-
自噬(autophagy)是进化上保守的细胞内物质的降解途径,在真核生物中广泛存在。与其他降解系统相比,自噬最显著的特征是能降解细胞中如生物分子、细胞器甚至入侵的微生物等几乎所有的物质[1]。自噬在饥饿条件下的营养循环中起着重要作用,能将营养物质重新分配进行物质循环[2-4]。LI等[5]对玉米Zea mays研究发现:自噬在缺氮环境下调控氮回收利用以增加农作物产量;LI等[6]在谷子Setaria italica中发现:自噬相关基因SiATG8在响应氮饥饿耐受中表达上调;AVIN-WITTENBERG等[7]发现:碳饥饿条件下自噬能通过降解蛋白质等生物大分子产生氨基酸作为替代性碳源,供植物体重新利用;在碳饥饿条件下,自噬会影响中枢代谢导致拟南芥Arabidopsis thaliana ATG(autophagy-related gene)突变体表型与野生型差异明显。BARROS等[8]发现:在碳饥饿情况下,自噬中的代谢物循环和呼吸的替代途径对维持正常的呼吸功能起作用。自噬在植物体自身生长和发育中也起着重要的作用[9-10],如参与根尖的细胞生长和分化[11-12]、减数分裂后花药的发育[13]、气管元素的分化[14]、对硝酸盐的利用率[9]、对光合作用产生淀粉粒的降解[15]、在衰老叶片中叶绿体的降解[16-18]、植物体衰老过程中能量的利用[19]以及植物夜间生长供能物质的利用[20]等等。GHIGLIONE等[21]对小麦Triticum aestivum的研究发现:自噬对其小花发育过程起到作用;KURUSU等[13]对水稻Oryza sativa研究发现:自噬缺陷Osatg7和Osatg9突变体在正常生长条件下表现出完全的孢子体雄性不育和有限的花药开裂。IZUMI等[19]对水稻的研究发现:自噬可在叶片变暗引起能量限制时被激活,对其具有重要的保护作用;TOYOOKA等[22]对发芽薇甘菊Mikania micrantha种子子叶的超微结构进行观察,认为微自噬机制可能参与储备淀粉的降解;YOSHIMOTO等[23]发现:当环境中的营养供应有限时,植物细胞中的养分调动需要自噬。WANG等[15]发现:细胞自噬能够在夜间对叶片淀粉的降解起作用。有实验室证实,自噬可以在晚间为植物提供能量[20]。毛竹茎秆生长发育过程的研究分析主要在解剖结构[24]、反射光谱[25]、光合酶[26-27]、碳水化合物代谢[28-30]、蛋白质组[31]、基因组学[32-33]等方面。对模式植物拟南芥和水稻中的研究表明:自噬对植物生长起重要作用[7, 15, 19-20],而在毛竹Phyllostachys edulis中关于自噬与其快速生长关系的研究鲜见报道。本研究探讨了毛竹茎秆快速生长过程中PeATG1和PeATG4基因的表达变化,以期为毛竹快速生长内在的自噬调控机制及分子生物学基础提供参考。
-
供试毛竹采自浙江省杭州市临安区(29°56′~30°23′N,118°51′~119°52′E)现代毛竹纯林示范园。该区属中亚热带季风气候,温暖湿润,四季分明,年平均气温15.8 ℃,具有春多雨,夏湿热,秋气爽,冬干冷的气候特点。全年平均日照时数1 847.3 h,年平均降水量1 628.6 mm, 无霜期234.0 d,森林覆盖率76.5%。毛竹林土壤属山地红壤,土层深度60 cm以上。
-
试验材料为2018年当年生毛竹笋竹,2018年5月初,在毛竹出笋后生长至(3.0 ± 0.2)m时,分别在6个时间点(10:00、14:00、18:00、22:00、2:00、6:00)选取生境条件一致,生长状况良好,基径约15 cm自然状态下的毛竹笋竹,从茎秆地上部分的基部将其伐倒,将节间按照从基部至顶部的顺序编号(1~N)。每间隔2个节间进行取样,从基部至顶部依次选取编号为1、4、7、10、13、16、19、22的节间。取茎秆节间下部外层绿色组织,取样厚度为2 mm,用于电镜观察样品后,取下,迅速将样品放进液氮中冷冻,存于-80 ℃备用。选取3株笋竹,每株作为1个独立实验,共3次重复。
-
将笋竹的第4、7、10、13节间的茎秆切成0.2 mm × 5.0 mm的小块置于体积分数2.5%戊二醛溶液中,4 ℃固定24 h。然后,将样品置于0.1 mol·L-1磷酸缓冲液(PBS, pH 7.0)中漂洗3次,15 min·次-1,再置于质量分数1%锇酸中固定2 h。之后,用PBS缓冲液漂洗3次固定材料后,使用各级梯度乙醇(体积分数30%、50%、70%、80%、90%、95%和100%)脱水处理15 min,过渡到纯丙酮处理20 min;在45 ℃下用环氧树脂(Epon 812)浸透包埋经脱水处理的材料25 h,之后在70 ℃条件下加热聚合48 h;使用LEICA EM UC7型超薄切片机修整包埋样品并切片,再分别置于柠檬酸铅溶液和醋酸双氧铀50%乙醇饱和溶液双染色10 min,在Hitachi H-65型(日本, HITACHI公司)电镜上观察,并拍照记录结果。
-
基于毛竹基因组数据库中PeATG1和PeATG4的基因序列设计基因全长和定量引物,选择毛竹PeNTB基因作为荧光定量的内参基因,并使用在线网站Primer 3设计引物(),由有康生物科技公司合成(表 1)。
表 1 本研究所用的引物名称
Table 1. PCR primers used in this study
基因名称 序列 用途 PeATG1-F TTCTACCTCACCCCTACCGA 基因全长扩增 PeATG1-R TTCTTCTTGTTGCGCTCGAG PeATG4-F CTGTCAAGCGTTGTCGTG 基因全长扩增 PeATG4-R AACCAATAGCCAGGGATG PeATG1-F CACCGACATCATCGTCCGCTAC 荧光定量聚合酶链式反应 PeATG1-R TCGCCCTCCCGTGCCTTATC PeATG4-F ATGGCTCTTTATGTTGTTTC 荧光定量聚合酶链式反应 PeATG4-R GTGGGATGTACCTTGGAT PeNTB-F TCTTGTTTGACACCGAAGAGGAG 荧光定量聚合酶链式反应 PeNTB-R AATAGCTGTCCCTGGAGGAGTTT -
分别取适量6个时间段的毛竹笋竹茎秆第7节间,使用改良Trizol法提取茎秆组织总RNA。使用NanoDrop® ND-1000分光光度计检测样品RNA的含量与纯度,并通过质量分数1.2%琼脂糖电泳检测样品RNA的完整性。
-
以上述RNA为模板,冰上配置20 μL RT-PCR体系:包括gDNA Eraser 1.0 μL,5×gDNA Eraser Buffer 2.0 μL,RT Primer Mix 1.0 μL,Prime Script RT Enzyme MixⅠ1.0 μL,5×PrimeScript Buffer 2(for Real Time)4.0 μL,总RNA 500 ng,加RNase Free dH2O至总体积20.0 μL。体系置于PCR仪中,37 ℃孵育15 min,85 ℃加热5 s。将通过反转录得到的cDNA存于-20 ℃下用于实时荧光定量分析。
-
根据毛竹数据库PeATG1和PeATG4基因序列设计基因全长引物(表 1),扩增这2个基因的系列片段,扩增程序分别为:94 ℃ 5 min;94 ℃ 30 s,52 ℃ 30 s,72 ℃ 70 s,共30个循环;72 ℃ 10 min。通过质量分数1.2%琼脂糖凝胶电泳检测PCR扩增产物,送往有康生物科技公司测序。
-
采用TaKaRa公司SYBR®Premix Ex TaqTM(Perfect Real Time)试剂盒,冰上配置20 μL反应体系:10 μL SYBR Premix Ex TaqTM,0.8 μL Forward Primer,0.8 μL Reverse Primer,2.0 μL反转录cDNA模版,6.4 μL灭菌蒸馏水。反应于Bio-Rad CFX manager 3.1 PCR仪上进行,采用两步法扩增标准程序:95 ℃预变性3 min,95 ℃下10 s、60 ℃下30 s,循环39次。以内参为对照,每个样品重复3次。反应完成后,得到含所有样品的记录点曲线,得出循环阈值(Ct值)。
-
所有数据均为3次重复,荧光定量数据按照公式计算:相对表达量=2-∆∆Ct[34],使用内参基因校正拷贝数,利用Origin 9.0软件进行统计分析和作图。统计方法采用单因素方差分析(one-way ANOVA),对笋竹PeATG1及PeATG4基因表达量分别进行Turkey比较。
-
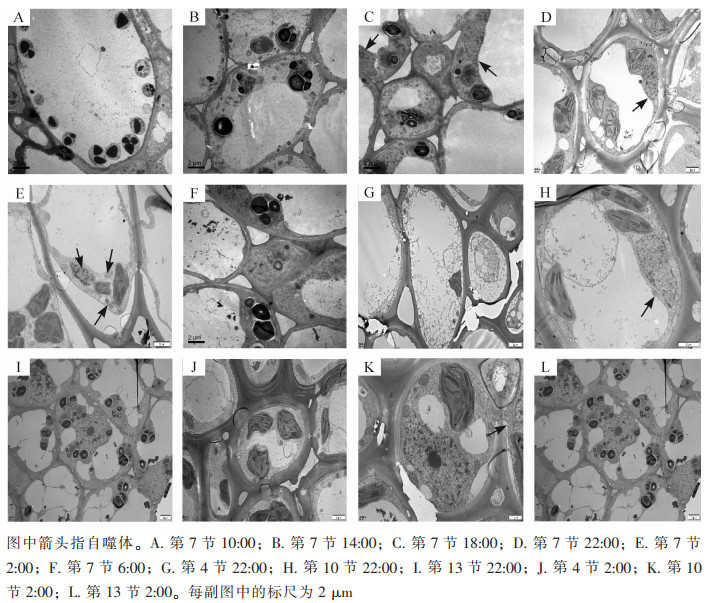
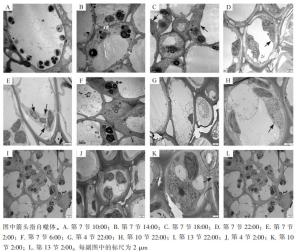
使用透射电子显微镜监测毛竹茎秆第7节间组织不同时间段的自噬活性(图 1),在10:00和14:00的节间中观察到大量淀粉粒存在,未发现自噬体(图 1A和图 1B);在18:00的节间观察到淀粉粒和自噬体(图 1C);22:00第7节中观察到包裹有淀粉粒的自噬体(图 1D);2:00第7节中观察到经典的双层膜自噬体(图 1E);6:00第7节中未观察到自噬体,有少量淀粉粒(图 1F)。
-
对毛竹茎秆快速生长节24 h自噬活性监测发现:自噬活性在22:00和2:00较强,进一步对22:00和2:00的毛竹茎秆不同节间组织第4、7、10、13节的自噬活性进行监测,22:00和2:00第4节中未观察到淀粉粒和自噬体(图 1G和图 1J),第7节和第10节中观察到淀粉粒和自噬体(图 1D、图 1E、图 1H和图 1K),第13节中均未观察到自噬体,有大量淀粉粒(图 1I和图 1L)。
-
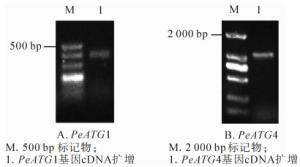
以确定良好纯度和完整度的毛竹茎秆总RNA的反转录产物cDNA为模板,通过普通PCR扩增PeATG1及PeATG4基因全长,并通过质量分数1.2%琼脂糖凝胶电泳检测扩增产物(图 2),条带清晰且单一,获得与预期片段大小一致、长度分别为392和1 087 bp扩增产物。
-
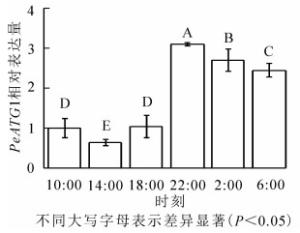
24 h内毛竹茎秆PeATG1表达变化量如图 3所示。PeATG1在22:00表达丰度最高,2:00和6:00的表达丰度次之,再次为10:00和18:00的表达量,14:00的表达量最低。从18:00开始,毛竹茎秆PeATG1表达量显著增加,到22:00达到最高水平,分别是14:00、10:00、18:00、6:00和2:00的4.8、3.1、3.0、1.3和1.1倍(P<0.05)。从10:00到6:00,表达量总体呈先下降后上升再下降趋势。PeATG1在夜间的表达量显著高于白天,在2:00及6:00的表达量较22:00有所下降,但仍显著高于白天。
-
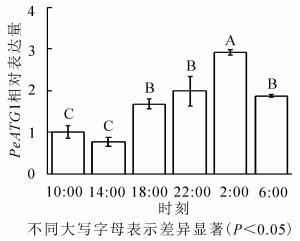
24 h内毛竹茎秆PeATG4表达变化量(图 4)显示:PeATG4在2:00表达丰度最高,其次为22:00、6:00和18:00,再次为10:00,14:00的表达量最低。毛竹茎秆PeATG4表达量从18:00开始增加,到2:00达最高水平,分别是14:00、10:00、18:00、6:00和22:00的3.8、2.9、1.7、1.6和1.5倍(P<0.05)。从10:00到6:00,表达量呈先下降后上升再下降的趋势。PeATG4在夜间的表达量显著高于白天,在6:00表达量较2:00显著下降,但仍显著高于白天的水平。
-
植物依靠光合作用获得生命活动所需的能量与物质。植物在黑暗条件下,光反应不再进行,但仍然需要营养用以维持生命活动、细胞的基本结构与功能[35]。此时,淀粉的降解便为细胞有序进行夜间活动提供了能量来源。除了淀粉降解的经典途径外,WANG等[15]研究发现:植物细胞中的自噬能够在叶片淀粉降解中发挥作用。IZUMI等[20]发现:自噬可以通过夜间的碳代谢提供能源替代物如游离氨基酸来提高夜间的能源供应,有助于植物生长,证实自噬可以在夜间为植物提供能量。本研究通过24 h自噬活性监测,在22:00和2:00毛竹竹笋第7节茎秆组织中观察到自噬体的存在,而白天在相同部位未观察到,表明自噬在夜间被激活。同时在该部位中也有淀粉粒,表明自噬可能与淀粉的分解有关,通过分解淀粉提供能源,促进毛竹夜间快速生长,这与WANG等[15]对拟南芥自噬体定量统计的研究相一致。在监测了24 h自噬活性,确认自噬在夜间被激活后,进一步在22:00和2:00对第4、7、10和13节进行夜间自噬监测,不同部位的自噬活性存在差别。毛竹茎秆下部表面包被的笋箨已逐渐脱落,其组织中无淀粉粒和自噬体,该部位已完成快速生长,发育较为成熟,为其上部组织的生长发育提供了营养物质运输通道,将竹篼中储存的碳水化合物向上运输至快速生长的部位,供其代谢消耗所需;中部正处于快速生长阶段,自噬体的存在为分解淀粉及其他胞内物质,从而促进该部位快速生长提供了快速供能;而茎秆上部较幼嫩,尚未进入快速生长阶段,未观察到自噬体。在毛竹笋竹茎秆尚未萌发枝叶前的快速生长期内,茎秆的光合能力较弱,营养物质和能量主要来源于周围的成竹[30]。本研究结果表明:在快速生长部位存在较多的自噬体,可能对分解由周围成竹通过竹鞭运输至新生笋竹的碳水化合物的分解利用起到积极作用。以上结果表明,毛竹快速生长期,茎秆中部较多的自噬体及其较强的分解淀粉能力可能是维持毛竹笋竹快速生长的原因之一。
自噬涉及广泛的生理过程,其生物学意义是通过关键基因组分之间的相互作用与各种细胞途径相互作用来解释的[36]。ATG1是细胞自噬过程中的重要蛋白,与ATG13形成ATG1/ATG13蛋白激酶复合体,响应营养物质的需求,起始自噬小体的形成;ATG4对ATG8的酶切加工是ATG8进入类泛素连接系统的前提[37]。因此,ATG1和ATG4是自噬过程的关键因子之一。到目前为止,在模式植物拟南芥和水稻中有关ATG1和ATG4基因的鉴定及其在细胞自噬过程中的功能研究比较集中。拟南芥中丝氨酸/苏氨酸蛋白激酶ATG1是调控自噬的关键组分ATG1/ATG13蛋白激酶复合体的核心组分,对自噬小体到液泡的转运十分重要。ATG8耦合系统是自噬体成膜过程中的一个类泛素化系统,ATG8蛋白在细胞自噬中发挥泛素化作用的前提是其蛋白前体的C端经半胱氨酸蛋白ATG4剪切后,暴露其C末端甘氨酸残基[38]。结果表明:PeATG1在夜间的表达加强,显著高于白天,诱导自噬小泡形成。PeATG4在夜间也增强表达,显著高于白天,使自噬体得以形成来分解茎秆组织细胞内的淀粉,满足毛竹茎秆进行夜间生长所需的营养供给。在白天毛竹主要进行光合作用合成营养物质供夜间持续生长所需,与诱导自噬小泡形成有关的PeATG1和调控自噬体形成的PeATG4表达水平均较低。PeATG1和PeATG4基因的表达水平在适应黑暗的前4 h内均有所上升,PeATG1表达量在22:00达到峰值,PeATG4在2:00达到最高表达水平,然后在白天下降到基础水平,这与WANG等[15]对烟草Nicotiana tabacum夜间不同时间点ATG基因的表达变化的研究相一致。以上结果表明:这2个ATG基因的转录水平在夜间显示出不同的动态变化,这表明潜在的细胞自噬机制是活跃的。
关于毛竹茎秆快速生长相关基因的研究,李丹丹等[29]发现:与海藻糖合成相关的PeTPS1在夜间表达增强,糖信号调控关键激酶PeSnRK1在夜间表达量先升高后降低,T6P/SnRK1信号共同调节笋竹夜间快速生长;祝巧鸣等[39]发现:与糖皮质激素代谢酶相关的11β-羟类固醇脱氢酶家族基因PeHSD1在毛竹快速生长时期表达量最低,与毛竹快速生长负相关;叶家其等[40]发现:大量的赤霉素类基因参与到毛竹的快速生长过程,且对赤霉素关键基因进行表达分析,发现GA20ox、GA3ox和GA2ox以及赤霉素受体GID1和正调控基因GID2都在竹笋的形态学上部有大量表达。本研究中对PeATG1及PeATG4进行定量分析发现,两者在总体上趋势一致,均在夜间毛竹快速生长阶段增强表达,白天恢复至基础水平,表明PeATG1和PeATG4共同协调参与自噬过程,进而调节毛竹茎秆快速生长。PeATG1与PeATG4在达到表达最高峰的时间点有所不同,PeATG1在22:00达最高表达水平,PeATG4则在2:00表达量最高,可能是由于它们在诱导自噬发生、调控自噬体形成过程中发挥的作用不同。
综上所述,处于快速生长阶段的毛竹茎秆内存在较高活性的自噬体和PeATG1的PeATG4基因表达量,这为此时期茎秆中存在细胞自噬提供了有力证据。自噬活性和PeATG1、PeATG4的表达量随时间由白天到黑夜呈现规律性变化,表明毛竹茎秆不同时间段的生长发育存在明显差异,白天生长相对缓慢,夜间生长迅速;且不同节间的自噬活性也不相同,表明毛竹茎秆不同节间的生长发育也存在明显差异,茎秆中部节间生长较快,下、上部节间生长相对缓慢;茎秆中具有细胞自噬途径,参与调节茎秆的快速生长过程。研究成果对明确毛竹速生生长机制具有参考价值。
Analysis of PeATG1/PeATG4 gene expression in Phyllostachys edulis during rapid growth
-
摘要:
目的 探讨毛竹Phyllostachys edulis茎秆快速生长与PeATG1和PeATG4基因表达的关系。 方法 以毛竹笋竹茎秆为材料,采用透射电镜监测笋竹快速生长期不同时间(10:00、14:00、18:00、22:00、2:00和6:00)和不同部位(第4、7、10、13节)的自噬活性,并用实时荧光定量聚合酶链式反应(qRT-PCR)分析技术测定第7节PeATG1和PeATG4基因表达量。 结果 对毛竹茎秆24 h自噬活性监测,22:00和2:00在第7节和第10节观察到自噬体;第4节和第13节没有观察到自噬体。在夜间,PeATG1和PeATG4转录水平表达增强,PeATG1表达量在22:00最高,分别是18:00和6:00的3.0倍和1.3倍(P < 0.05);PeATG4表达量在2:00最高,分别是18:00和6:00的1.7和1.6倍(P < 0.05)。 结论 毛竹茎秆不同时间段的生长发育存在显著差异,夜间有自噬体形成,PeATG1和PeATG4基因表达量较高,茎秆生长迅速。 Abstract:Objective The aim is to discuss the relationship between rapid growth of stems of Phyllostachys edulis and gene expression of PeATG1 and PeATG4. Method Stems of P. edulis shoots were used as materials, the autophagic activity at different periods (10:00, 14:00, 18:00, 22:00, 2:00, and 6:00) and in different parts (internodes 4, 7, 10 and 13) was monitored by transmission electron microscope (TEM), and the expression of PeATG1 and PeATG4 genes in the 7th internode was determined by real-time fluorescence quantitative polymerase chain reaction (qRT-PCR). Result The autophagosomes were observed at 22:00 and 2:00 in internodes 7 and 10 during 24 h monitoring, while no autophagosome was observed in internodes 4 or 13. The expression of PeATG1 and PeATG4 enhanced at night. The expression of PeATG1 was highest at 22:00, which was 3.0-fold and 1.3-fold of 18:00 and 6:00 respectively (P < 0.05). The expression of PeATG4 was highest at 2:00, which was 1.7-fold and 1.6-fold of 18:00 and 6:00 respectively (P < 0.05). Conclusion The research showed significant differences in growth and development of P. edulis at different stages. Autophagosomes were formed at night, and the expressions of PeATG1 and PeATG4 were high, which resulted in rapid growth of stems. -
Key words:
- botany /
- Phyllostachys edulis /
- stem /
- autophagy /
- PeATG1 /
- PeATG4 /
- rapid growth
-
表 1 本研究所用的引物名称
Table 1. PCR primers used in this study
基因名称 序列 用途 PeATG1-F TTCTACCTCACCCCTACCGA 基因全长扩增 PeATG1-R TTCTTCTTGTTGCGCTCGAG PeATG4-F CTGTCAAGCGTTGTCGTG 基因全长扩增 PeATG4-R AACCAATAGCCAGGGATG PeATG1-F CACCGACATCATCGTCCGCTAC 荧光定量聚合酶链式反应 PeATG1-R TCGCCCTCCCGTGCCTTATC PeATG4-F ATGGCTCTTTATGTTGTTTC 荧光定量聚合酶链式反应 PeATG4-R GTGGGATGTACCTTGGAT PeNTB-F TCTTGTTTGACACCGAAGAGGAG 荧光定量聚合酶链式反应 PeNTB-R AATAGCTGTCCCTGGAGGAGTTT -
[1] SUZUKI H, OSAWA T, FUJIOKA Y, et al. Structural biology of the core autophagy machinery[J]. Curr Opin Struct Biol, 2017, 43:10-17. [2] THOMPSON A R, DOELLING J H, SUTTNGKAKUL A. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways[J]. Plant Physiol, 2005, 138(4):2097-2110. [3] TOYOOKA K, MORIYASU Y, GOTO Y, et al. Protein aggregates are transported to vacuoles by macroautophagic mechanism in nutrient-starved plant cells[J]. Autophagy, 2006, 2(2):96-106. [4] HOFIUS D, SCHULTZ-LARSEN T, JOENSEN J, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis[J]. Cell, 2009, 137(4):773-783. [5] LI Faqiang, CHUNG T, PENNINGTON J G, et al. Autophagic recycling plays a central role in maize nitrogen remobilization[J]. Plant Cell, 2015, 27(5):1389-1408. [6] LI W W, CHEN M, LIU J M, et al. Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setariaitalica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis[J]. Biochem Biophys Res Commun, 2015, 468(4):800-806. [7] AVIN-WITTENBERG T, BAJDZIENKO K, WITTENBERG G, et al. Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation[J]. Plant Cell, 2015, 27(2):306-322. [8] BARROS J A S, CAVALCANTI J H, MEDEIROS D B, et al. Autophagy deficiency compromises alternative pathways of respiration following energy deprivation in Arabidopsis thaliana[J]. Plant Physiol, 2017, 175(1):62-76. [9] GUIBOILEAU A, OSPINA L A, YOSHIMOTO K, et al. Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability[J]. New Phytol, 2013, 199(3):683-694. [10] MARSHALL R S, VIERSTRA R D. Autophagy:the master of bulk and selective recycling[J]. Ann Rev Plant Biol, 2018, 69(1):173-208. [11] INOUE Y, SUZUKI T, HATTORI M, et al. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells[J]. Plant Cell Physiol, 2006, 47(12):1641-1652. [12] YANO K, SUZUKI T, MORIYASU Y. Constitutive autophagy in plant root cells[J]. Autophagy, 2007, 3(4):360-362. [13] KURUSU T, KOYANO T, HANAMATA S, et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development[J]. Autophagy, 2014, 10(5):878-888. [14] KWON S I, CHO H J, JUNG J H, et al. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis[J]. Plant J, 2010, 64(1):151-164. [15] WANG Yan, YU Bingjie, ZHAO Jinping, et al. Autophagy contributes to leaf starch degradation[J]. Plant Cell, 2013, 25(4):1383-1399. [16] ISHIDA H, YOSHIMOTO K, IZUMI M, et al. Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process[J]. Plant Physiol, 2008, 148(1):142-155. [17] WADA S, ISHIDA H, IZUMI M, et al. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves[J]. Plant Physiol, 2009, 149(2):885-893. [18] IZUMI M, WADA S, MAKINO A, et al. The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis[J]. Plant Physiol, 2010, 154(3):1196-1209. [19] IZUMI M, HIDEMA J, WADA S. et al. Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation[J]. Plant Physiol, 2015, 167(4):1307-1320. [20] IZUMI M, HIDEMA J, MAKINO A, et al. Autophagy contributes to nighttime energy availability for growth in Arabidopsis[J]. Plant Physiol, 2013, 161(4):1682-1693. [21] GHIGLIONE H O, GONZALEZ F G, SERRAGO R, et al. Autophagy regulated by day length determines the number of fertile florets in wheat[J]. Plant J, 2008, 55(6):1010-1024. [22] TOYOOKA K, OKAMOTO T, MINAMIKAWA T. Cotyledon cells of Vigna mungo seedlings use at least two distinct autophagic machineries for degradation of starch granules and cellular components[J]. Cell Biol, 2001, 154(5):973-982. [23] YOSHIMOTO K, TAKANO Y, SAKAI Y. Autophagy in plants and phytopathogens[J]. FEBS Lett, 2010, 584(7):1350-1358. [24] 陈登举, 高培军, 吴兴波, 等.毛竹茎秆叶绿体超微结构及其发射荧光光谱特征[J].植物学报, 2013, 48(6):635-642. CHEN Dengju, GAO Peijun, WU Xingbo, et al. Chloroplast ultrastructure and emission fluorescence spectrum characteristics for stems of Phyllostachys edulis[J]. Chin Bull Bot, 2013, 48(6):635-642. [25] 刘琳, 王玉魁, 王星星, 等.毛竹出笋后快速生长期茎秆色素含量与反射光谱的相关性[J].生态学报, 2013, 33(9):2703-2711. LIU Lin, WANG Yukui, WANG Xingxing, et al. Correlation between pigment content and reflectance spectrum of Phyllostachys edulis stems during its rapid growth stage[J]. Acta Ecol Sin, 2013, 33(9):2703-2711. [26] 王星星, 刘琳, 张洁, 等.毛竹出笋后快速生长期内茎秆中光合色素和光合酶活性的变化[J].植物生态学报, 2012, 36(5):456-462. WANG Xingxing, LIU Lin, ZHANG Jie, et al. Changes of photosynthetic pigment and photosynthetic enzyme activity in stems of Phyllostachys pubescens during rapid growth stage after shooting[J]. Chin J Plant Ecol, 2012, 36(5):456-462. [27] 孙建飞, 翟建云, 马元丹, 等.毛竹快速生长期茎秆不同节间光合色素和光合酶活性的差异[J].植物学报, 2018, 53(6):773-781. SUN Jianfei, ZHAI Jianyun, MA Yuandan, et al. Differences in photosynthetic pigments and photosynthetic enzyme activities in different internodes of Phyllostachys edulis during rapid growth stage[J]. Chin Bull Bot, 2018, 53(6):773-781. [28] 程路芸, 温星, 马丹丹, 等.毛竹快速生长过程中碳水化合物的时空变化[J].浙江农林大学学报, 2017, 34(2):261-267. CHENG Luyun, WEN Xing, MA Dandan, et al. Spatial and temporal change of carbohydrates during rapid growthprocesses of Phyllostachys edulis[J]. J Zhejiang A&F Univ, 2017, 34(2):261-267. [29] 李丹丹, 许馨露, 翟建云, 等.毛竹笋竹快速生长期可溶性糖质量分数与PeTPS1/PeSnRK1基因表达分析[J].浙江农林大学学报, 2017, 34(6):1016-1023. LI Dandan, XU Xinlu, ZHAI Jianyun, et al. Soluble sugar content and PeTPS1/PeSnRK1 gene expression in Phyllostachys edulis during rapid growth[J]. J Zhejiang A&F Univ, 2017, 34(6):1016-1023. [30] 翟建云, 孙建飞, 马元丹, 等.毛竹快速生长期茎秆不同节间碳水化合物代谢的变化[J].竹子学报, 2018, 37(1):42-48. ZHAI Jianyun, SUN Jianfei, MA Yuandan, et al. Changes of carbohydrates metabolism in differentinternodes of Phyllostachys edulis during rapid growth period[J]. J Bamboo Res, 2018, 37(1):42-48. [31] CUI Kai, HE Caiyun, ZHANG Jianguo, et al. Temporal and spatial profiling of internode elongation-associated protein expression in rapidly growing culms of bamboo[J]. J Proteome Res, 2012, 11(4):2492-2507. [32] ZHOU Mingbing, YANG Ping, GAO Peijun, et al. Identification of differentially expressed sequence tags in rapidly elongating Phyllostachys pubescens internodes by suppressive subtractive hybridization[J]. Plant Mol Biol Rep, 2011, 29(1):224-231. [33] PENG Zhenhua, LU Ying, LI Lubing, et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla)[J]. Nat Gen, 2013, 45(4):456-461. [34] LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method[J]. Methods, 2001, 25(4):402-408. [35] 任晨霞, 龚清秋.细胞自噬在植物碳氮营养中作用的研究进展[J].中国细胞生物学学报, 2014, 36(4):407-414. REN Chenxia, GONG Qingqiu. Progress on the involvement of plant autophagy incarbon and nitrogen utilization[J]. Chin J Cell Biol, 2014, 36(4):407-414. [36] HAN Shaojie, WANG Yan, ZHENG Xiyin, et al. Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana[J]. Plant Cell, 2015, 27(4):1316-1331. [37] 刘洋, 张静, 王秋玲, 等.植物细胞自噬研究进展[J].植物学报, 2018, 53(1):5-16. LIU Yang, ZHANG Jing, WANG Qiuling, et al. Research progress in plant autophagy[J]. Chin Bull Bot, 2018, 53(1):5-16. [38] KIRISAKO T, ICHIMURA Y, OKADA H, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway[J]. J Cell Biol, 2000, 151(2):263-276. [39] 祝巧鸣, 娄帅通, 杨勇, 等.毛竹茎秆快速生长相关基因PeHSD1的功能探究[J].分子植物育种, 2018, 16(19):6269-6275. ZHU Qiaoming, LOU Shuaitong, YANG Yong, at al. The preliminary functional study of bamboo culm rapid growth related gene PeHSD1[J]. Mol Plant Breeding, 2018, 16(19):6269-6275. [40] 叶家其, 张毓婷, 傅鹰, 等.毛竹茎秆伸长过程中赤霉素生物合成、降解和信号转导关键基因的鉴定及表达分析[J].生物工程学报, 2019, 35(6):1-20. YE Jiaqi, ZHANG Yuting, FU Ying, et al. Genome-wide identification and expression analysis of gibberellin biosynthesis, metabolism and signaling familygenes in Phyllostachys edulis[J]. Chin J Biotechnol, 2019, 35(6):1-20. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.2020.01.006






 下载:
下载: