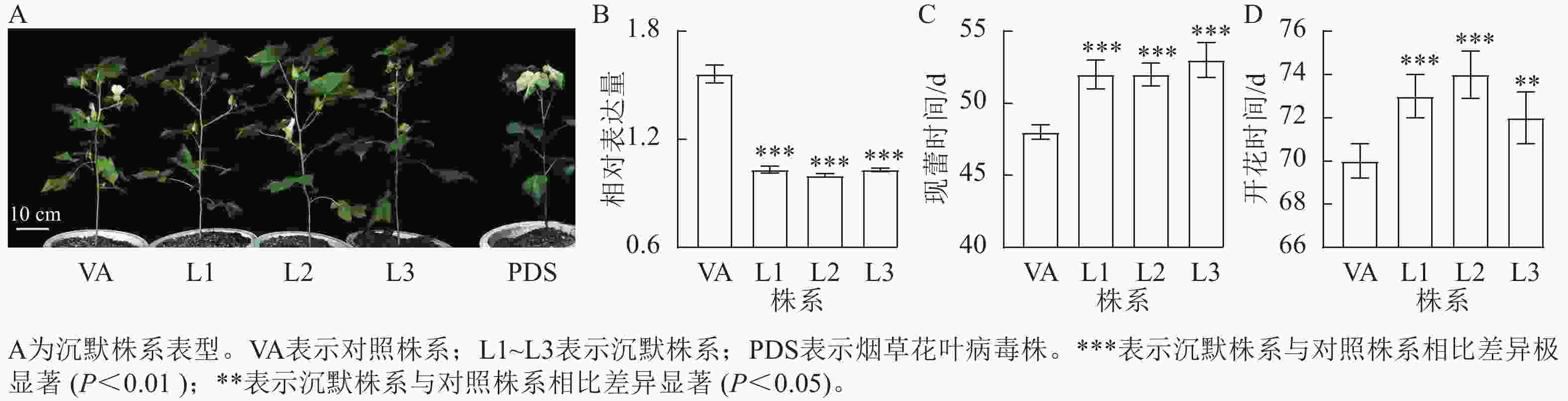
对VIGS沉默株系进行表达量检测发现:沉默株系中,GhPDS株系出现白化表型(图5A),且GhPRR9的表达量极显著降低(图5B,P<0.01),表明GhPRR9基因成功得到了沉默。与对照株系相比,沉默株系现蕾时间延迟约3~4 d,开花时间延迟约2~5 d,表明沉默GhPRR9可推迟开花时间,反向证明其调节陆地棉早花的功能。
-
本研究共鉴定出陆地棉14个GhPRR亚家族成员,成员含有CCT和REC保守结构域,说明其行使转录因子功能。转录组分析显示:大部分成员主要在开花前的茎叶、纤维发育后期和胚珠发育中期发挥作用,表明大部分成员可能存在功能冗余或协同拮抗作用。启动子元件分析显示:陆地棉GhPRR亚家族可能频繁地参与光感效应相关的生理过程,这与在拟南芥的结果中一致,据此可推测其与拟南芥同源基因作用相似。进化分析表明:陆地棉GhPRR亚家族成员基因数量多于拟南芥。前人研究也发现:棉花基因组进化加倍使该家族基因得到了扩增[52]。
对过表达株系研究发现:抽薹日期、开花日期都稍有提前,抽薹高度显著高于同期野生型植株,证明GhPRR9可以使拟南芥花期提前。构建GFP表达载体侵染烟草叶片,表明GhPRR9蛋白定位于细胞核,与生物信息学分析相互印证,进一步确认该基因行使转录因子的功能。对GhPRR9基因1 d内表达水平分析显示:光暗交替条件下基因表达量存在着周期性变化,按照其表达模式推断该基因在光照开始后积累,中午达到顶峰后慢慢下降,这与拟南芥同源基因的表达模式[53]相似,据此推测,陆地棉早花基因GhPRR9可能与拟南芥中的同源基因行使着类似的功能。陆地棉三叶期叶片的基因表达量结果显示:GhPRR9基因在早熟种中表达量高于晚熟种,据此可推断其与早熟性状有正向关联。构建VIGS株系发现:沉默株系高度降低,生育期推迟,反向证明了其促进生育期的功能。启动子分析显示:光和赤霉素可能影响该基因的转录。GUS染色结果显示:启动子上游500~2 000 bp可能存在关键调控元件。有研究显示:陆地棉转录因子与通路主要基因启动子的结合可随温度产生变化[54],且同源转录因子可能存在相互调控的作用[55]。
-
本研究预测了陆地棉GhPRR亚家族的功能和作用模式,找到拟南芥早花基因AtTOC1的陆地棉同源基因GhPRR9,并成功克隆,构建遗传转化株系对其功能进行验证显示:GhPRR9对早花性状存在正向促进作用。
Functional analysis and validation of early flowering gene GhPRR9 in Gossypium hirsutum
-
摘要:
目的 研究早花基因GhPRR9在陆地棉Gossypium hirsutum生长发育过程中的功能,为培养早熟棉品种提供理论依据。 方法 从全基因组筛选鉴定陆地棉GhPRR亚家族成员并研究其结构和表达特征;使用实时荧光定量PCR (RT-qPCR)分析GhPRR9在不同品种、不同组织及1 d内的表达差异;在烟草Nicotiana tabacum叶片中采用瞬时转化法进行亚细胞定位;分段克隆GhPRR9基因启动子并构建β-葡萄糖苷酶基因(GUS)植株进行染色实验;构建拟南芥Arabidopsis thaliana过表达株系,统计开花表型及相关基因表达量;进行病毒诱导沉默(VIGS)实验并观察开花时间差异。 结果 生物信息学分析显示:陆地棉GhPRR亚家族14个成员多数定位于细胞核,结构较为保守且主要在开花前的茎叶中集中表达。RT-qPCR分析显示:陆地棉GhPRR9表达量与品种早熟性呈正向相关,在花器官中更高,且在1 d内呈周期变化。烟草瞬时转化表明:陆地棉GhPRR9分布在细胞核。GUS染色结果显示:启动子上游500~2 000 bp内可能存在关键调控元件。拟南芥过表达株系可观察到早花。VIGS实验显示:基因沉默导致开花时间延迟。 结论 基因GhPRR9对陆地棉早花性状有正向调控作用,在一定程度上能促进陆地棉生长。图5表2参55 Abstract:Objective The objective is to study the function of early flowering gene GhPRR9 in the growth and development of Gossypium hirsutum (cotton), so as to provide theoretical basis for cultivating early maturing cotton cultivars. Method Members of GhPRR subfamily were identified by genome-wide screening and the structure and expression characteristics were studied. Real-time quantitative PCR (RT-qPCR) was used to analyze the expression of GhPRR9 among different varieties, tissues and within a single day. Subcellular localization was performed in Nicotiana tabacum (tobacco) leaves by transient transformation method. The promoter of the gene was cloned and β-D-glucosidase (GUS) plants were constructed for staining experiment. The overexpression lines of Arabidopsis thaliana were constructed to analyze the flowering phenotype data and related gene expression. Virus induced silencing (VIGS) experiment was performed to observe the difference in flowering time. Result Bioinformatics analysis showed that most of the 14 members of the GhPRR subfamily were located in the nucleus, with a relatively conserved structure and mainly expressed in stems and leaves before flowering. RT-qPCR analysis indicated that the expression level of GhPRR9 was positively correlated with early maturity of cultivars, higher in flower organs, and exhibited a periodic change within one day. Transient transformation of tobacco revealed that GhPRR9 was distributed in the nucleus. GUS staining suggested that there might be key regulatory elements in the promoter within 500−2 000 bp upstream. Early flowering was observed in A. thaliana overexpressed strains. VIGS experiment revealed that gene silencing led to delayed flowering. Conclusion Gene GhPRR9 has a positive regulatory effect on the early flowering of upland cotton and can promote its growth and development. [Ch, 5 fig. 2 tab. 55 ref.] -
Key words:
- Gossypium hirsutum /
- family analysis /
- GhPRR9 /
- functional verification /
- early blossoming
-
表 1 GhPRR亚家族蛋白理化性质
Table 1. Physicochemical properties of protein in GhPRR subfamily
蛋白名称 染色体位置 等电点 分子量/kDa 氨基酸/个 亚细胞定位 亲水性 GhPRR1 ChrA03 5.49 53.53 487 细胞核 −0.876 GhPRR2 ChrA05 7.32 76.62 696 细胞核 −0.725 GhPRR3 ChrA05 8.07 81.80 743 叶绿体 −0.738 GhPRR4 ChrA05 8.55 60.76 552 细胞核 −0.834 GhPRR5 ChrA09 6.33 73.66 669 内质网 −0.592 GhPRR6 ChrA11 6.53 68.72 625 细胞核 −0.565 GhPRR7 ChrA11 5.16 73.11 665 细胞核 −0.688 GhPRR8 ChrA11 6.84 82.54 750 细胞核 −0.689 GhPRR9 ChrD03 5.66 61.96 563 细胞核 −0.743 GhPRR10 ChrD09 7.11 85.30 775 叶绿体 −0.677 GhPRR11 ChrD11 7.56 70.20 638 细胞核 −0.692 GhPRR12 ChrD11 5.62 72.77 661 细胞核 −0.622 GhPRR13 ChrD11 7.91 76.08 691 细胞核 −0.680 GhPRR14 ChrD12 6.67 70.92 645 细胞核 −0.742 表 2 GhPRR9启动子顺式元件预测
Table 2. Cis-acting element prediction of GhPRR9 promoter
名称 起始位置/bp 所在链 功能 名称 起始位置/bp 所在链 功能 ARE 43 − 厌氧胁迫响应 TATA-box 635 − 核心元件 P-box 1 121 − 赤霉素响应 TATA-box 636 − 核心元件 G-box 167 + 光响应 Sp1 1 057 − 光响应 G-box 1 070 + 光响应 G-Box 1 009 − 光响应 A-box 882 − 顺式调节 ABRE 168 + 脱落酸响应 TCCC-motif 871 + 光响应 TGACG-motif 878 + 茉莉酸响应 CAAT-box 249 + 增强区域 TGACG-motif 1 991 − 茉莉酸响应 CAAT-box 354 + 增强区域 Box Ⅱ 1 007 − 光响应 AE-box 535 − 光响应 Box 4 419 + 光响应 GATA-motif 710 + 光响应 MRE 1 513 − 光响应MYB结合 ATCT-motif 1 343 − 光响应 CGTCA-motif 878 − 茉莉酸响应 TATA-box 634 − 核心元件 -
[1] 喻树迅. 中国棉花产业百年发展历程[J]. 农学学报, 2018, 8(1): 85−91. YU Shuxun. 100 years of development of China’s cotton industry [J]. Journal of Agriculture, 2018, 8(1): 85−91. [2] 喻树迅, 王寒涛, 魏恒玲, 等. 棉花早熟性研究进展及其应用[J]. 棉花学报, 2017, 29(增刊1): 1−10. YU Shuxun, WANG Hantao, WEI Hengling, et al. Research progress and application of early maturity in upland cotton [J]. Cotton Science, 2017, 29(suppl 1): 1−10. [3] 李培良, 雷亚平, 李亚兵, 等. 中国棉花产业发展现状与未来展望[J]. 农业展望, 2016, 12(12): 38−45. LI Peiliang, LEI Yaping, LI Yabing, et al. Development status quo of China’s cotton industry and its outlook [J]. Agricultural Outlook, 2016, 12(12): 38−45. [4] 喻树迅. 我国棉花生产现状与发展趋势[J]. 中国工程科学, 2013, 15(4): 9−13. YU Shuxun. Present situation and development trend of cotton production in China [J]. Strategic Study of CAE, 2013, 15(4): 9−13. [5] BÄURLE I, DEAN C. The timing of developmental transitions in plants [J]. Cell, 2006, 125(4): 655−664. [6] YU Sha, CAO Li, ZHOU Chuanmiao, et al. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants [J/OL]. eLife, 2013, 2 : e00269[2024-03-20]. DOI: 10.7554/eLife.00269. [7] 刘永平, 杨静, 杨明峰. 植物开花调控途径[J]. 生物工程学报, 2015, 31(11): 1553−1566. LIU Yongping, YANG Jing, YANG Mingfeng. Pathways of flowering regulation in plants [J]. Chinese Journal of Biotechnology, 2015, 31(11): 1553−1566. [8] HELLIWELL C A, WOOD C C, ROBERTSON M, et al. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex [J]. The Plant Journal, 2006, 46(2): 183−192. [9] SRIKANTH A, SCHMID M. Regulation of flowering time: all roads lead to Rome [J]. Cellular and Molecular Life Sciences, 2011, 68(12): 2013−2037. [10] ANDRÉS F, COUPLAND G. The genetic basis of flowering responses to seasonal cues [J]. Nature Reviews Genetics, 2012, 13(9): 627−639. [11] CHOWDHURY Z, MOHANTY D, GIRI M K, et al. Dehydroabietinal promotes flowering time and plant defense in Arabidopsis via the autonomous pathway genes FLOWERING LOCUS D, FVE, and RELATIVE OF EARLY FLOWERING 6 [J]. Journal of Experimental Botany, 2020, 71(16): 4903−4913. [12] TURCK F, FORNARA F, COUPLAND G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage [J]. Annual Review of Plant Biology, 2008, 59(1): 573−594. [13] LI Xu, MA Dingbang, LU S X, et al. Blue light- and low temperature-regulated COR27 and COR28 play roles in the Arabidopsis circadian clock [J]. The Plant Cell, 2016, 28(11): 2755−2769. [14] ROSAS U, MEI Yu, XIE Qiguang, et al. Variation in Arabidopsis flowering time associated with cis-regulatory variation in CONSTANS [J/OL]. Nature Communications, 2014, 5 (1): 3651[2024-03-20]. DOI: 10.1038/ncomms4651. [15] VALVERDE F, MOURADOV A, SOPPE W, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering [J]. Science, 2004, 303(5660): 1003−1006. [16] LEE K, MAS P, SEO P J. The EC-HDA9 complex rhythmically regulates histone acetylation at the TOC1 promoter in Arabidopsis [J/OL]. Communications Biology, 2019, 2 : 143[2024-03-20]. DOI: 10.1038/s42003-019-0377-7. [17] SOY J, LEIVAR P, GONZÁLEZ-SCHAIN N, et al. Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters [J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(17): 4870−4875. [18] ARANA M V, TOGNACCA R S, ESTRAVIS-BARCALÁ M, et al. Physiological and molecular mechanisms underlying the integration of light and temperature cues in Arabidopsis thaliana seeds [J]. Plant, Cell & Environment, 2017, 40(12): 3113−3121. [19] DELIS C, KROKIDA A, TOMATSIDOU A, et al. AtHESPERIN: a novel regulator of circadian rhythms with poly(A)-degrading activity in plants [J]. RNA Biology, 2016, 13(1): 68−82. [20] HEMMES H, HENRIQUES R, JANG I C, et al. Circadian clock regulates dynamic chromatin modifications associated with Arabidopsis CCA1/LHY and TOC1 transcriptional rhythms [J]. Plant & Cell Physiology, 2012, 53(12): 2016−2029. [21] FARINAS B, MAS P. Histone acetylation and the circadian clock: a role for the MYB transcription factor RVE8/LCL5 [J]. Plant Signaling & Behavior, 2011, 6(4): 541−543. [22] LEGNAIOLI T, CUEVAS J, MAS P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought [J]. The EMBO Journal, 2009, 28(23): 3745−3757. [23] ITO S, KAWAMURA H, NIWA Y, et al. A genetic study of the Arabidopsis circadian clock with reference to the TIMING OF CAB EXPRESSION 1 (TOC1) gene [J]. Plant and Cell Physiology, 2009, 50(2): 290−303. [24] PARA A, FARRÉ E M, IMAIZUMI T, et al. PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock [J]. The Plant Cell, 2007, 19(11): 3462−3473. [25] FUJIWARA S, WANG Lei, HAN Linqu, et al. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins [J]. The Journal of Biological Chemistry, 2008, 283(34): 23073−23083. [26] THAIN S C, VANDENBUSSCHE F, LAARHOVEN L J J, et al. Circadian rhythms of ethylene emission in Arabidopsis [J]. Plant Physiology, 2004, 136(3): 3751−3761. [27] WANG Cuiling, WANG Leili, LIU Qingqing, et al. Genome-wide identification and characterization of PRR gene family and their diurnal rhythmic expression profile in maize [J/OL]. International Journal of Genomics, 2022, 2022 : 6941607[2024-03-20]. DOI: 10.1155/2022/6941607. [28] WANG Peiguo, WANG Liwei, ZHANG Lixin, et al. Genomic dissection and diurnal expression analysis reveal the essential roles of the PRR gene family in geographical adaptation of soybean [J/OL]. International Journal of Molecular Sciences, 2022, 23 (17): 9970[2024-03-20]. DOI: 10.3390/ijms23179970. [29] 王浩, 卢银, 顾爱侠, 等. 大白菜lcc-1突变体生物钟核心基因在不同光周期下的表达分析[J]. 河北农业大学学报, 2019, 42(4): 21−28. WANG Hao, LU Yin, GU Aixia, et al. Expression analysis of key clock genes of lcc-1 mutant from Chinese cabbage under different photoperiods [J]. Journal of Hebei Agricultural University, 2019, 42(4): 21−28. [30] 甘卓然, 石文茜, 黎永力, 等. 大豆生物钟基因GmLNK1/2、GmRVE4/8和GmTOC1 CRISPR/Cas9组织表达分析及敲除靶点的鉴定[J]. 作物学报, 2020, 46(8): 1291−1300. GAN Zhuoran, SHI Wenqian, LI Yongli, et al. Identification of CRISPR/Cas9 knockout targets and tissue expression analysis of circadian clock genes GmLNK1/2, GmRVE4/8, and GmTOC1 in soybean [J]. Acta Agronomica Sinica, 2020, 46(8): 1291−1300 [31] 刘璇, 张丽, 巩檑, 等. 生物钟对植物非生物胁迫应答调控的进展[J]. 基因组学与应用生物学, 2019, 38(9): 4160−4166. LIU Xuan, ZHANG Li, GONG Lei, et al. Progress of circadian clock regulation of abiotic stress response in plants [J]. Genomics and Applied Biology, 2019, 38(9): 4160−4166. [32] LI Xiao, WU Yuanlong, CHI Huabin, et al. Genome-wide identification and characterization of the genes involved in the flowering of cotton [J/OL]. International Journal of Molecular Sciences, 2022, 23 (14): 7940[2024-03-20]. DOI: 10.3390/ijms23147940. [33] YIN Xiaoyu, LIU Ye, ZHAO Hang, et al. GhCOL2 positively regulates flowering by activating the transcription of GhHD3A in upland cotton (Gossypium hirsutum L.) [J/OL]. Biochemical Genetics, 2024[2024-03-20]. DOI: 10.1007/s10528-024-10727-3. [34] 柳晔. 陆地棉CO蛋白对FT的调控研究[D]. 曲阜: 曲阜师范大学, 2021. LIU Ye. Regulation of CO Protein on FT in Upland Cotton [D]. Qufu: Qufu Normal University, 2021. [35] 张盼, 范术丽, 宋美珍, 等. 陆地棉开花相关基因GhFLP1的克隆与功能验证[J]. 棉花学报, 2016, 28(3): 199−207. ZHANG Pan, FAN Shuli, SONG Meizhen, et al. Cloning and functional analysis of the flowering-related gene GhFLP1 from upland cotton (Gossypium hirsutum L. ) [J]. Cotton Science, 2016, 28(3): 199−207. [36] 李洁, 范术丽, 宋美珍, 等. 陆地棉GhSPL3基因的克隆、亚细胞定位及表达分析[J]. 棉花学报, 2012, 24(5): 414−419. LI Jie, FAN Shuli, SONG Meizhen, et al. Cloning, subcellular localization and expression analysis of GhSPL3 gene in Gossypium hirsutum L. [J]. Cotton Science, 2012, 24(5): 414−419. [37] 张爱, 王彩香, 宿俊吉, 等. 陆地棉MADS-box家族基因鉴定及组织特异性表达分析[J]. 棉花学报, 2020, 32(5): 404−417. ZHANG Ai, WANG Caixiang, SU Junji, et al. Identification of MADS-box family and analysis of tissue specific expression in Gossypium hirsutum L. [J]. Cotton Science, 2020, 32(5): 404−417. [38] CHENG Xiaoqian, WANG Hantao, WEI Hengling, et al. The MADS transcription factor GhAP1.7 coordinates the flowering regulatory pathway in upland cotton (Gossypium hirsutum L. ) [J/OL]. Gene, 2021, 769 : 145235[2024-03-20]. DOI: 10.1016/j.gene.2020.145235. [39] 孟超敏, 耿翡翡, 卿桂霞, 等. 陆地棉磷高效基因GhMGD3的克隆与表达分析[J]. 浙江农林大学学报, 2022, 39(6): 1203−1211. MENG Chaomin, GENG Feifei, QING Guixia, et al. Cloning and expression of phosphorus efficient gene GhMGD3 in Gossypium hirsutum [J]. Journal of Zhejiang A&F University, 2022, 39(6): 1203−1211. [40] 孟超敏, 耿翡翡, 卿桂霞, 等. 陆地棉低磷胁迫应答基因GhGDPD1的克隆与表达分析[J]. 浙江农林大学学报, 2023, 40(4): 723−730. MENG Chaomin, GENG Feifei, QING Guixia, et al. Cloning and expression analysis of low phosphorus stress response gene GhGDPD1 in Gossypium hirsutum [J]. Journal of Zhejiang A&F University, 2023, 40(4): 723−730. [41] LI Libei, ZHAO Shuqi, SU Jinji, et al. High-density genetic linkage map construction by F2 populations and QTL analysis of early-maturity traits in upland cotton (Gossypium hirsutum L. ) [J/OL]. PLoS One, 2017, 12 (8): e0182918[2024-03-20]. DOI: 10.1371/journal.pone.0182918. [42] LI Libei, ZHANG Chi, HUANG Jianqing, et al. Genomic analyses reveal the genetic basis of early maturity and identification of loci and candidate genes in upland cotton (Gossypium hirsutum L. ) [J]. Plant Biotechnology Journal, 2021, 19(1): 109−123. [43] YU Jing, JUNG S, CHENG Chunhuai, et al. CottonGen: the community database for cotton genomics, genetics, and breeding research [J/OL]. Plants, 2021, 10 (12): 2805[2024-03-20]. DOI: 10.3390/plants10122805. [44] SWARBRECK D, WILKS C, LAMESCH P, et al. The Arabidopsis information resource (TAIR): gene structure and function annotation [J]. Nucleic Acids Research, 2007, 36(D1): D1009−D1014. [45] GOODSTEIN D M, SHU Shengqiang, HOWSON R, et al. Phytozome: a comparative platform for green plant genomics [J]. Nucleic Acids Research, 2012, 40(D1): D1178−D1186. [46] LU Shennan, WANG Jiyao, CHITSAZ F, et al. CDD/SPARCLE: the conserved domain database in 2020 [J]. Nucleic Acids Research, 2020, 48(D1): D265−D268. [47] ARTIMO P, JONNALAGEDDA M, ARNOLD K, et al. ExPASy: SIB bioinformatics resource portal [J]. Nucleic Acids Research, 2012, 40(W1): W597−W603. [48] KUMAR S, STECHER G, LI M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms [J]. Molecular Biology and Evolution, 2018, 35(6): 1547−1549. [49] CHEN Chengjie, WU Ya, LI Jiawei, et al. TBtools-Ⅱ: A “one for all, all for one” bioinformatics platform for biological big-data mining [J]. Molecular Plant, 2023, 16(11): 1733−1742. [50] BAILEY T L, JOHNSON J, GRANT C E, et al. The MEME Suite [J]. Nucleic Acids Research, 2015, 43(W1): W39−W49. [51] LESCOT M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Research, 2002, 30(1): 325−327. [52] WANG Jingjing, DU Zhaohai, HUO Xuehan, et al. Genome-wide analysis of PRR gene family uncovers their roles in circadian rhythmic changes and response to drought stress in Gossypium hirsutum L. [J/OL]. PeerJ, 2020, 8 : e9936[2024-03-20]. DOI: 10.7717/peerj.9936. [53] RONALD J, DAVIS S J. Making the clock tick: the transcriptional landscape of the plant circadian clock [J/OL]. F1000Research, 2017, 6 : 951[2024-03-20]. DOI: 10.12688/f1000research.11319.1. [54] LIU Lingyun, JIA Mingzhu, WANG Shengnan, et al. Identification and characterization of cotton PHYTOCHROME-INTERACTING FACTORS in temperature-dependent flowering [J]. Journal of Experimental Botany, 2023, 74(12): 3765−3780. [55] ZHANG Xiaohong, REN Zhongying, HU Genhai, et al. Functional divergence of GhAP1.1 and GhFUL2 associated with flowering regulation in upland cotton (Gossypium hirsutum L. ) [J/OL]. Journal of Plant Physiology, 2022, 275 : 153757[2024-03-20]. DOI: 10.1016/j.jplph.2022.153757. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.20240267






 下载:
下载: