-
榧树Torreya grandis是红豆杉科Taxaceae榧属Torreya中国特有植物,为第三纪孑遗植物,零星片状分布于浙江、安徽、福建、江西、湖南等省的丘陵地带[1]。榧树雌雄异株,稀同株。雄性榧树是香榧T. grandis ‘Merrillii’的父本,为雌性香榧提供花粉,并在在维持榧树多样性中具有重要地位。目前,生产中大面积栽培的仅香榧一个栽培类型,因而榧树天然杂交产生的多样性在优良种质选育及新品种培育方面潜力巨大,其中雄性榧树起着重要的作用。以前由于雄株不结果,榧农不了解它对香榧生产的重要性,雄性榧树被严重破坏,种群数量及范围越来越小,雌雄比例严重失调[2]。近年来,随着香榧产业的发展,人们逐渐认识到充分授粉对种实产量的影响,日益重视香榧造林中授粉树的配置。研究发现:雄性榧树单株间在花期、花粉得率与生活力等表型指标上存在丰富的变异[3]。因为变异是选育的基础,因此,从DNA标记分析研究雄性榧树的多样性,可从遗传物质DNA水平了解雄性榧树表型丰富变异的内在基础,从根本上为榧树雄株选育提供理论基础。遗传多样性是生物多样性的重要组成部分。随着生物技术的发展和研究层次的深入,DNA分子标记成为检测遗传多样性的主要方法[4]。简单序列重复(SSR)是基于聚合酶链式反应(PCR)的分子标记技术,具有重复性好,多态性高,共显性遗传,操作方便等优点,已广泛应用于植物基因定位、遗传育种、遗传图谱构建和遗传多样性研究等方面[5-8]。在榧树遗传多样性研究方面,闵会等[9]利用扩增片段长度多态(AFLP)分子标记对香榧天然群体进行分析,发现香榧群体的遗传多样性丰富,且居群内的遗传变异大于居群间;吴昊等[10]依据个体间的差异现象,建立了基于序列相关扩增多态(SRAP)标记的榧树核心种质确定方法;刘浩凯[11]则用SRAP分子标记研究榧树雄性居群的遗传多样性,结果表明:榧树种内变异十分复杂。前人的研究揭示了榧树种质资源复杂的遗传背景和丰富的遗传多样性,但他们均使用的是显性标记。显性标记不能区分纯合显性与杂合显性的基因型,而SSR标记则反之,它是共显性标记。有关雄性榧树遗传多样性的研究尚未见报道。本研究以5个雄性榧树居群为研究对象,用荧光SSR分子标记分析榧树雄株的遗传多样性和遗传结构,旨在为榧树雄株后续的培育、雄株种质资源的保护与可持续利用提供理论基础。
HTML
-
詹利云等[3]对浙江省淳安县半夏村(淳安居群)、杭州市临安区洪岭村(临安居群)、杭州市富阳区洞桥村(富阳居群)、浙江嵊州市榆树村(嵊州居群)及安徽省黄山市呈坎村(黄山居群)等5个野生雄性榧树居群叶片及雄球花相关性状进行了研究。本研究的叶片材料来自上述研究相同居群相同单株,121份叶片材料包括来源于淳安17份、临安24份、富阳24份、嵊州24份和黄山32份。每株采集的新鲜嫩叶放入装有硅胶的自封袋中为1份,带回实验室后置于−80 ℃冰箱保存备用。
-
采用改良的CTAB法提取基因组DNA[12],用紫外分光核酸测定仪(GENEQUANT, Eppen-dorf, 德国)测定DNA浓度和光密度[D(λ)]值,用质量分数为1.2%的琼脂糖凝胶电泳检测,并在AlphaImager成像系统(Alpha Innotech Corporation, 美国)中拍照储存电泳结果。检测合格的DNA样品用无菌水稀释到50 mg·L−1,备用。
-
从榧属物种(长叶榧T. jackii和榧树)研究中选取55对SSR引物[13-16]。SSR反应体系由北京睿博兴科生物技术有限公司提供,即采用2步法,其中第2步扩增时的正向引物加有M13序列(5′-TGTAAAACGACGGCCAGT-3′)(M13-F),并在合成时分别加有蓝 (FAM)、绿 (HEX)、红(ROX)、黑 (TAMRA)荧光,以方便PCR产物的毛细管电泳检测。引物均由北京睿博兴科生物技术有限公司合成。
PCR第1步反应体系为50 mg·L−1的DNA 1.0 μL、2×Taq PCR MasterMix(诺唯赞生物科技有限公司)5.0 μL、10 μmol·L−1正反向引物各0.5 μL、灭菌去离子水补足10.0 μL;PCR扩增程序为95 ℃预变性5 min,后35个循环,每个循环95 ℃变性30 s,60 ℃退火30 s,72 ℃延伸30 s,最后72 ℃延伸10 min,4 ℃保存。PCR产物用质量分数为1.2%的琼脂糖凝胶电泳检测。第2步PCR反应体系为第1步的PCR产物1.0 μL、2×Taq PCR MasterMix 10.0 μL,M13-F及反向引物各0.5 μL,灭菌去离子水补足20.0 μL;PCR扩增程序同第1次PCR,仅退火温度降低了5 ℃。PCR产物用质量分数为1.2%的琼脂糖凝胶电泳检测,条带清晰的样送北京睿博兴科生物技术有限公司毛细管电泳检测。
随机选取5株雄性榧树样品进行PCR扩增的引物筛选,并用筛选得到的引物对样品进行SSR分析。
-
在Excel中统计扩增的等位基因位点信息,并用DataFormater软件[17]进行格式转换。利用GenALEx 6.5软件[18]计算多态信息含量(PIC)、多态位点百分比(PPL)、近交系数(Fis)、遗传相似度(GD)和遗传距离(GI),并进行分子方差分析(analysis of molecular variance, AMOVA)和主坐标分析(principal co-ordinates analysis, PCoA),用Popgene Ver.1.3.2软件计算等位基因数(Na)、有效等位基因数(Ne)、观测杂合度(Ho)、期望杂合度(He)、Nei’s遗传多样性指数(H)、Shannon’s信息指数(I)、Nei’s总基因多样度(Ht)、遗传分化系数(Fst)、基因流(Nm)等遗传参数。
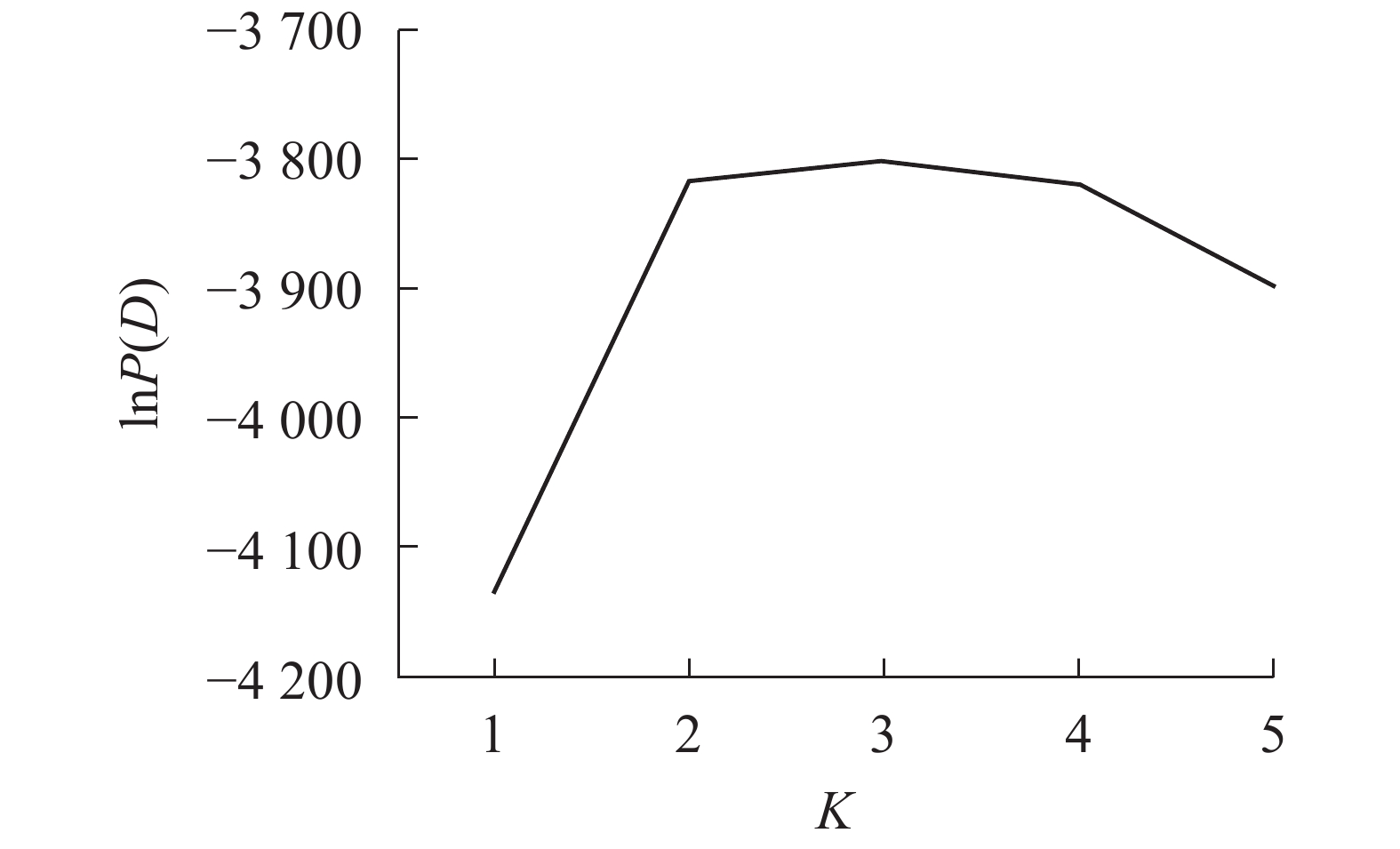
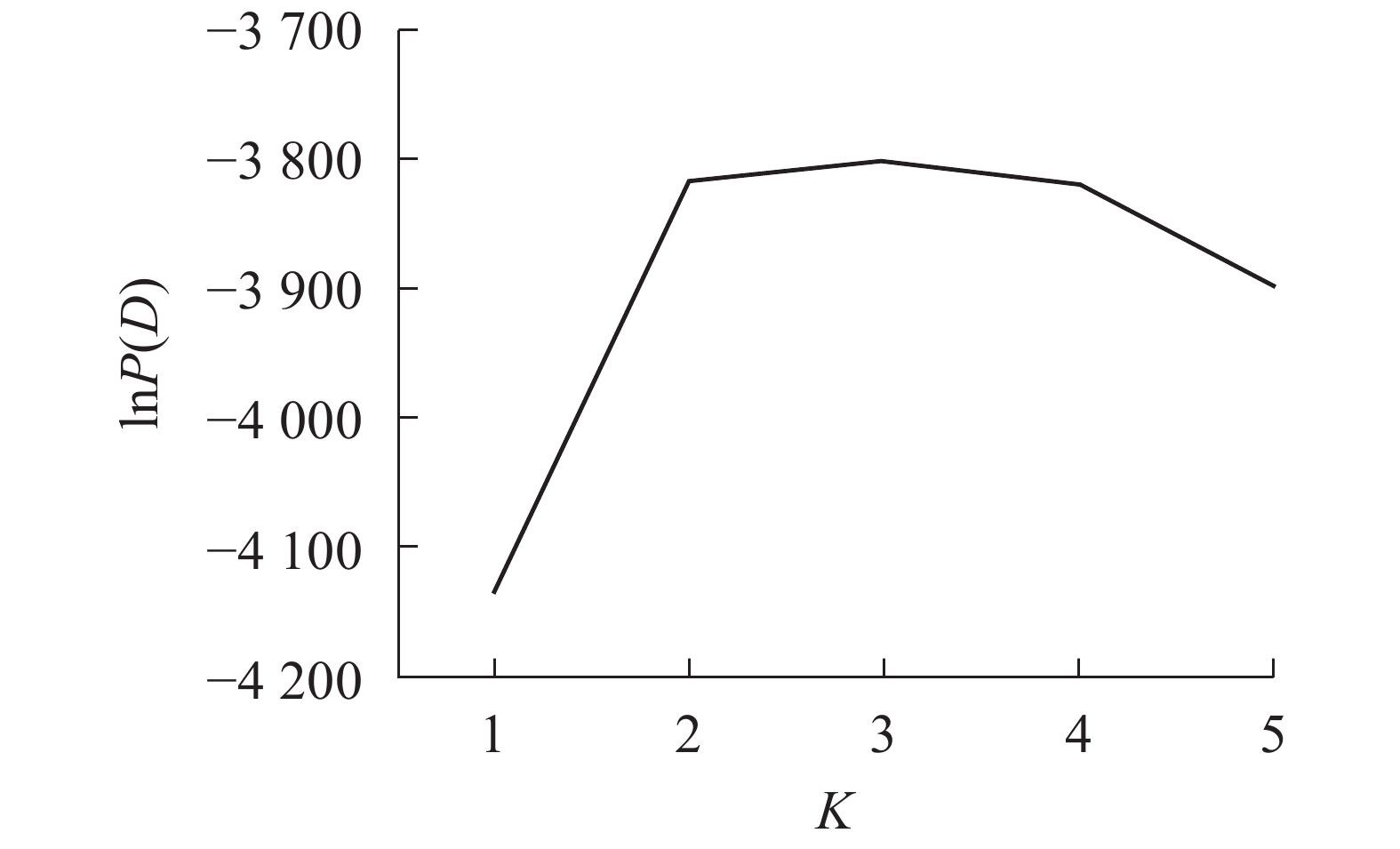
采用NTSYS 2.10e软件[19]计算Nei’s遗传相似性。采用混合模型和Structure 2.3.4软件分析居群的遗传结构和lnP(D)[20]。lnP(D)为Delta K和似然值的对数函数,针对基因库数目建模,以确定最佳群体数K值,设置K为1~5,每个参数运行10次,每次运行的丢弃迭代时间和重复次数都为10000。
1.1. 试验材料
1.2. 试验方法
1.2.1. 基因组DNA提取
1.2.2. SSR分析
1.2.3. 数据统计与分析
-
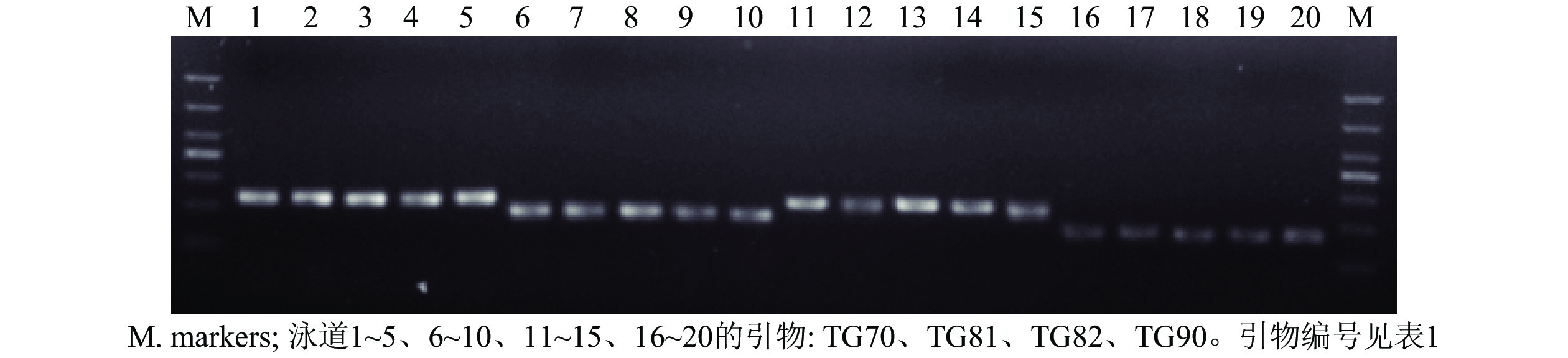
从55对引物中筛选出了24对扩增稳定、条带清晰的引物对(图1),并将其用于榧树雄株遗传多样性分析。引物信息见表1。
引物编号 SSR基序及重复数 正、反向引物序列 退火温
度/℃PCR扩增片段长度/ bp 来源 荧光
标记ZAFU-1 (TG)9 F: GGCTATGCTACACCCAAAGAA
R: GGGGCACCACCTATTGTATG59.0 160~200 张敏等[16] HEX ZAFU-2 (TG)9 F: TCAAAGTGCAACCGGTACAA
R: CAACAGGCCAACATGGAGTA59.0 110~140 张敏等[16] FAM ZAFU-3 (CAG)8 F: GGGTTACCCCCTTGCTTTAT
R: CCCTACTTTATTTCCGTGCG59.0 250~280 张敏等[16] ROX ZAFU-4 (TAA)8 F: GAATTCCCATTCCCATTGTG
R: ACCCCCTTCTGCTCTGATTT59.0 140~170 张敏等[16] TAMRA ZAFU-5 (TTC)8 F: AATGAATGCGTGTTACGCTG
R: TTGGAGCGGAAGGAATAATG59.0 170~200 张敏等[16] HEX ZAFU-6 (TTCT)5 F: CCAATTTGTGGAGCGTTTCT
R: TGTGGAAAGGTGGTGAACAA59.0 190~210 张敏等[16] FAM ZAFU-7 (TATT)5 F: TTTTCCAACTCCAACCCTTG
R: ATGTTTGGGGTTGACGTGTT59.0 170~190 张敏等[16] ROX ZAFU-8 (TAGCA)5 F: AATTGGCCCTTCATTCAACA
R: CTAGTGGGTGCATTTGAGCC60.0 250~700 张敏等[16] TAMRA ZAFU-11 (AT)8 F: ACATCTGCAAGGCAAGGTTC
R: TTGAATTTTCACCAGGCTCC59.0 170~180 张敏等[16] FAM ZAFU-16 (TGAGCC)7 F: AAGGTTGCCACCTCAGTCAC
R: ACAGAACGTCTCCAACCGAC60.0 220~260 张敏等[16] TAMRA GR12 (ATTT)6 F: GCTGTCGAAGCGTTGGAGAA
R: TCTGAAACCTCGCTCGAACC56.0 204~216 LI等[15] HEX GR48 (CA)11 F: TTTTAGAACTGCTTGCCCGT
R: CATGTACATGCACCATCATGC58.0 197~205 LI等[15] ROX GR67 (TCC)12 F: TCCAGTCAGCGCGAATAGTC
R: AGTAGAGGAGTCCATGGCGT58.0 141~162 LI等[15] TAMRA GR81 (CCT)7 F: GGCTCAGTACTCCCAAACCC
R: TCGGCTCCTTTATACGACGC57.0 211~226 LI等[15] HEX GR98 (ATCT)5 F: TATTCGAGACGCGCATTCGA
R: CTCGCATTGAAGCTGTCTGC58.0 161~173 LI等[15] FAM TG19 (CAT)7 F: GGACGTCTCAGCAATGTCAA
R: GCAAAGAAAAGGATTGCCAC53.8 100~250 YI 等[14] TAMRA TG32 (GAA)6GT(AGA)6 F: GGCCGTGAGAGTAGCATAGC
R: AGGTCCCTCACCATGAGCTT58.5 100~250 YI 等[14] HEX TG55 (AAC)7 F: AGTCAAGAGCAGAAGGAGCG
R: TATTGGTGTTGGTGGTGGTG56.3 100~250 YI 等[14] ROX TG65 (TTG)8 F: GCTTTCACTCGGGTTTGTCT
R: AGCAGCAGCAGCAATAACAA55.3 200~300 YI等[14] TAMRA TG70 (AAG)7 F: AGCCTCCGATGAATCCTCTT
R: AACATCTGCTTTTCCATGCC54.5 200~300 YI 等[14] HEX TG81 (TGC)6(TTC)5 F: AGTTGACGCAGCGCTTTAAT
R: GGTTTTGTGGGGAGTTTCAA54.0 180~250 YI 等[14] FAM TG82 (CAG)5(GAG)5 F: AACACCACACCACCTGATGA
R: TACCGCTACAGCAACACCTG57.0 200~300 YI 等[14] ROX TG88 (TG)9 F: GCACAAACATCCATGCAAAC
R: AACAAGGGTCCAGGGAGAGT55.6 200~300 YI 等[14] TAMRA TG90 (CTG)7(ATT)6 F: CACTAGGGCTTCCTGCACTC
R: AGAACAAATATGCCCCGTTG55.7 150~250 YI 等[14] HEX Table 1. SSR primers used in male T. grandis
-
24对SSR引物在121株榧树雄株中共获得85个等位基因,每对引物扩增的等位基因数为2~7个,平均每对引物扩增出3.54个等位基因,其中引物对ZAFU-4检测到7个等位基因;有效等位基因数为45.69个,平均每个位点有有效等位基因数为1.92个;平均观测杂合度(0.461)略高于平均期望杂合度(0.400);Shannon’s信息指数为0.10~1.31,平均为0.70,其中GR98位点的Shannon’s信息指数最高(1.310);多态信息含量为0.040~0.699,平均为0.400,其中GR98位点的多态信息含量最高(0.699),GR48位点最低(0.040) (表2)。
引物编号 平均等位基因 有效等位基因 观测杂合度 期望杂合度 Shannon’s信息指数 多态信息含量 ZAFU-1 2 1.584 0.422 0.370 0.555 0.369 ZAFU-2 6 2.111 0.570 0.529 0.962 0.526 ZAFU-3 2 1.113 0.107 0.102 0.209 0.102 ZAFU-4 7 1.790 0.554 0.443 0.855 0.441 ZAFU-5 3 2.149 0.488 0.537 0.826 0.535 ZAFU-6 2 1.339 0.281 0.254 0.421 0.254 ZAFU-7 2 1.141 0.116 0.124 0.244 0.123 ZAFU-8 3 2.290 1.000 0.566 0.903 0.563 ZAFU-11 2 1.821 0.504 0.453 0.643 0.451 ZAFU-16 6 2.361 0.777 0.579 1.158 0.576 GR12 3 2.396 0.488 0.585 0.967 0.583 GR48 2 1.042 0.041 0.041 0.101 0.041 GR67 5 2.165 0.554 0.540 0.100 0.539 GR81 3 2.721 0.934 0.635 1.044 0.632 GR98 5 3.320 0.760 0.702 1.310 0.699 TG19 3 1.095 0.074 0.088 0.199 0.087 TG32 3 1.086 0.083 0.080 0.186 0.080 TG55 5 3.038 0.703 0.674 1.255 0.671 TG65 2 1.563 0.356 0.362 0.546 0.361 TG70 3 2.394 0.901 0.585 0.945 0.583 TG81 3 1.069 0.066 0.064 0.158 0.064 TG82 3 1.095 0.091 0.088 0.199 0.086 TG88 6 2.842 0.603 0.651 1.205 0.648 TG90 4 2.439 0.546 0.593 1.010 0.590 平均 3.542 1.915 0.459 0.402 0.704 0.400 Table 2. Genetic information of SSR loci amplified from male T. grandis
平均等位基因数淳安居群最高,为2.292个,嵊州居群最少,为2.792个,平均为2.508个。平均有效等位基因数为1.798个;平均观测杂合度为0.459,平均期望杂合度为0.365。5个居群的平均多态位点比率为82.50%,其中嵊州居群最高(95.83%),黄山居群和临安居群分别为87.50%和79.17%,富阳居群和淳安居群最低(75.00%)。Nei’s遗传多样性指数,嵊州居群最高,为0.431,淳安居群最低,为0.332,平均为0.365;Shannon’s信息指数嵊州居群最高,为0.720,淳安居群最低,为0.541,平均为0.608;各居群的Shannon’s信息指数高于Nei’s遗传多样性指数,且两者变化趋势一致。以多样性指数为评价指标,则嵊州居群的遗传多样性最高(Nei’s遗传多样性指数为0.431,Shannon’s信息指数为0.720),淳安居群的遗传多样性较低(Nei’s遗传多样性指数为0.332,Shannon’s信息指数为0.541) (表3)。
居群 样本数
量/株平均等位基
因数平均有效等位
基因数观测杂合度 期望杂合度 Nei’s遗传多样性
指数Shannon’s信息
指数多态位点百
分比/%淳安居群 17 2.292 1.690 0.409 0.342 0.332 0.541 75.00 临安居群 24 2.667 1.857 0.464 0.379 0.371 0.627 79.17 富阳居群 24 2.375 1.822 0.453 0.360 0.352 0.587 75.00 嵊州居群 24 2.792 1.899 0.556 0.441 0.431 0.720 95.83 黄山居群 32 2.417 1.723 0.414 0.346 0.340 0.566 87.50 平均 24.2 2.508 1.798 0.459 0.374 0.365 0.608 82.50 Table 3. Genetic diversity of male populations in male T. grandis
-
5个雄性榧树居群各位点的近交系数为−0.923(ZAFU-8)~0.143(GR12),平均近交系数为−0.227,绝大数位点(22个)近交系数为负值,说明绝大数位点表现杂合子过剩[21]。各位点遗传分化系数为0.006(GR81)~0.286(ZAFU-7),平均遗传分化系数为0.096。由 Nei’s遗传多样性指数估算的雄性榧树Nei’s总基因多样度为0.427。基因流的变化范围较大,平均基因流为4.172(表4)。AMOVA结果表明:榧树雄株居群间差异极显著(P<0.01),居群内的遗传变异远大于居群间,居群内占79%,而居群间则只占21% (表5),表明遗传变异主要集中在居群内。
引物编号 Nei’s总基因多样度 近交系数 遗传分化系数 基因流 引物编号 Nei’s总基因多样度 近交系数 遗传分化系数 基因流 ZAFU-1 0.355 −0.186 0.043 5.616 GR81 0.628 −0.525 0.026 9.520 ZAFU-2 0.526 −0.158 0.068 3.445 GR98 0.687 −0.174 0.058 4.065 ZAFU-3 0.101 −0.165 0.093 2.430 TG19 0.086 −0.011 0.160 1.316 ZAFU-4 0.465 −0.474 0.121 1.820 TG32 0.080 −0.234 0.158 1.336 ZAFU-5 0.635 −0.015 0.084 2.719 TG55 0.670 −0.197 0.114 1.947 ZAFU-6 0.255 −0.145 0.029 8.345 TG65 0.368 0.031 0.009 26.235 ZAFU-7 0.124 −0.313 0.286 0.625 TG70 0.579 −0.818 0.184 1.106 ZAFU-8 0.564 −0.928 0.080 2.882 TG81 0.650 −0.174 0.122 1.793 ZAFU-11 0.460 −0.218 0.083 2.769 TG82 0.089 −0.113 0.062 3.799 ZAFU-16 0.580 −0.425 0.046 5.242 TG88 0.650 −0.045 0.101 2.226 GR12 0.581 0.143 0.082 2.791 TG90 0.549 −0.108 0.155 1.360 GR48 0.031 −0.085 0.064 3.688 平均 0.427 −0.227 0.096 4.172 GR67 0.542 −0.101 0.076 3.048 Table 4. Genetic differentiation of SSR loci in male T. grandis
变异来源 自由度 平方和 均方 方差分量 变异百分比/% P 居群间 4 199.526 49.882 1.797 21 <0.01 居群内 116 791.118 6.820 6.820 79 总计 120 990.645 56.702 8.617 100 Table 5. Analysis of molecular variance (AMOVA) of male populations in male T. grandis
-
5个榧树雄株居群的遗传相似度为0.865~0.978,平均遗传相似度为0.932,相似性很高,但也存在着一定的遗传变异。淳安居群与嵊州居群的遗传距离最大(0.145),而遗传相似度最小(0.865),说明它们的遗传变异最大。临安居群和黄山居群的遗传距离最小(0.022),遗传相似度最大(0.978),表明两者之间的遗传差异最小(表6)。
居群 淳安居群 临安居群 富阳居群 嵊州居群 黄山居群 淳安居群 0.957 0.964 0.865 0.952 临安居群 0.044 0.976 0.895 0.978 富阳居群 0.037 0.024 0.877 0.967 嵊州居群 0.145 0.111 0.132 0.873 黄山居群 0.049 0.022 0.034 0.136 说明:对角线左下角为遗传距离,右上角为遗传相似度 Table 6. Genetic distance and genetic identity among the 5 populations in male T. grandis
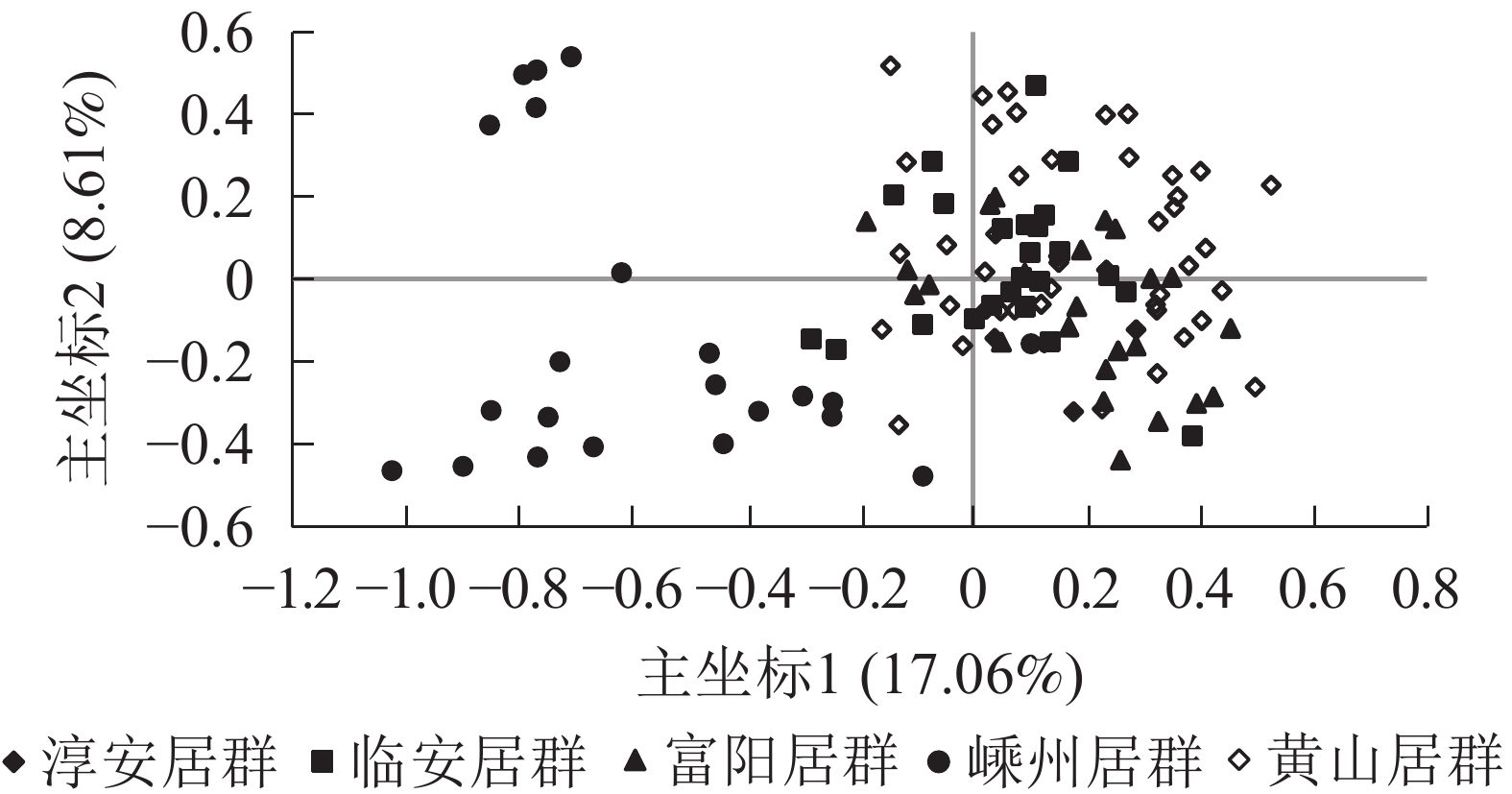
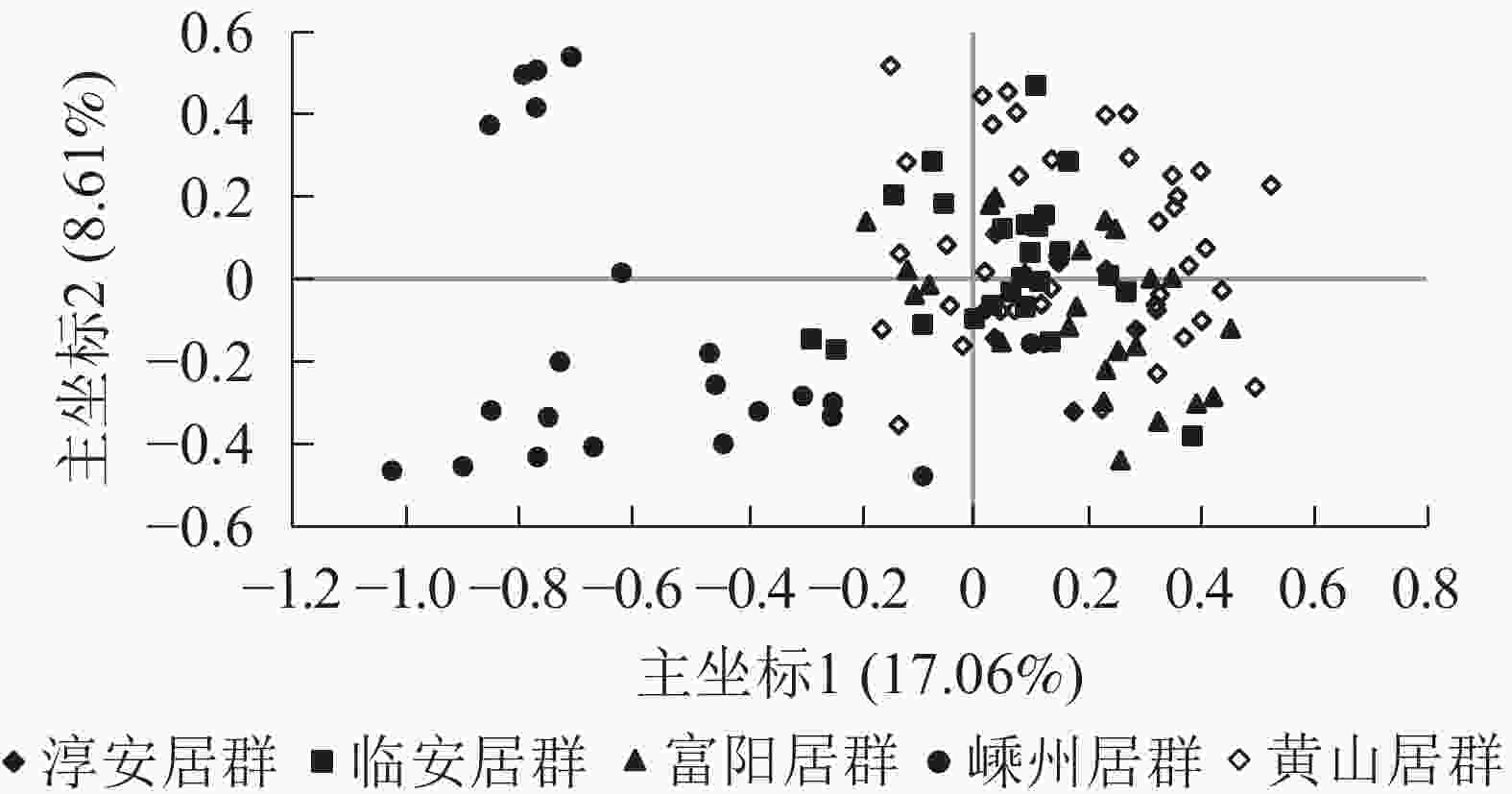
主坐标分析结果表明:5个野生榧树居群主要分成2大类群,其中第1类群包括淳安居群、富阳居群、临安居群、黄山居群,且彼此间混合在一起,难以区分,而嵊州居群单独聚为一类(图2)。
-
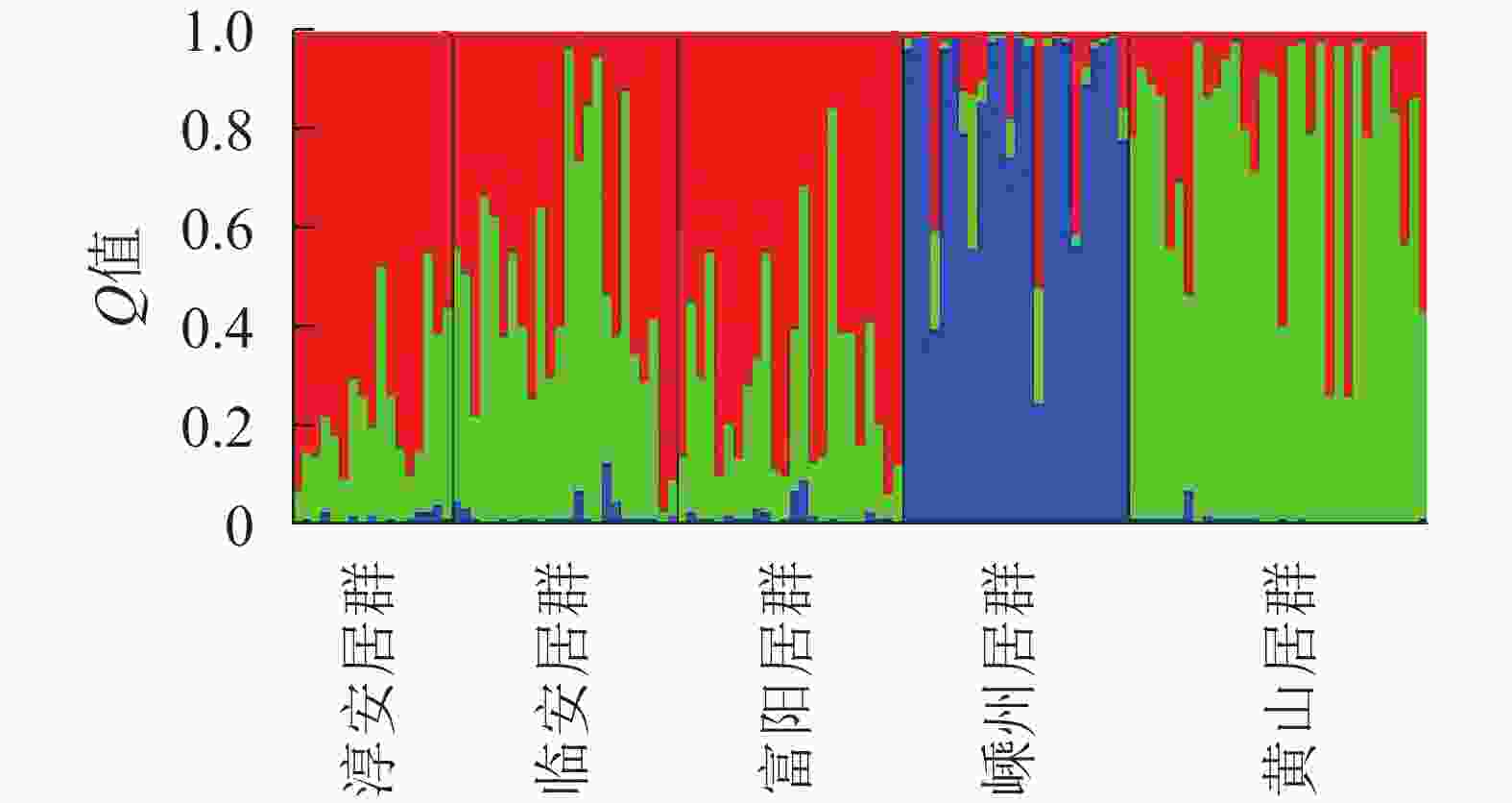
Structure软件分析结果表明:K为2~4 (图3)。供试的所有样品如果分成2个群体,则嵊州居群的雄株单独成1个群体,其余4个居群组合成1个群体。如果将所有样品归属于3个群体,则嵊州居群和黄山居群分别各自形成1个群体,另外3个居群形成1个群体。如果所有样品归属于4个群体,则结果会和聚类的结果一样,临安居群和黄山居群组合成1个群体,另3个居群则各自形成1个群体。结合主坐标分析(图2)的结果,将供试材料分成2~3个群体比较合理。当K为3时,lnP (D)最大,故将所有个体分成3个群体(图3)。由群体遗传结构图(图4)可知:每个颜色表示一种遗传组成,同一聚类亚群中相同的遗传组成占据主要成分,即同一颜色所占比率最大的居群聚类为同一亚群。因此5个野生榧树居群可以划分为3大亚群,淳安居群和富阳居群聚为一类(红色基因池),临安居群和黄山居群聚为一类(绿色基因池),嵊州居群单独聚为一类(蓝色基因池)。
2.1. SSR引物筛选
2.2. 遗传多样性分析
2.3. 遗传分化分析
2.4. 居群聚类分析
2.5. 遗传结构分析
-
本研究表明:尽管SSR标记具有物种的特异性,但近缘种的SSR引物在同属物种中具有一定的通用性。这也从一个侧面反映了物种的亲缘关系。SSR标记的开发基于DNA序列,利用近缘种的SSR引物进行目标物种SSR引物的筛选不失为一种经济的方法。理论上,同一物种的SSR引物在具体的研究中应该都是适用的,但各研究所使用的PCR仪在循环过程中升降温不同,会不同程度地影响引物与模板的结合,进而影响扩增结果,因而实验过程需进行引物筛选。本研究专门研究雄性榧树,这有别于雌性榧树。这也可能造成已有的榧树SSR引物不适用于雄株的现象。
本研究的SSR位点显示:雄性榧树存在多个等位基因的现象(表1)。除多态信息含量以外,不管是观察杂合度还是期望杂合度,雄性榧树的杂合度都较高,这也体现在Shannon’s信息指数上。BOSTSEIN等[22]认为:Shannon’s信息指数大于0.5时表明遗传多样性较高。这与榧树雌雄异株天然杂交产生杂种的特性有关。同样,居群水平观察杂合度、Shannon’s信息指数、多态位点百分比都不低,但观察杂合度要高于期望杂合度。与刘浩凯[11]利用SRAP标记研究雄性榧树居群的结果相比,本研究结果(多态位点百分比、Nei’s遗传多样性指数、Shannon’s信息指数)要高于显性标记的研究结果,可见共显性SSR标记要优于显性标记,在茶树Camellia sinensis[23]、甜菜Beta vulgaris[24]中也有类似的结果。
本研究所得的平均Shannon’s信息指数为0.704,均高于同属植物巴山榧T. fargesii居群(Shannon’s信息指数为0.370)[25]和长叶榧居群(Shannon’s信息指数为0.309)[26]以及同科植物南方红豆杉Taxus wallichiana (Shannon’s信息指数0.376)[27]和白豆杉Pseudotaxus chienii (Shannon’s信息指数0.236)[28]研究所得的结果。WRIGHT[25]研究认为:遗传分化系数大于0.25时,表明群体间有很大的遗传分化。本研究雄性榧树居群的遗传分化程度很小(遗传分化系数为0.096 <0.25),说明居群间基因交流较频繁。基因流是影响居群内部和居群间遗传变异程度的重要因素[27],其大小反映居群遗传结构的变异,它能防止基因分化。WRIGHT[25]认为:基因流>1时说明居群间存在一定的基因流动。本研究5个雄性榧树居群间的基因流为4.172,不仅居群间存在较大的基因流动,且大于SRAP标记的分析结果(基因流为2.192)[11],但低于异交、风媒植物基因流的平均水平(5.380)[29]。这与榧树雌雄异株风媒的特性相符。频繁的基因交流导致居群间遗传的均质化,使各居群间遗传相似度较高。这在遗传分化系数上有所体现,与刘浩凯[11]的研究结果一致,也与HAMRICK[29]研究异交风媒植物的结论相符,即遗传变异主要集中在居群内。因此,选育可基于居群内个体的表现进行,且要注意对优良单株的保护。
从主坐标分析结果及遗传结构来看,嵊州居群均相对单独地聚为一类。嵊州是香榧的主产地,当地不泛上千年的古香榧[30],也是榧树古老的原生地,而另4个居群则非也。从主坐标分析结果来看:除嵊州居群外,其余居群的个体混杂在一起,这也说明居群间分化较小。
遗传多样性与生物自身的生存和竞争能力息息相关,其丰富程度决定物种对环境的适应能力和进化潜力[31]。遗传多样性越高,该物种对环境变化的适应能力越强,有利于资源进一步的选择和保存[32]。本研究表明:雄性榧树的遗传多样性丰富。雄性榧树的多样性为雌性榧树的多样性及榧树物种水平的多样性作出了重要贡献,为优良种质的选择及杂交育种提供了丰富的物质基础,但同时也是造成栽培类型香榧内部分化的原因[30],因此,雄性榧树资源值得大力保护与利用。









 DownLoad:
DownLoad: