-
光合作用在绿叶中进行,也可在非同化器官叶柄,绿花,花萼,绿色水果,球果,茎组织,甚至根中发生[1-4]。非同化器官光合作用,无论表现为净光合作用还是内部二氧化碳(CO2)再固定,都是增加碳获取的重要方式[5]。蔡锡安等[6]研究发现植物茎秆组织能同化树干向生境排放的60%~90%CO2。LIU等[7]研究发现茎光合作用能够促进旱柳Salix matsudana扦插幼苗的器官发育。在干旱林地或沙漠中发现一些常见木本植物依赖绿茎作为碳同化器官[8]。ÁVILA-LOVERA等[9]发现霍金斯树Cercidium praecox在雨季主要由叶片进行光合作用,在旱季叶片完全脱落,主要由绿茎发生光合作用。植物碳同化方式多种多样,花环结构与3种不同细胞类型被认为是C4光合碳同化途径关键特征[10]。HIBBERD等[11]发现在典型C3植物烟草Nicotiana tabacum和芹菜Apium graveolens中,木质部和韧皮部周围的茎和叶柄细胞表现出NADP-苹果酸酶(NADP-ME)型细胞类型特征,这些细胞需要的碳来自于呼吸作用。SHEN等[12]发现水稻Oryza sativa叶片中脉具有C4光合特性。王莹等[13]发现木本植物丁香Syringa meyeri,银白杨Populus alba,落叶松Larix gmelinii绿色茎秆组织中均表现出典型C4光合生化特征。毛竹Phyllostachys edulis属禾本科Gramineae亚热带竹种,自然分布于中国南方省份,具有生长快,产量高,固碳能力强等特点。目前,关于毛竹光合作用前人已做了一些工作,温星等[14]发现毛竹幼叶叶绿素质量分数和净光合速率随其生长发育不断增加,从幼叶生长至第15天完全展叶,叶片就具备正常的光合生理功能。陈登举等[15]发现毛竹茎秆中光合色素含量较高,叶绿体发育完整,其类囊体垛叠程度高于叶片并含有淀粉粒。同时,毛竹在糖代谢[16]、激素调节[17]、非生物胁迫[18]、基因组鉴定与表达分析[19−20]等方面研究取得了较好的进展,但毛竹笋快速生长期茎秆光合碳同化分子特征的研究鲜有报道。本研究以快速生长阶段毛竹笋竹为研究对象,测定毛竹茎秆不同节间C3光合碳同化途径关键酶核酮糖-1,5-二磷酸羧化酶/加氧酶(Rubisco)、C4光合碳同化关键酶磷酸烯醇式丙酮酸羧化酶(PEPC)、丙酮酸磷酸双激酶(PPDK)、NADP-ME、NADP-苹果酸脱氢酶(NADP-MDH)、磷酸烯醇式丙酮酸羧激酶(PEP-CK)活性及各个关键酶基因相对表达量,探讨毛竹快速生长期笋竹茎秆光合碳同化规律,以期为进一步揭示毛竹茎秆快速生长机制提供参考。
HTML
-
供试毛竹来自浙江省杭州市临安区青山湖街道毛竹高效示范园。2018年4月中旬至5月下旬,选取生长状况良好,生境相同,株高6.0±0.2 m,基径约15 cm的当年自然生毛竹笋竹,在10:00−12:00从茎秆基部将其伐倒,由基部至顶部方向第1节间起顺序编号,依次取编号为7、10、13、16、19节间,切取各节间1/3处下部的绿色外层组织,取样厚度2~3 mm。成熟叶片样品采自成竹冠层,在同一生境和采样时间段选取生长状况良好的自然生成竹,取其冠层无斑点、无病害、延展性良好、大小均一的深绿色成熟叶片,去叶脉。节间组织和叶片样品均液氮冷冻,−80 ℃冰箱保存备用。取5株毛竹笋竹,5次重复,每株为1个独立实验。
-
酶液提取参照BERVEILLER等的方法[20],稍有改动。取待测毛竹笋竹绿色茎秆和叶片各0.5 g分置于研磨机中,加入5 mL预冷的提取液,研磨至匀浆。12 000 r·min−1离心30 min,取上清液即为粗酶液。Rubisco酶活性的测定参照姜振升等[21]的方法。PEPC和NADP-ME活性测定参照BERVEILLER等的方法[20]。NADP-MDH活性测定参照JOHNSON等的方法[22]。 PEPCK活性测定参照BURNELL的方法[23]。 PPDK活性测定参照HATCH等的方法[24]。
-
毛竹笋竹茎秆和叶片总RNA的提取使用宝生物工程(大连)有限公司(TaKaRa) RNAiso Plus (Code No.9109)*试剂盒,按其说明书的方法提取。使用Nano-100微量分光光度计检测RNA浓度与质量,用琼脂糖(添加量为12 g·L−1)凝胶电泳筛选可用RNA。反转录使用宝生物工程(大连)有限公司(TaKaRa)PrimeScriptTMRT reagent Kit with gDNA Eraser (Perfect Real Time)试剂盒,按其说明书的方法合成cDNA,反转录后的cDNA放于−20 ℃保存。
-
通过美国国家生物信息中心(NCBI, https://www.ncbi.nlm.nih.gov/)和PLAZ 4.0 Monocots(https://bioinformatics. psb.ugent.be/plaza/versions/plaza_v4_monocots/)搜索毛竹光合关键酶基因CDS序列,所有用于目的基因表达的定量引物序列(表1)均利用NCBI引物设计工具Primer-BLAST设计,同源比对后由浙江有康生物科技有限公司合成。各个基因表达分析使用宝生物工程(大连)有限公司(TaKaRa)TB Green™ Premix Ex Taq ™ Ⅱ (Tli RNaseH Plus)试剂盒进行实时荧光定量聚合酶链式反应(qPCR)。反应体系20 μL,采用两步法PCR扩增标准程序在Bio-Rad CFX manager 3.1 PCR仪上扩增。定量结果采用2-ΔΔCt的方法计算[25],选用毛竹PeNTB作为内参基因,每个样品3次重复。
基因名 5′→3′ 3′→5′ PeNTB TCTTGTTTGACACCGAAGAGGAG AATAGCTGTCCCTGGAGGAGTT Percbl ATCGTGCTCGCGGTATCTTT ATTTCGGTCAGAGCTGGCAT PePEPC TCGAGGGTTCGGACTGTTTGG GAGTTGGCTGAGTTCTTCGGA PeNADP-MDH CTGGATTTGGCCTTGGTGTTG TGTGCGCGGATATTTTTCTG PeNADP-ME TTTAGTGCAGAAGATCGTGGGG ATGCCAATACCTTGCACTCCC PePEP-CK CAGTACCACCACCACTACACCAC CTGGACGTGATGAACGAACCC PePPDK GAAACGTACGGGCAAAAGTG GCTCTACCATCTTGATCGCTG Table 1. Primers used in this study
-
所有数据均为5次重复的平均值±标准差。利用Origin 9.0软件(OriginLab公司, 美国)进行统计分析和作图。采用one-way ANOVA进行Tukey多重比较(P<0.01)。
1.1. 材料
1.2. 方法
1.2.1. 酶液提取与酶活测定
1.2.2. 毛竹笋竹茎秆和叶片总RNA的提取、质量检测及反转录
1.2.3. 荧光定量PCR
1.3. 数据分析
-
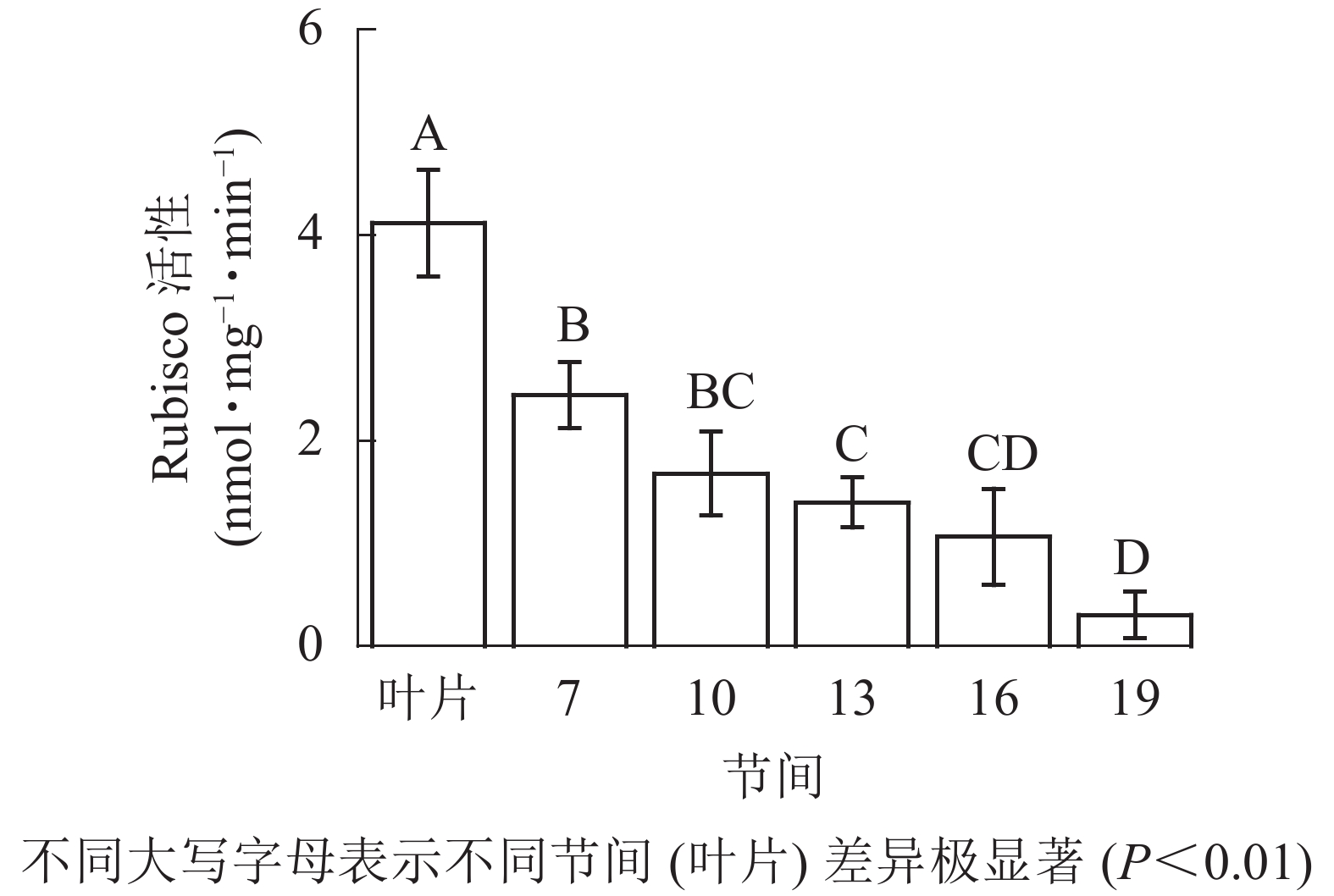
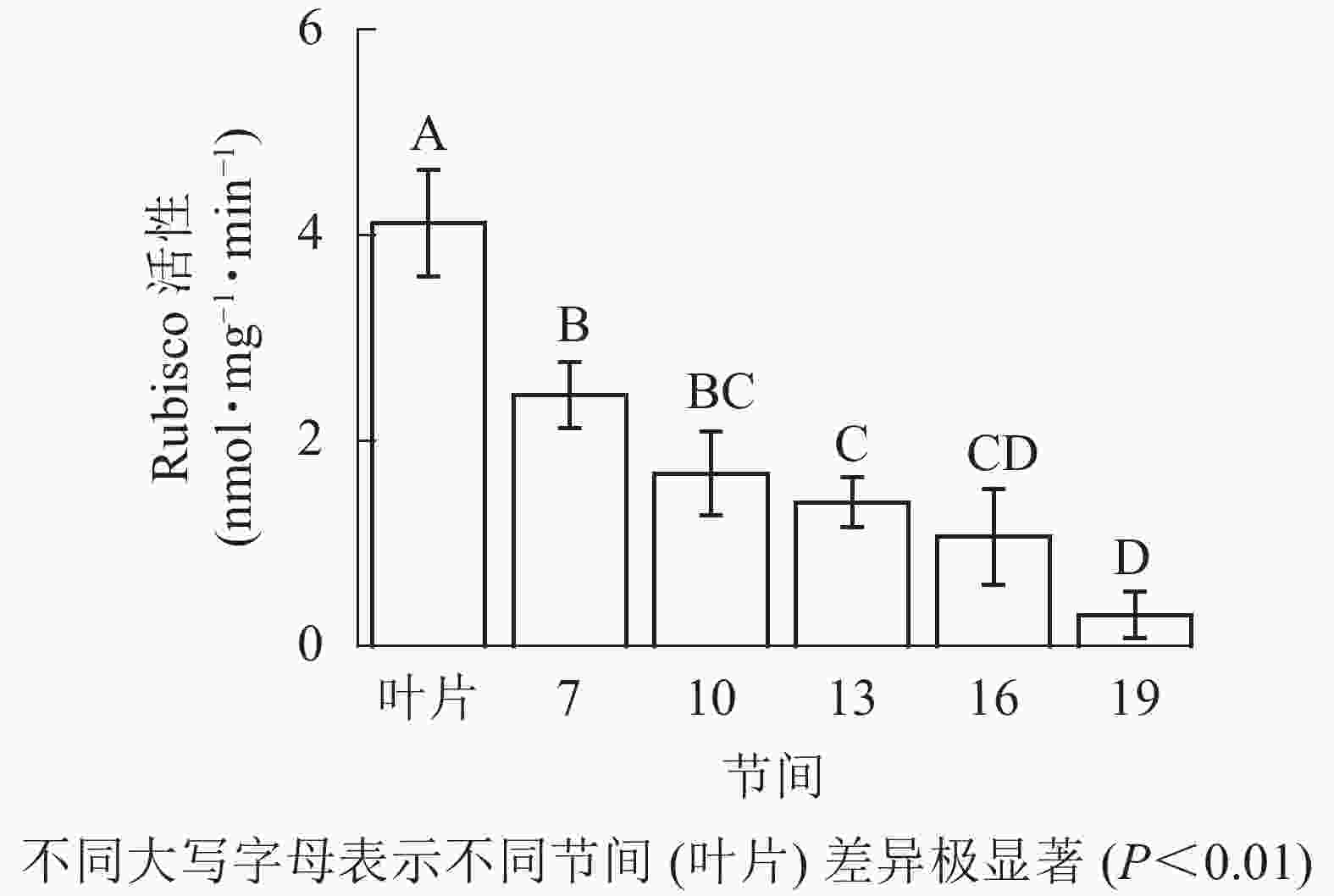
由图1可知:毛竹叶片Rubisco初始活性最高,极显著高于茎秆快速生长节间(P<0.01),分别是第7、10、13、16、19节间的1.69、2.45、2.95、3.87、13.40倍;随着节间的升高,Rubisco初始活性呈下降趋势,第7节间Rubisco初始活性高于其他节间。第19节间Rubisco活性最低,极显著低于叶片和第7节间(P<0.01),分别下降了92.5%和87.4%。
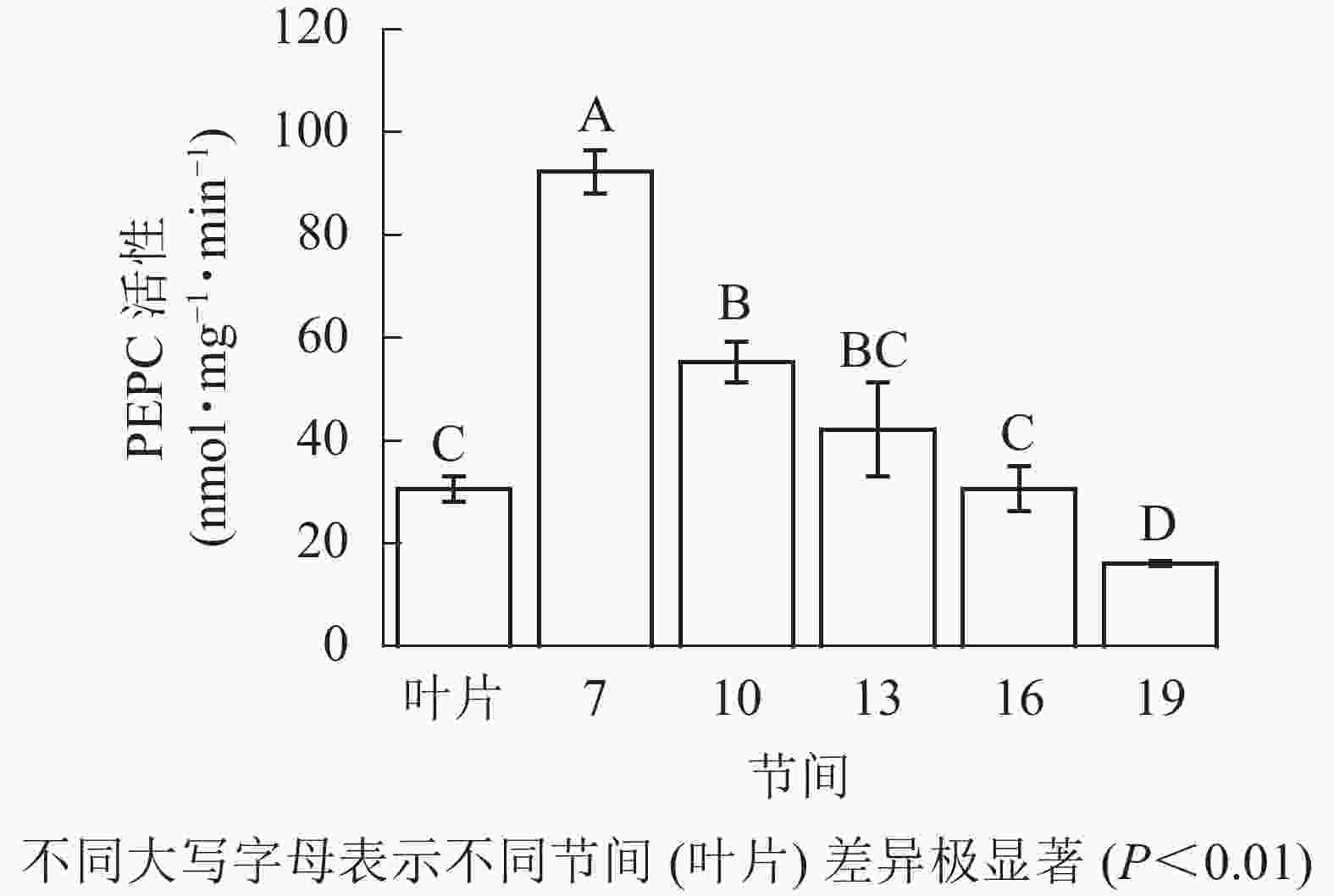
由图2可知:茎秆第7节间PEPC活性最高,极显著高于茎秆其他节间及叶片(P<0.01),分别是第10、13、16、19节间和叶片的1.70、2.19、3.01、5.70和3.01倍;随着节间的升高,PEPC活性呈下降趋势,第19节间PEPC活性最低,与第7节间相比下降了82.5%(P<0.01)。毛竹茎秆第7~19节间PEPC活性整体呈先极显著下降(P<0.01)而后在低水平趋于平稳的趋势。
-
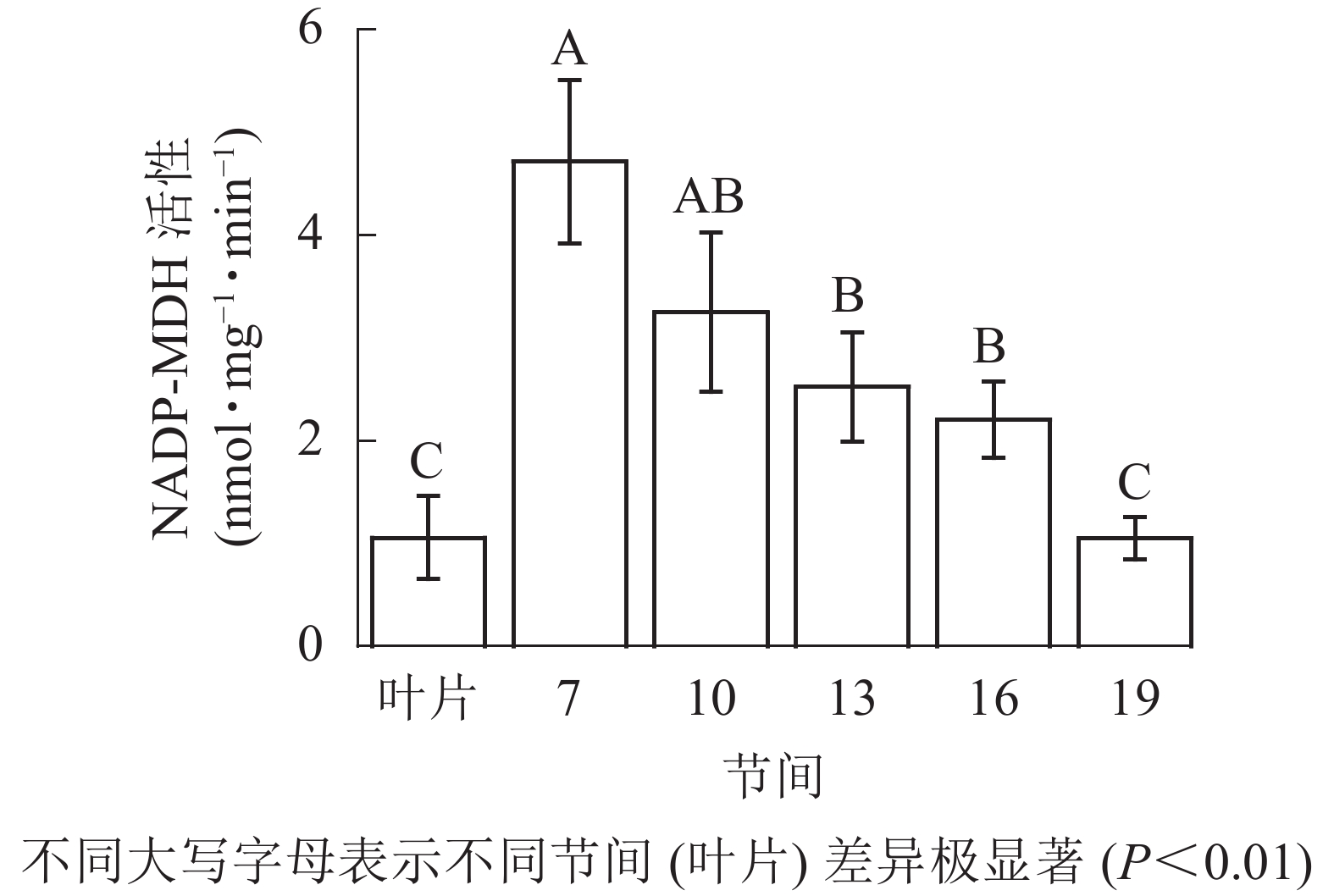
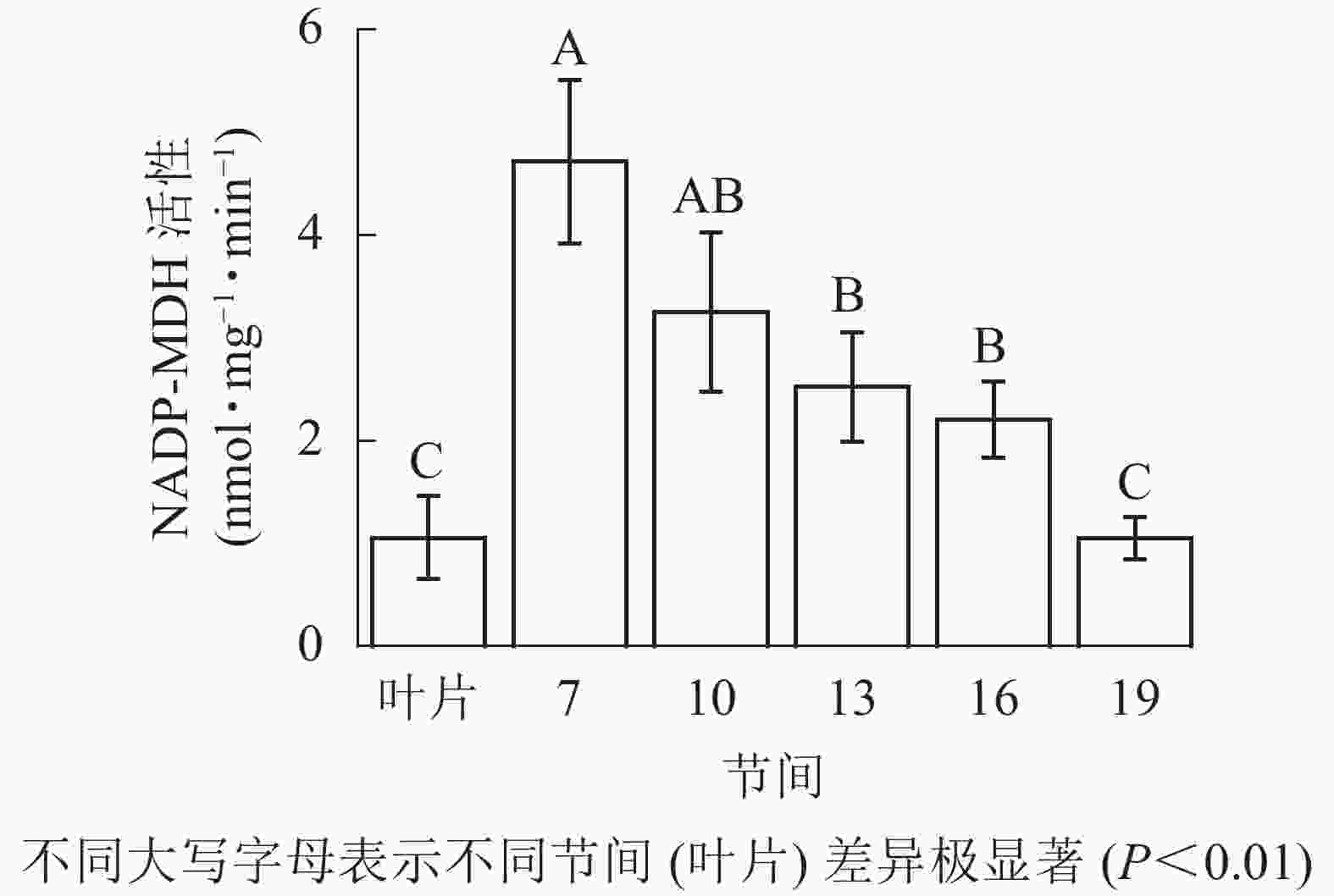
由图3可知:茎秆第7节间NADP-MDH酶活性最高,极显著高于13~19节间及叶片(P<0.01),分别是第10、13、16、19节间和叶片的1.45、1.87、2.14、4.50和4.46倍;随着节间的升高,NADP-MDH活性呈下降趋势,第19节间NADP-MDH活性最低,与第7节间相比下降了77.8%(P<0.01)。毛竹茎秆第7~19节间NADP-MDH酶活性整体呈先下降而后在低水平趋于平稳的变化规律。
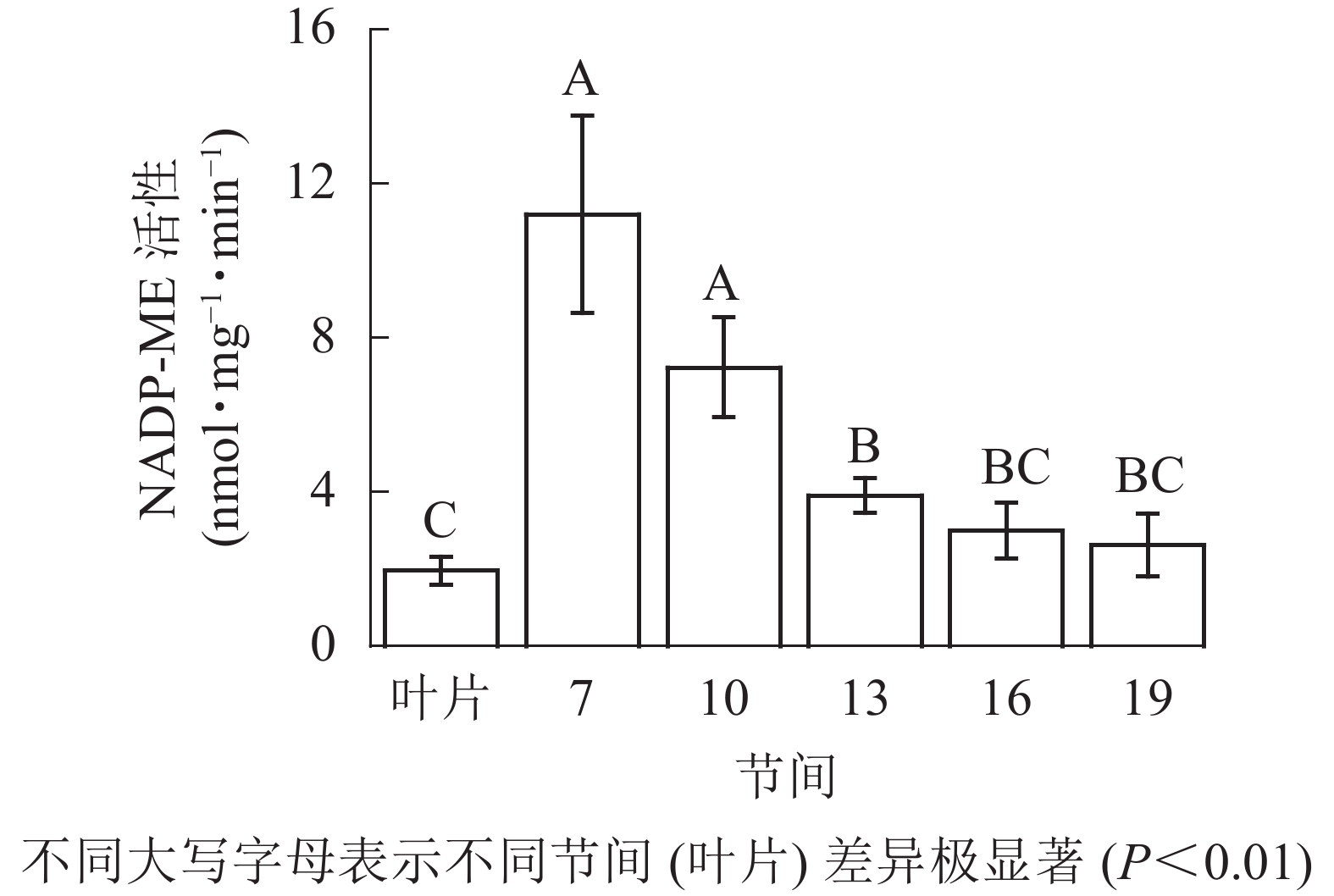
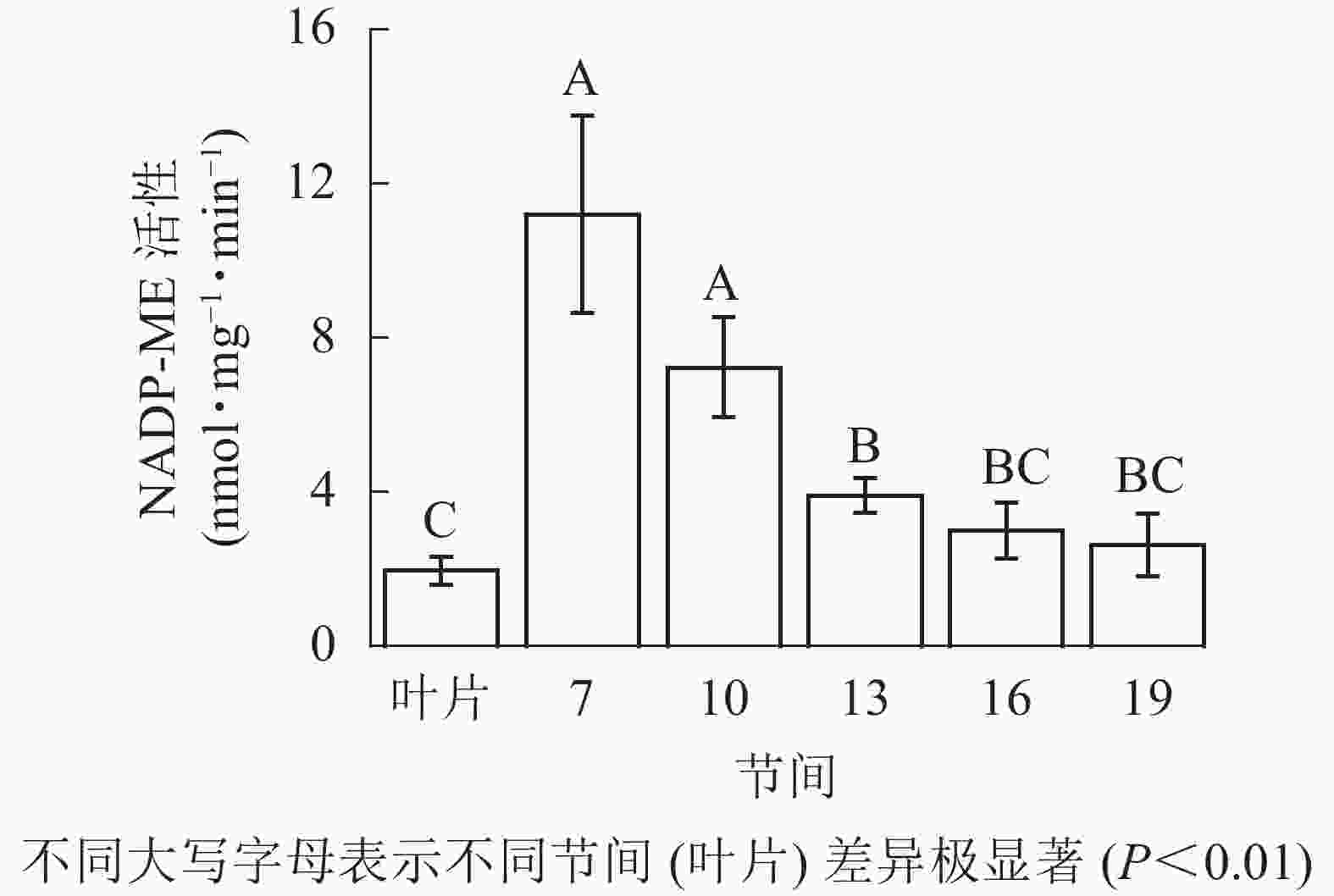
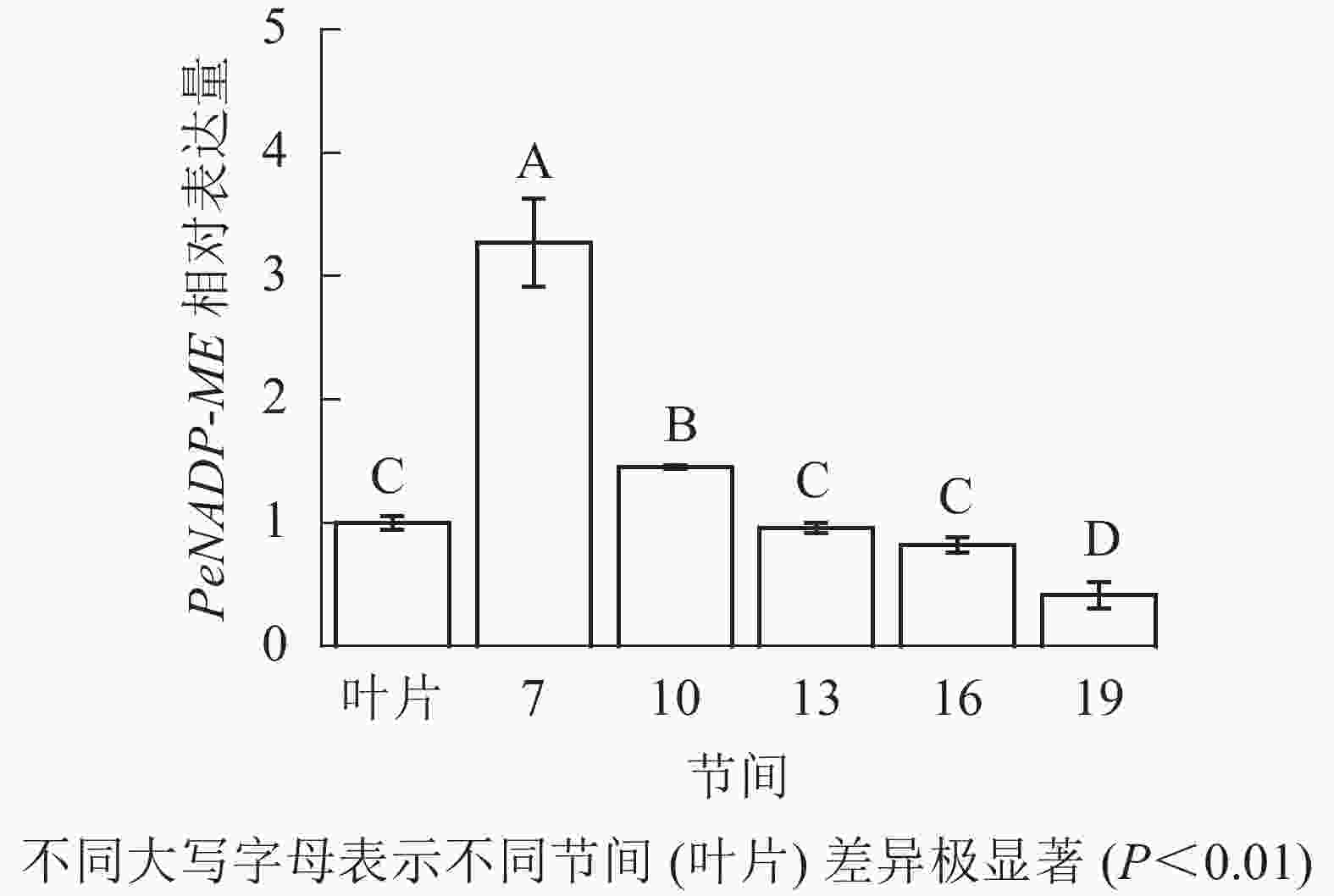
由图4可知:茎秆第7节间NADP-ME酶活性最高,极显著高于13~19节间及叶片(P<0.01),分别是第10、13、16、19节间和叶片的1.55、2.86、3.73、4.27和5.69倍;随着节间的升高,NADP-ME活性呈下降趋势,第19节间NADP-ME活性最低,与第7节间相比下降了76.6%(P<0.01)。毛竹茎秆第7~19节间NADP-ME酶活性整体呈下降,而后趋于平稳的变化规律。
-
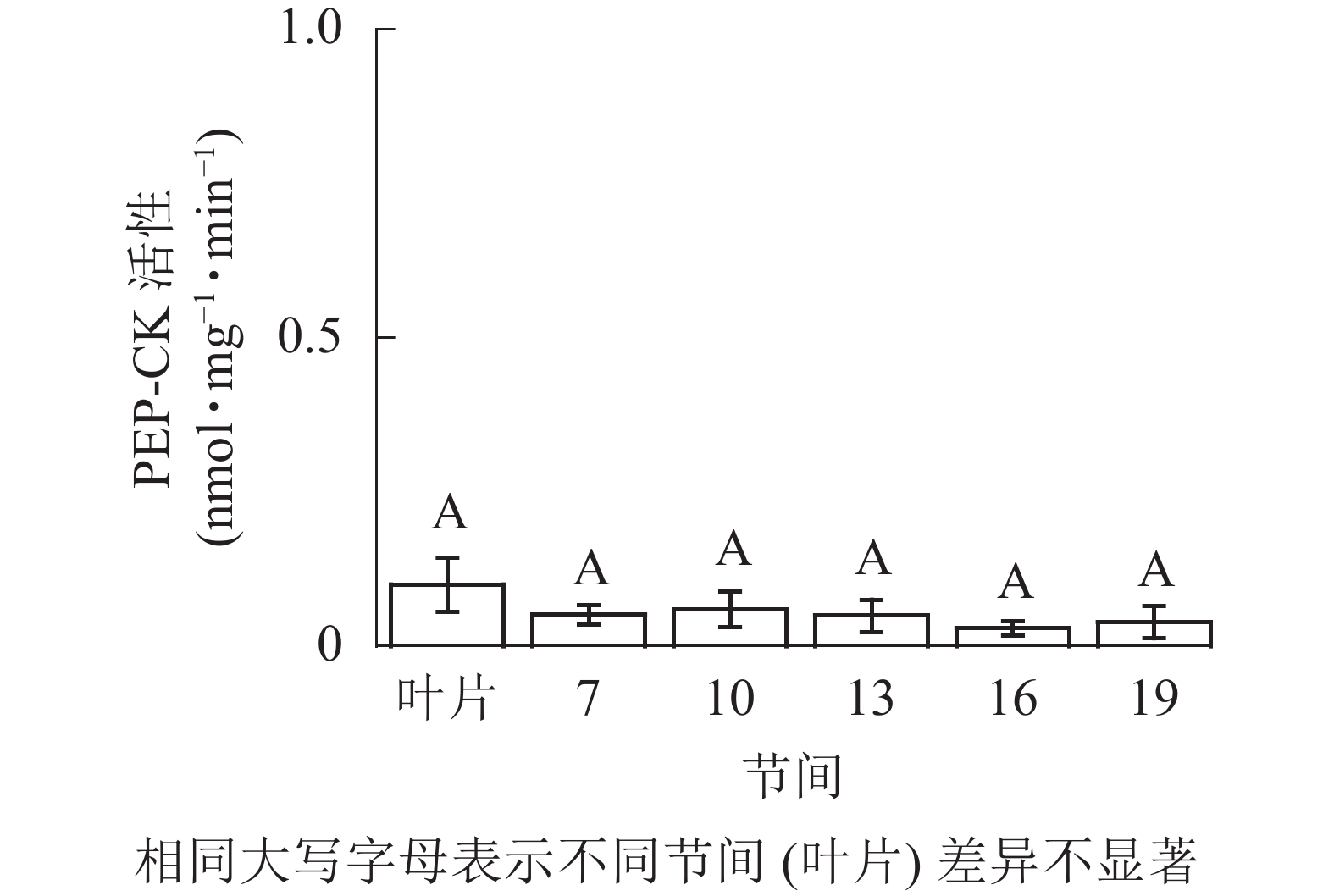
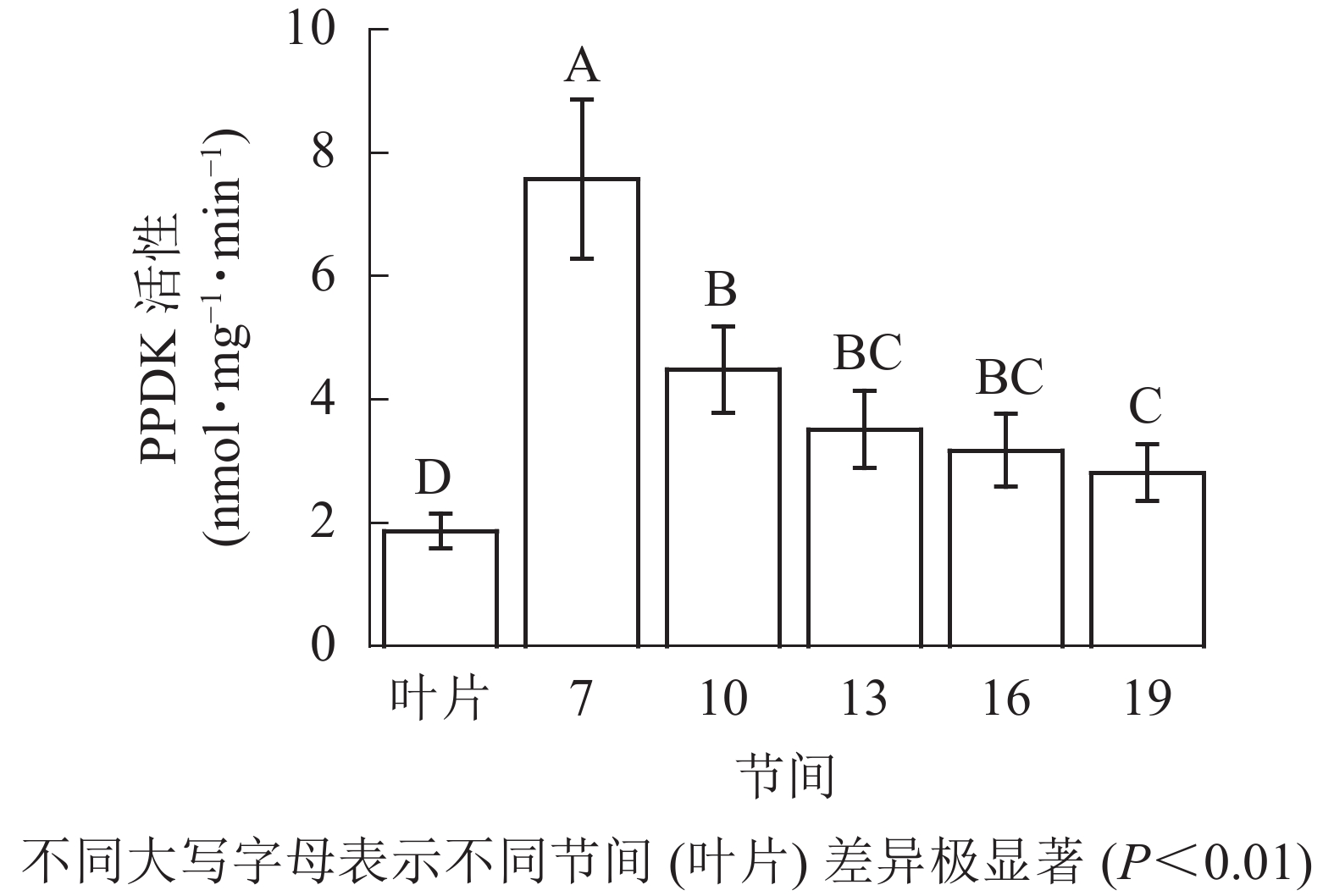
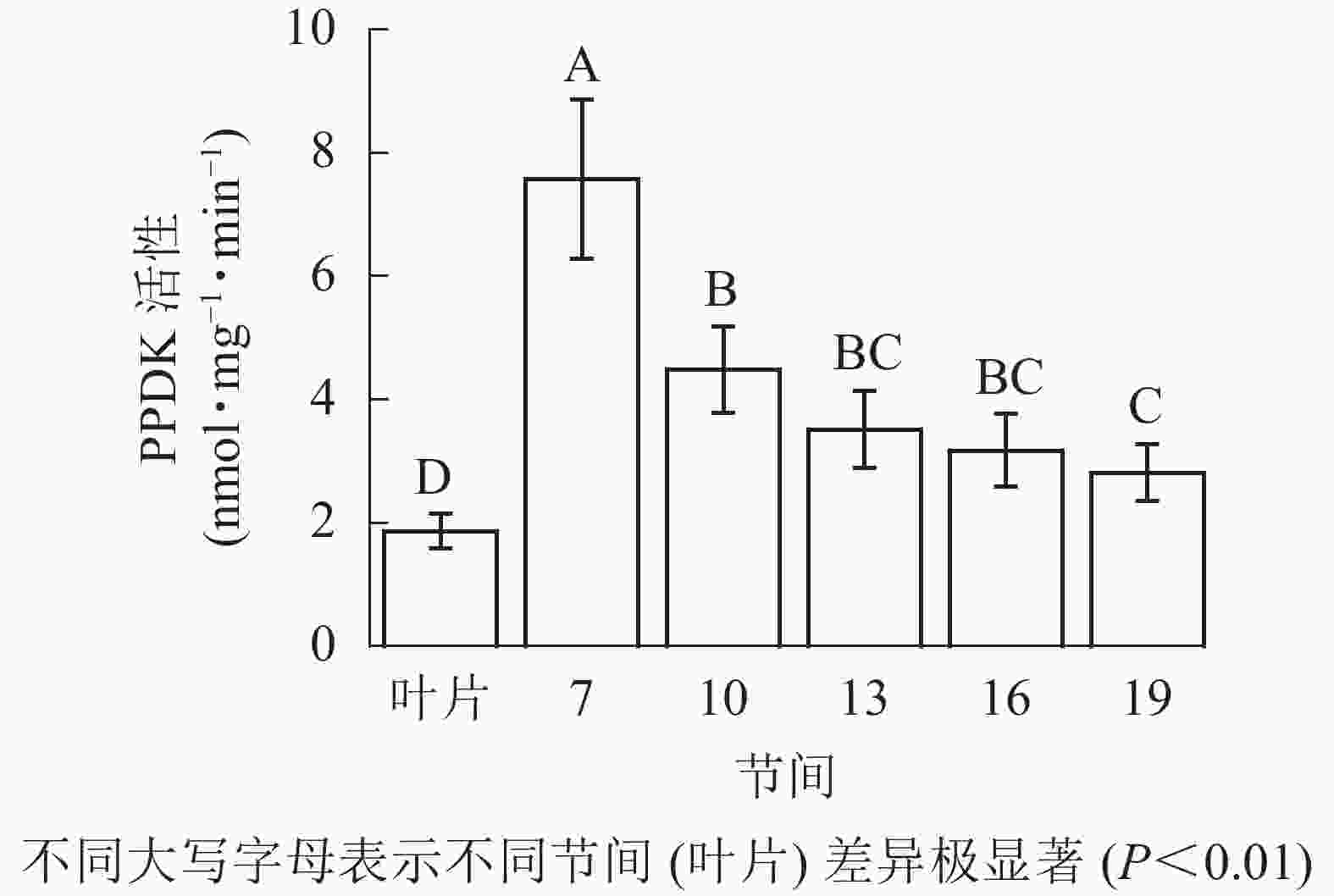
由图5可知:茎秆各节间与毛竹叶片PEP-CK活性无显著性差异。由图6可知:毛竹茎秆第7节间PPDK酶活性最高,极显著高于其他节间及叶片(P<0.01),分别是第10、13、16、19节间和叶片的1.69、2.15、2.38、2.69和4.05倍;随着节间的升高,PPDK活性呈下降趋势,第19节间PPDK活性最低,与第7节间相比下降了62.9%(P<0.01)。毛竹茎秆第7~19节间PPDK活性整体呈极显著下降(P<0.01),而后趋于平稳的变化规律。
-
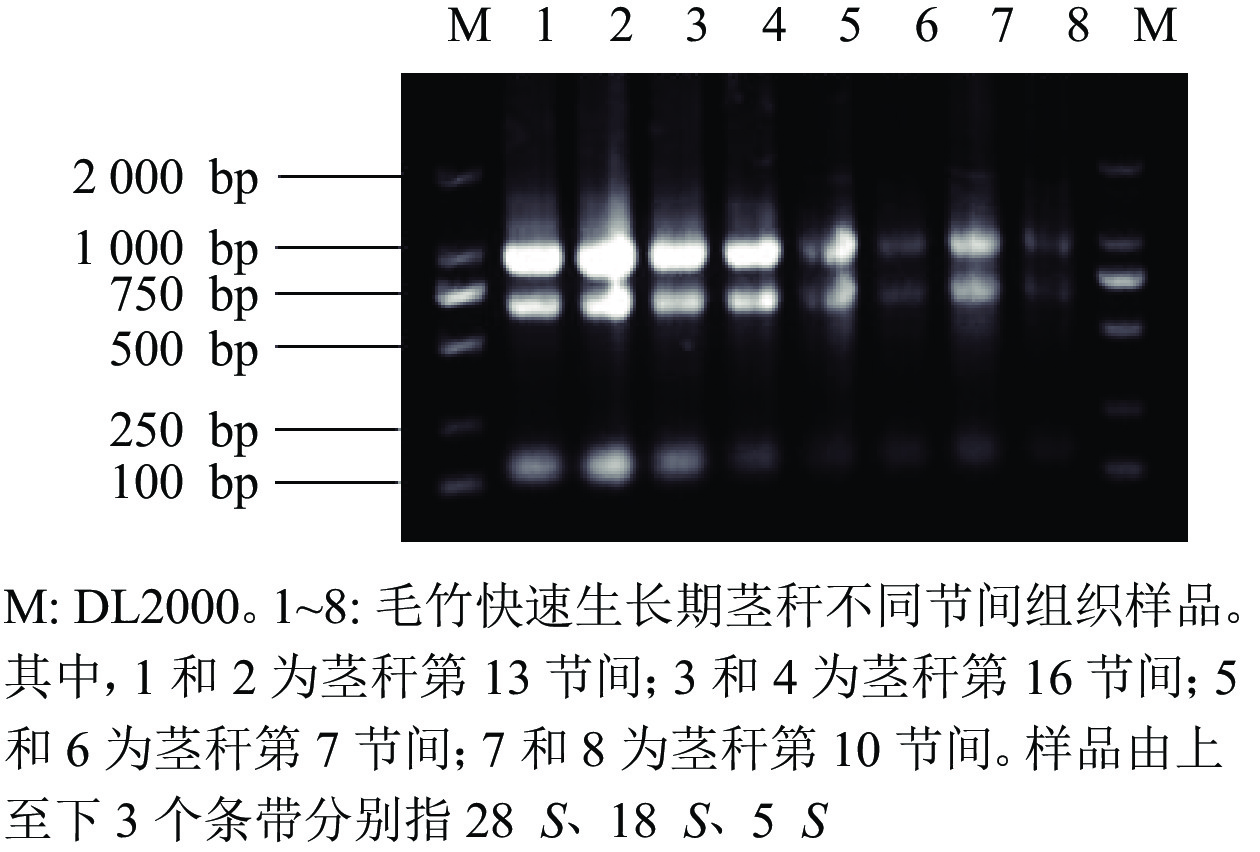
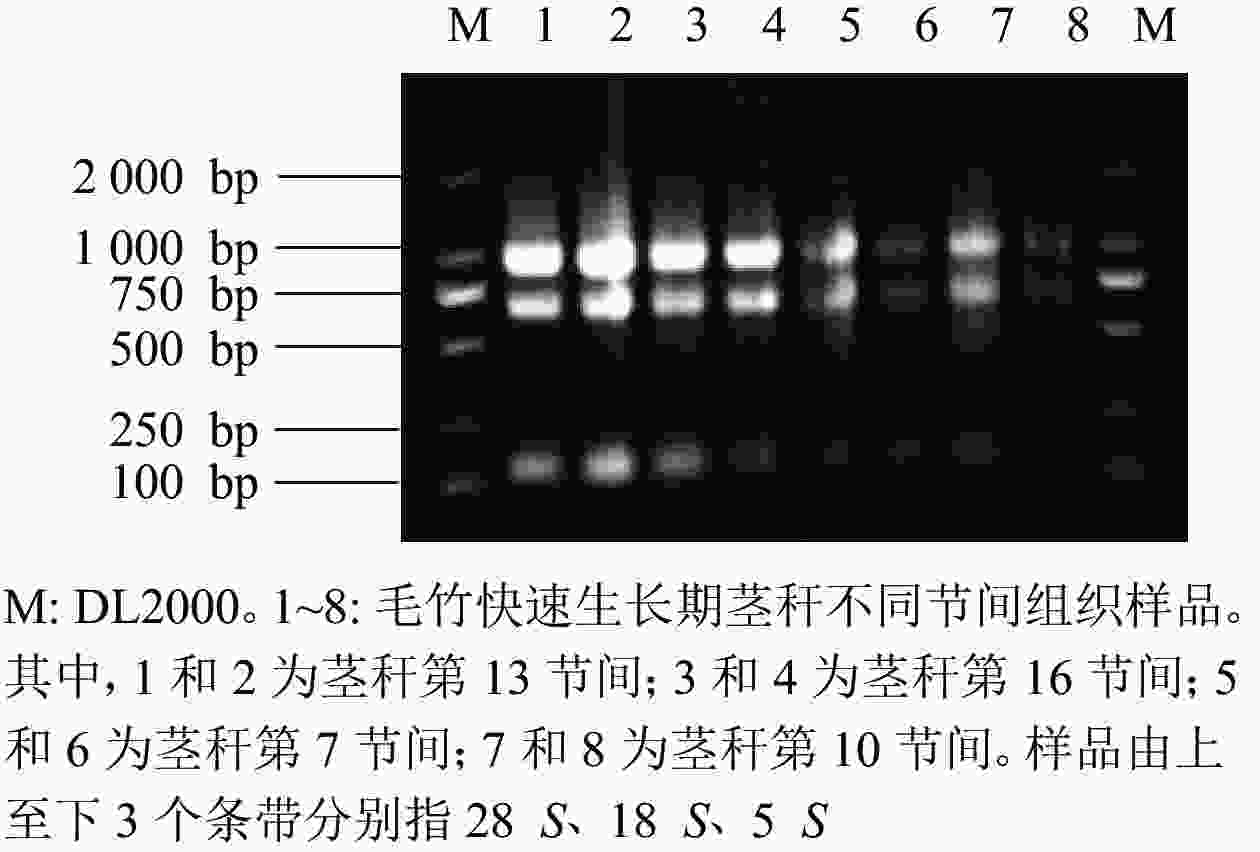
由图7可知:提取毛竹茎秆总RNA的琼脂糖凝胶电泳条带清晰、明亮,分离明显,28 S和18 S处的条带亮度是5 S处条带亮度的2倍,可见样品总RNA完整,未降解。
-
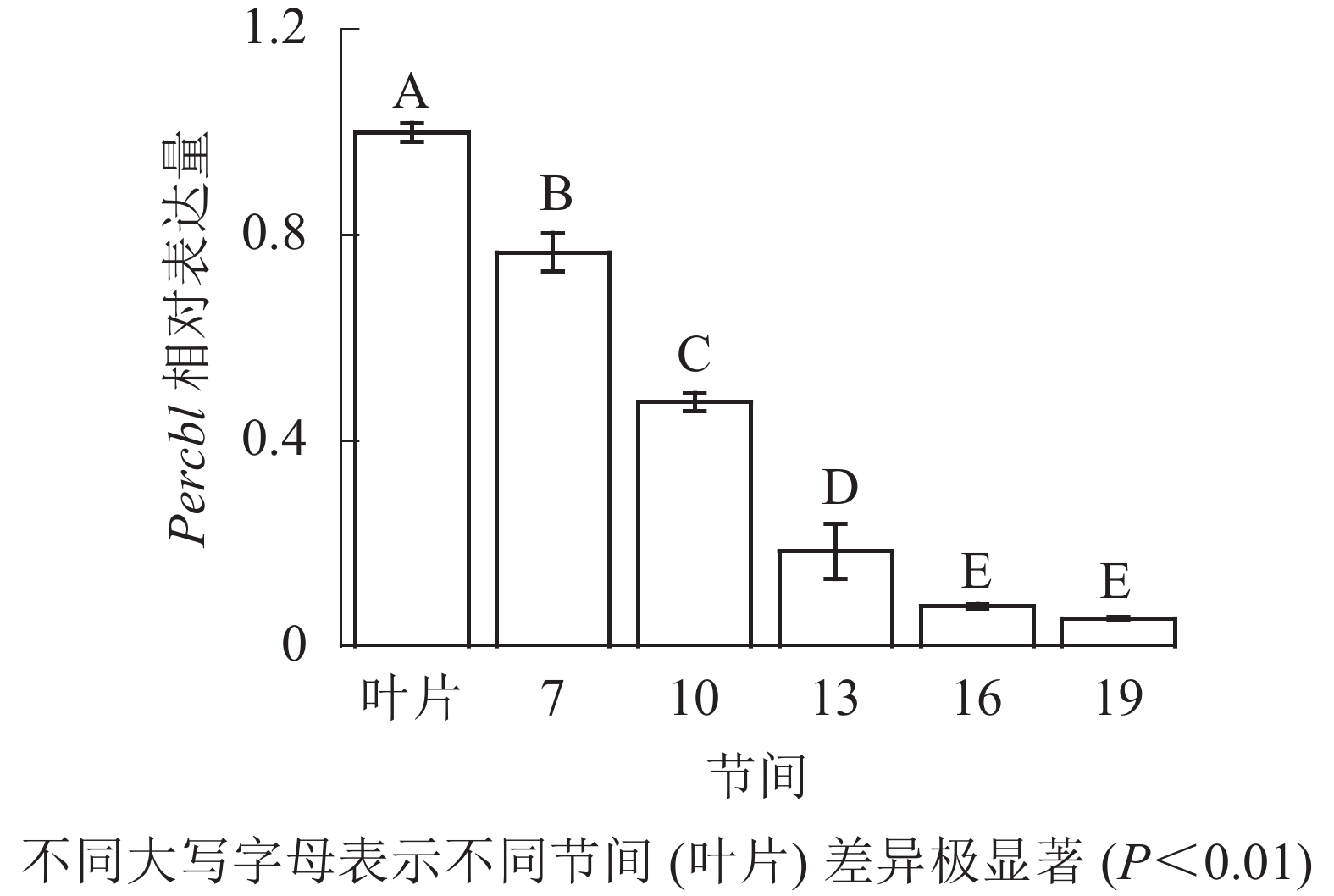
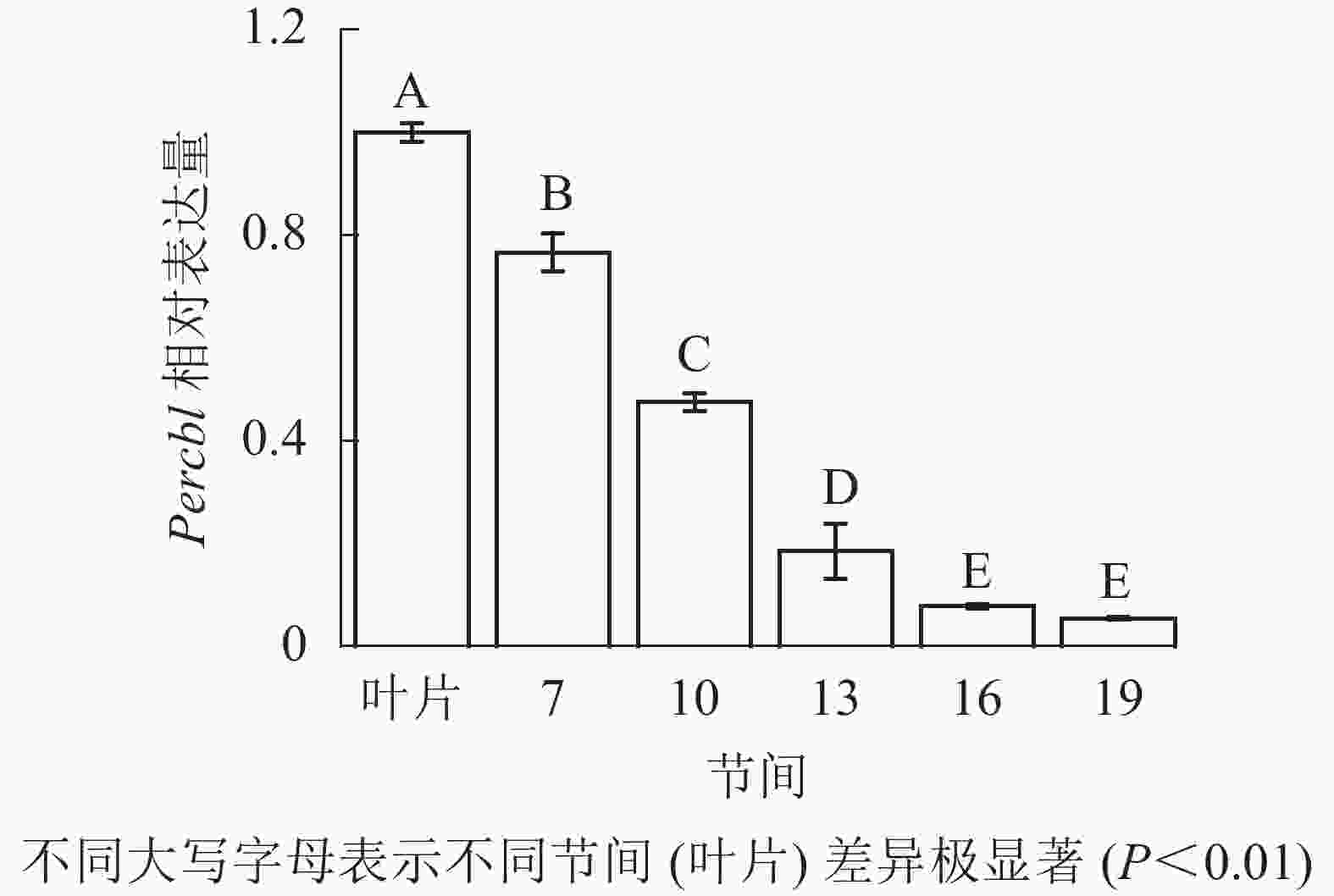
由图8可知:茎秆7~19节间和叶片Percbl基因表达丰度不同,毛竹叶片Percbl基因相对表达丰度最高,极显著高于茎秆快速生长节间(P<0.01),分别是第7、10、13、16、19节间的1.30、2.13、5.41、12.94、18.51倍。随着节间的升高,Percbl基因相对表达量极显著降低(P<0.01),第19节间Percbl基因相对表达量最低,极显著低于叶片和第7节间(P<0.01),相比分别下降了94.6%和93.0%。
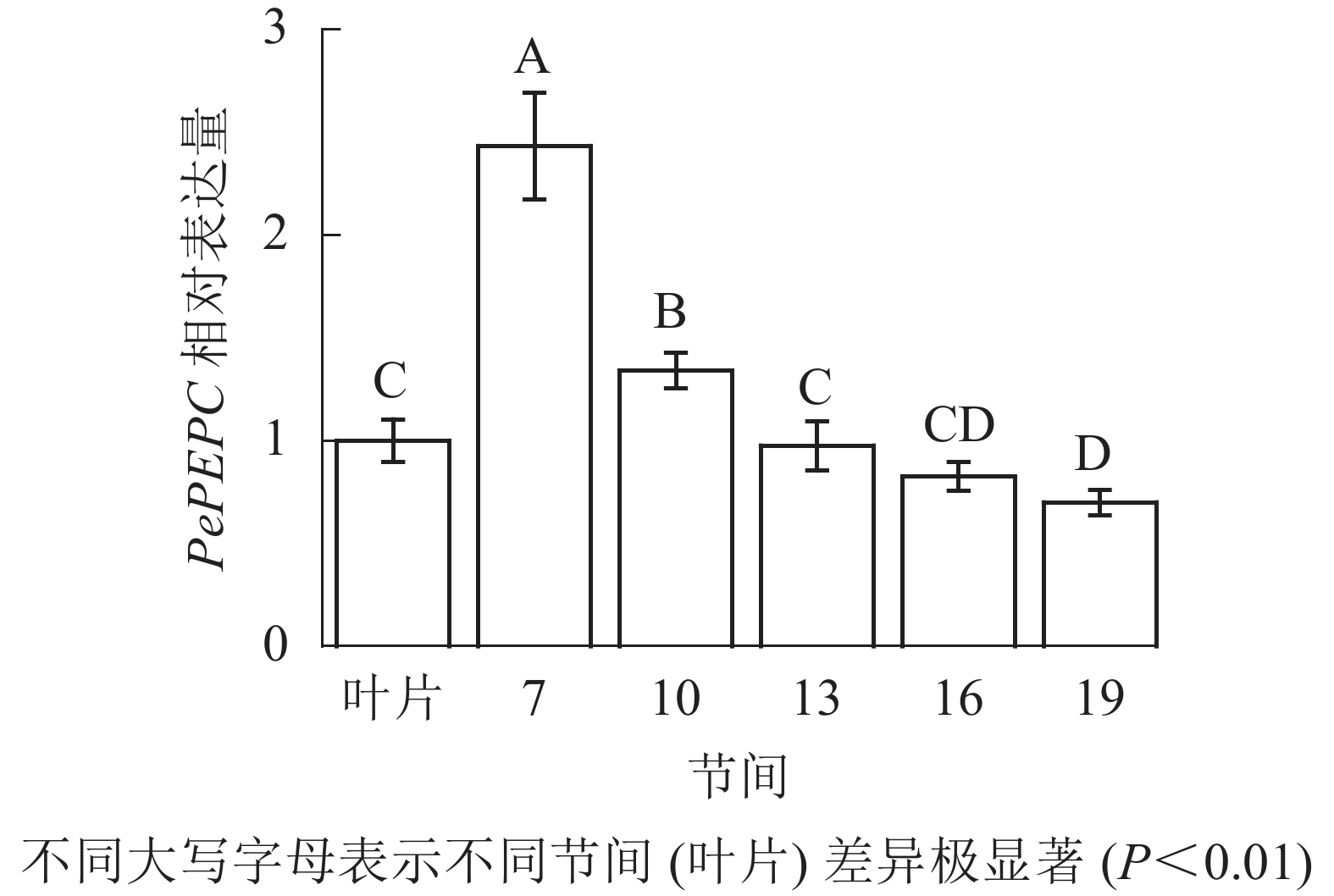
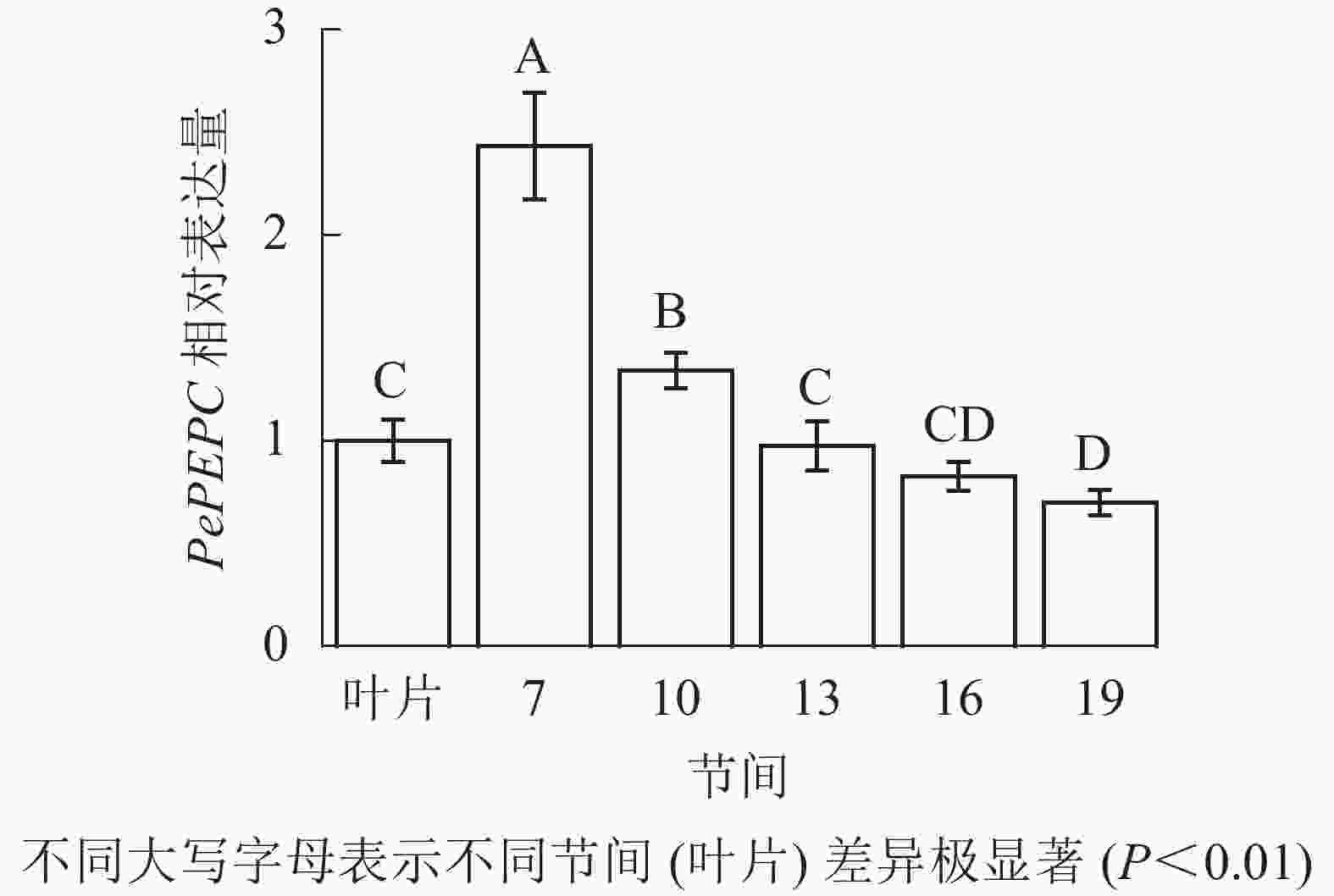
由图9可知:茎秆7~19节间和毛竹叶片PePEPC基因相对表达量不同,茎秆第7节间PePEPC基因相对表达量最高,极显著高于叶片及各个节间(P<0.01),分别是叶片、第10、13、16、19节间的2.43、1.82、2.50、2.95、3.48倍。随着节间的升高,PePEPC基因相对表达量极显著降低(P<0.01),第19节间PePEPC基因相对表达量最小,极显著低于叶片和第7节间(P<0.01),分别下降了30.1%和71.3%。
-
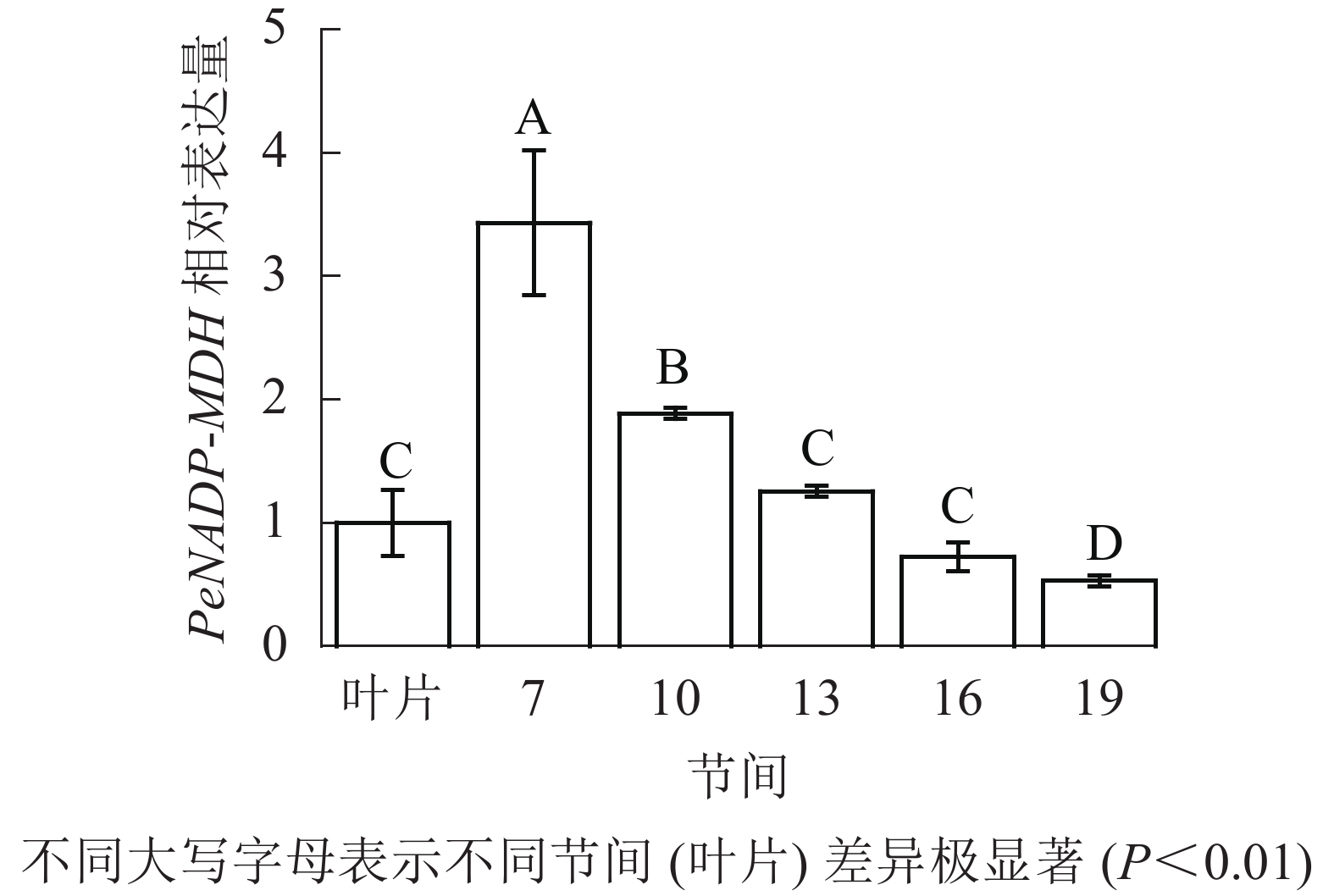
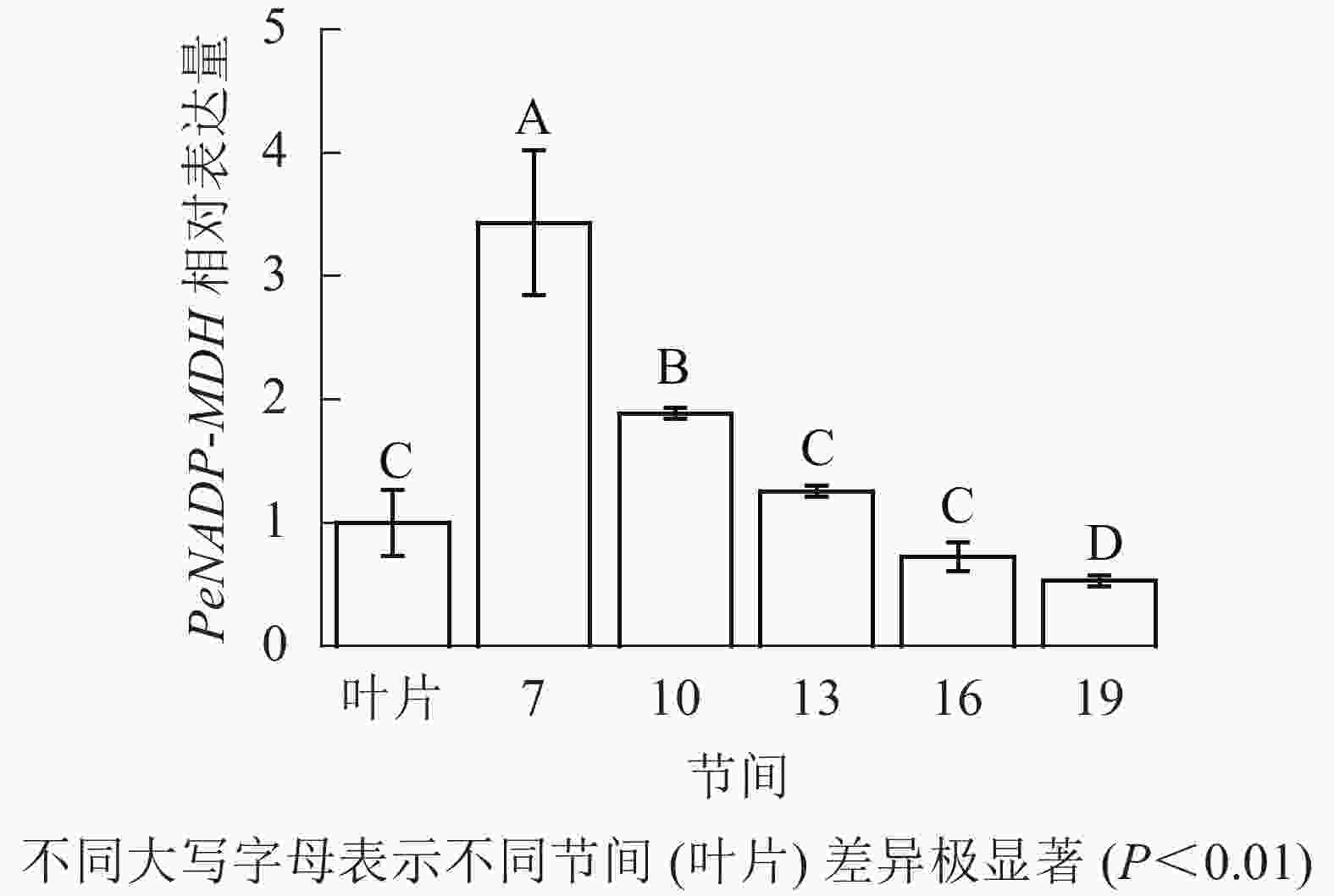
由图10可知:茎秆7~19节间和毛竹叶片PeNADP-MDH基因相对表达量不同,第7节间PeNADP-MDH基因相对表达量最高,极显著高于叶片及各个节间(P<0.01),分别是叶片、10、16、19、22节间的3.43、1.82、2.73、4.73、6.48倍。随着节间的升高,PeNADP-MDH基因相对表达量极显著降低(P<0.01),第19节间PeNADP-MDH基因相对表达量最低,极显著低于叶片和第7节间(P<0.01),相比分别降低了47.0%和84.6%。
由图11可知:茎秆7~19节间和毛竹叶片PeNADP-ME基因相对表达量不同,第7节间PeNADP-ME基因相对表达量最高,极显著高于叶片及各个节间(P<0.01),分别是叶片、10、13、16、19节间的3.27、2.25、3.41、3.99、7.89倍。随着节间的升高,PeNADP-ME基因相对表达量极显著降低(P<0.01),第19节间PeNADP-ME基因相对表达量最低,极显著低于叶片和第7节间(P<0.01),分别降低了58.5%和87.3%。
-
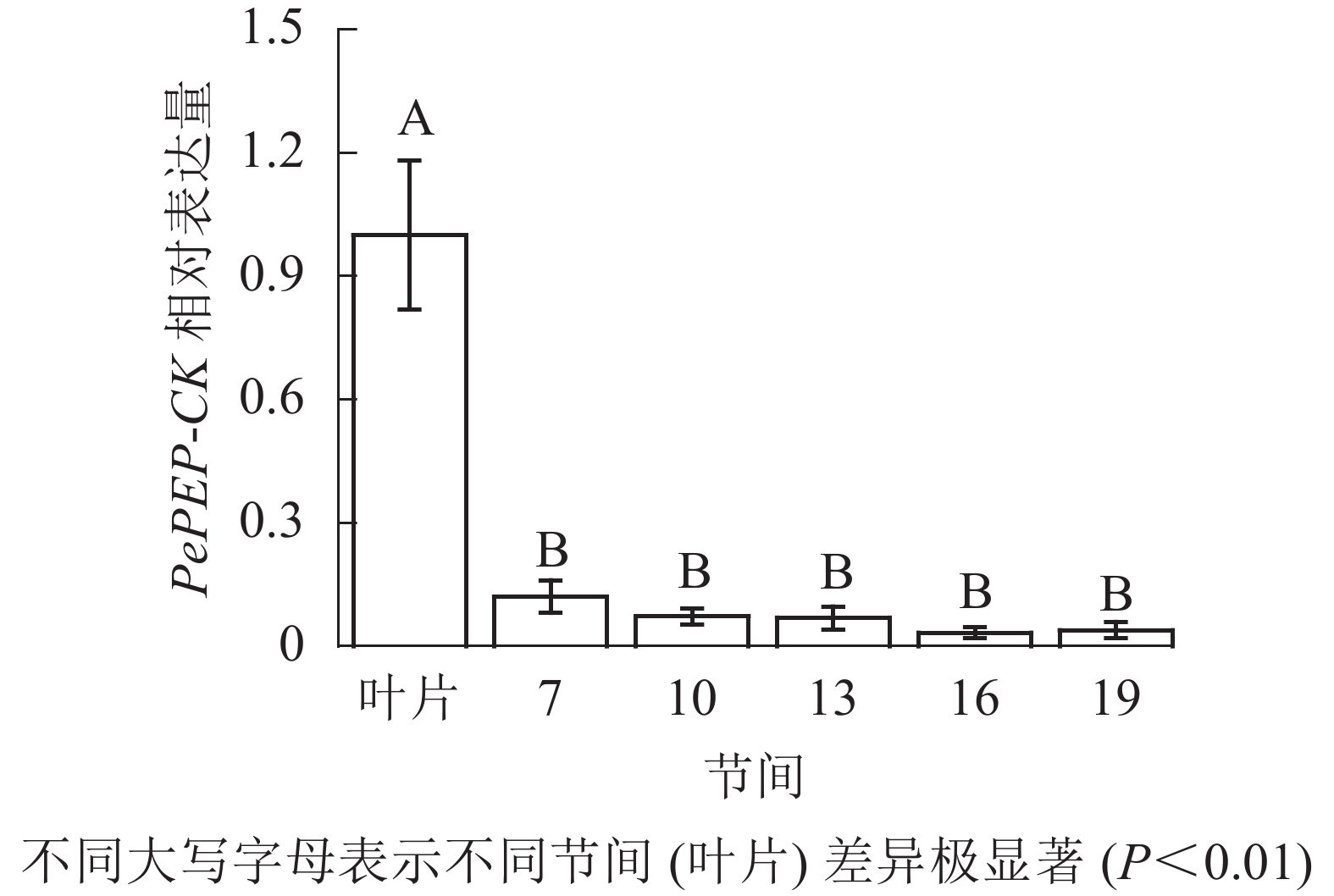
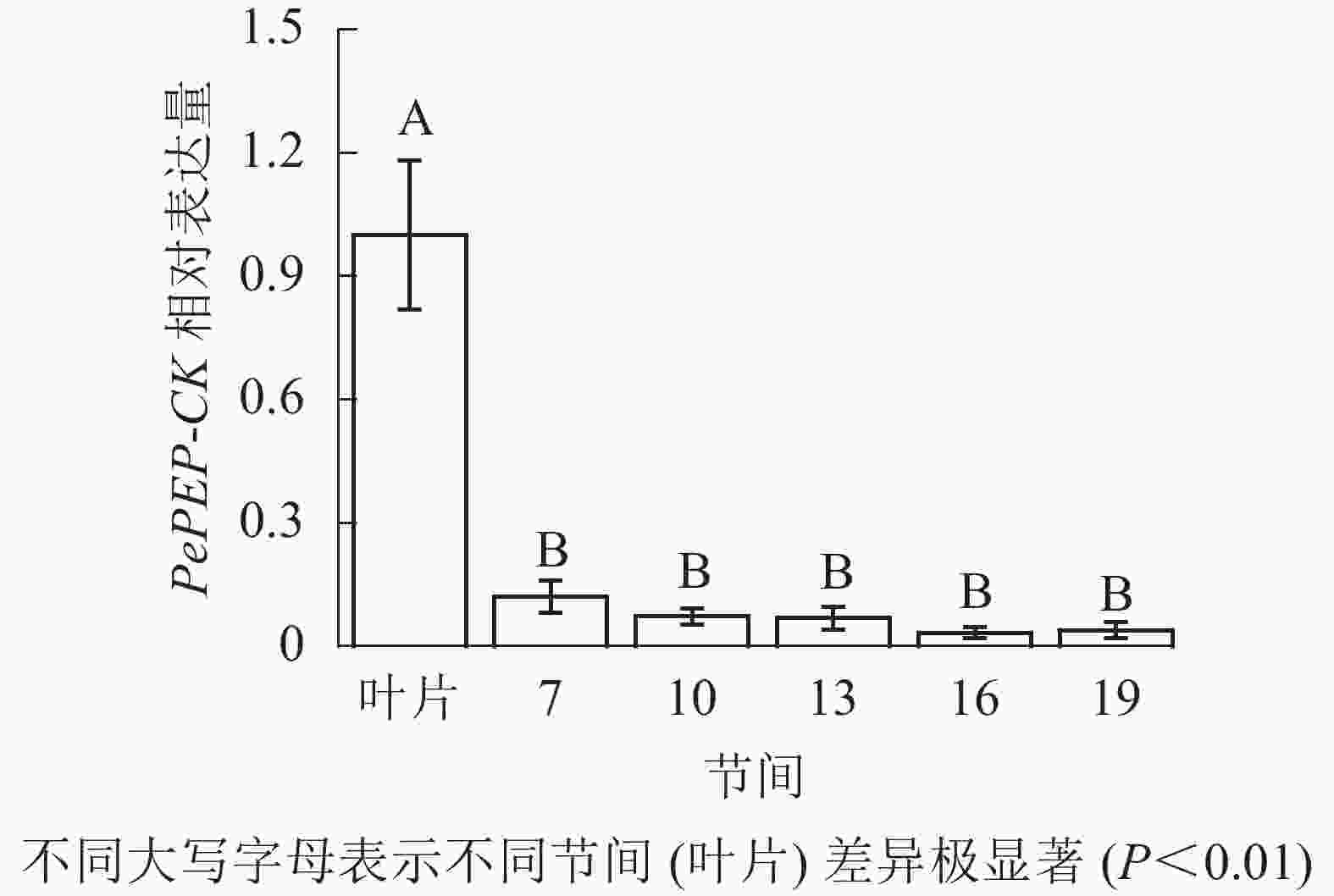
由图12可知:茎秆7~19节间和毛竹叶片PePEP-CK基因相对表达量不同,各个节间PePEP-CK基因相对表达量极显著低于叶片(P<0.01),第7~19节间PePEP-CK基因相对表达量无显著性差异。
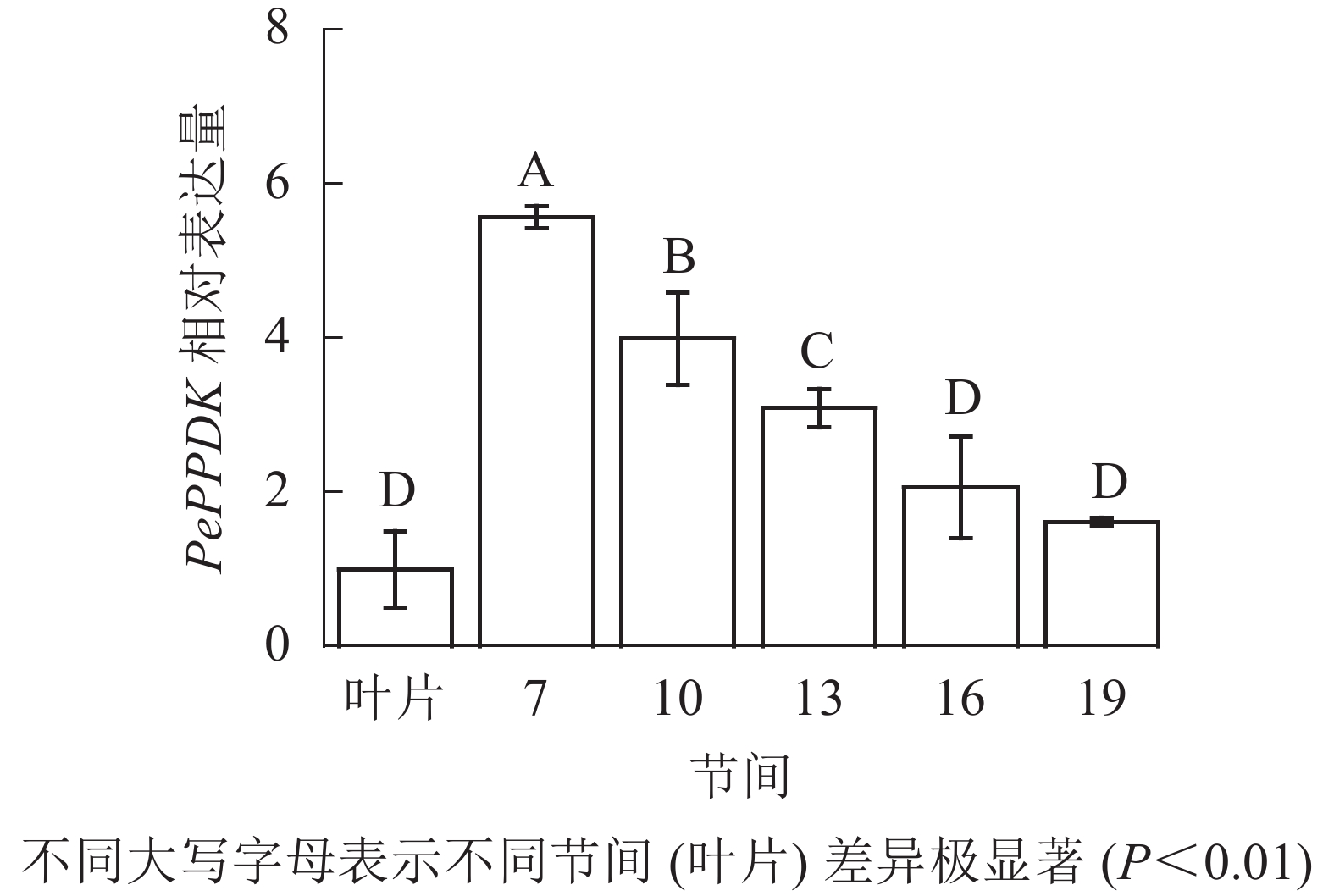
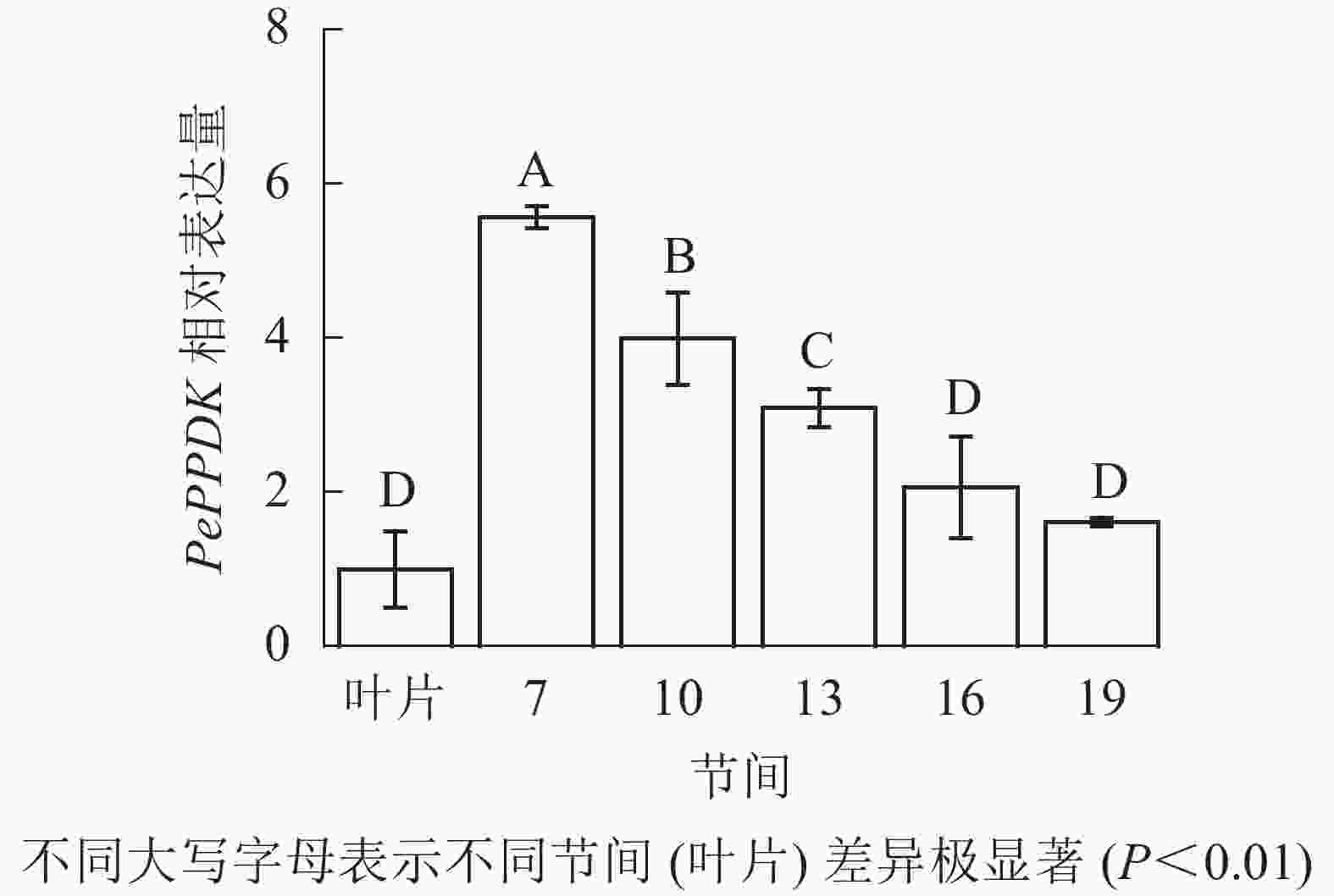
由图13可知:茎秆7~19节间和毛竹叶片PePPDK基因相对表达量不同,第7节间PePPDK基因相对表达量最高,极显著高于叶片及各个节间(P<0.01),分别是叶片、10、13、16、19节间的5.57、1.39、1.80、2.70、3.46倍。随着节间的升高,PePPDK基因相对表达量逐渐降低,第19节间PePPDK基因相对表达量最低,与第7节相比降低了71.1%(P<0.01)。
2.1. 不同节间和叶片Rubisco和PEPC活性变化
2.2. 不同节间和叶片NADP-MDH和NADP-ME活性变化
2.3. 不同节间和叶片PEP-CK和PPDK活性变化
2.4. 毛竹茎秆提取总RNA完整性及纯度检测
2.5. 不同节间和叶片Percbl及PePEPC基因相对表达量的变化
2.6. 不同节间和叶片PeNADP-MDH及PeNADP-ME基因相对表达量的变化
2.7. 不同节间和叶片PePEP-CK及PePPDK基因相对表达量的变化
-
植物光合碳固定主要通过C3途径进行,也可由C4途径进行[26-27]。IVANOV等[28]研究发现欧洲赤松Pinus sylvestris树皮和针叶NAD-ME、NADP-ME和PEPC活性较低,认为茎秆光合碳固定主要是C3途径;BERVEILLER等[29]研究发现成年欧洲山毛榉Fagus sylvatica树干和叶片Rubisco及PEPC动力学和免疫学特征与C3植物一致。HIBBERD等[11]研究发现烟草茎和叶柄周围细胞存在类似C4植物的花环束鞘结构,这些细胞NADP-ME、NADP-MDH和PEP-CK活性较高,认为此部位主要以C4途径固定无机碳。本研究中,毛竹茎秆快速生长节间PEPC、NADP-ME、NADP-MDH、PPDK活性均极显著高于叶片,茎秆Rubisco活性均极显著低于叶片,这与王莹等[13]对不同木本植物枝条中C4酶活性普遍高于叶片的研究结果相一致。结合毛竹茎秆存在与C4植物相类似的花环结构[30],推断毛竹茎秆快速生长阶段茎秆主要通过C4途径固定CO2。除此之外,本研究发现茎秆节位越高C4途径关键酶活性和基因表达越低。这是因为茎秆下部节间已发育成熟竹箨脱落,节间裸露于有光环境中,光合作用活跃;上部节间竹箨未脱落,光不能被茎秆接收,光合作用无法进行,这与王珂杨等[31]对毛竹茎秆中下部节间PSⅡ反应中心活性强,光能转换效率高,上部节间光合功能弱生长缓慢的研究结果相对应。
非同化器官光合固碳方式可能与相关光合酶基因选择性表达密切相关。黑杨Populus nigra中存在与玉米高同源性的PEPC基因,其茎组织有明显高水平选择性表达,对其碳代谢有潜在积极作用[32]。烟草茎和叶柄光合细胞通过优先、特异性地表达高活性NADP-ME、NADP-MDH和PEP-CK固定无机碳[11]。本研究中,毛竹茎秆快速生长节间PeNADP-ME基因和PeNADP-MDH基因相对表达量均极显著高于叶片,茎秆PePEPC基因和PePPDK基因相对表达量均极显著高于叶片,茎秆快速生长节间Rubisco基因Perbcl相对表达量极显著低于叶片,与其酶活性水平具有一致性,这与LI等[32]对杂种黑杨Populus simonii × P. nigra茎组织PsnPEPC2基因高水平表达的研究结果类似。表明毛竹茎秆由C4途径固定无机碳可能受基因水平的调控,茎秆内PEPC催化无机碳羧化形成草酰乙酸,之后主要通过NADP-ME型和NAD-ME型通路由NADP-ME和NADP-MDH进行转化,这与烟草叶柄和芹菜中C4同化途径类似[11]。毛竹茎秆速生时期以C4途径固定无机碳可能是其能进行高效生长的重要原因之一。
植物组织用于光合作用的CO2主要来源于大气,一些木本植物茎秆、树干组织气孔较少,外皮层组织间排列紧密,气体通透性差,大气CO2难以扩散进茎秆皮层绿色组织[6]。木本植物树枝和茎秆光合作用普遍被认为利用内部代谢及呼吸产生的CO2[5]。根系吸收的无机碳盐和自身组织呼吸作用释放的CO2会被装载到木质部液流供茎秆光合细胞重新固定利用[11]。本研究中,毛竹茎秆表皮不具气孔,大气CO2不能由气孔扩散进皮层组织[30],茎秆内部测得高浓度CO2(数据未显示),这与ZACHARIAH等[33]对竹腔中空部具高浓度CO2和气体正压的研究结果相一致。表明茎秆处于高CO2低氧气(O2)环境,这与C4植物维管束细胞所处环境类似[6,34],PEPC在高CO2环境下往往活性更强[35],茎秆内部组织呼吸作用所产生的CO2被其羧化吸收重新固定。一方面避免了呼吸碳外排损耗,碳利用更加节约成本[5,34,36];另一方面,其二次积累产生的糖类支持茎秆进行快速生长,释放的O2在一定程度上缓解了茎秆组织内部缺氧状态和潜在酸化威胁[37],毛竹茎秆这种特殊固碳机制可能是其能进行快速生长的重要原因之一。
综上所述,毛竹快速生长期茎秆主要以C4途径固定CO2,NADP-ME和NAD-ME型是茎秆进行C4途径的主要通路;茎秆C4途径主要用于竹腔内部高浓度CO2再固定,形成的碳水化合物被茎秆快速生长再利用,这在一定程度上减少了自身碳损耗,为进一步解析毛竹茎秆快速生长机制提供理论依据。










 DownLoad:
DownLoad: