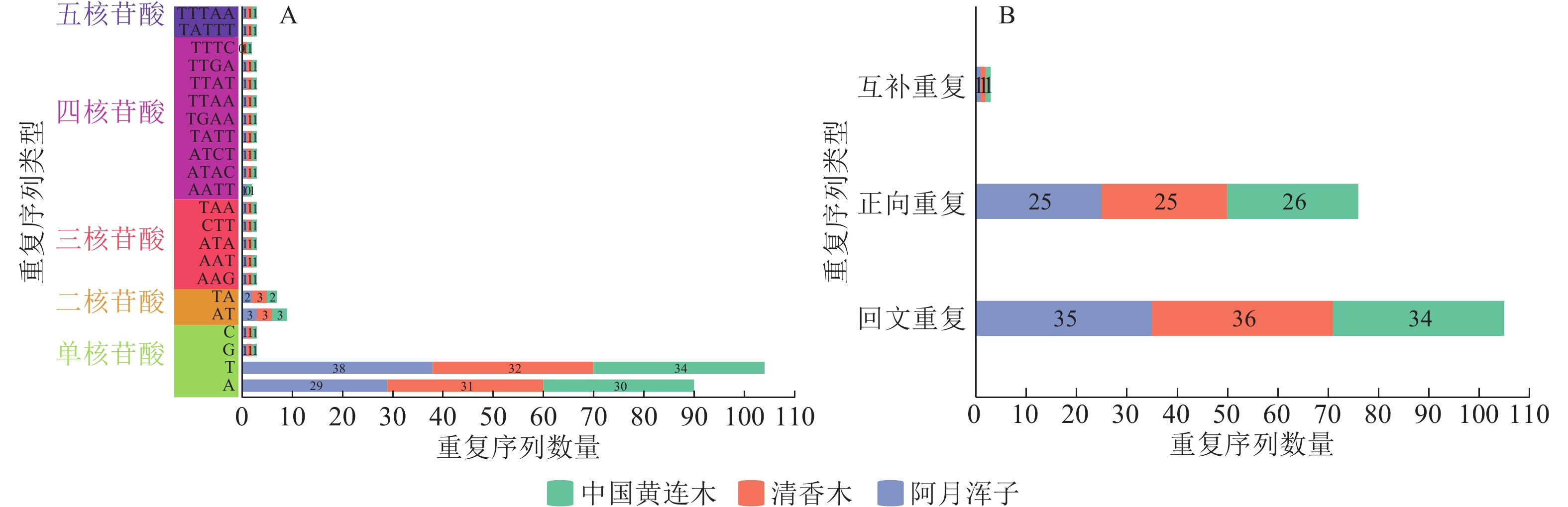
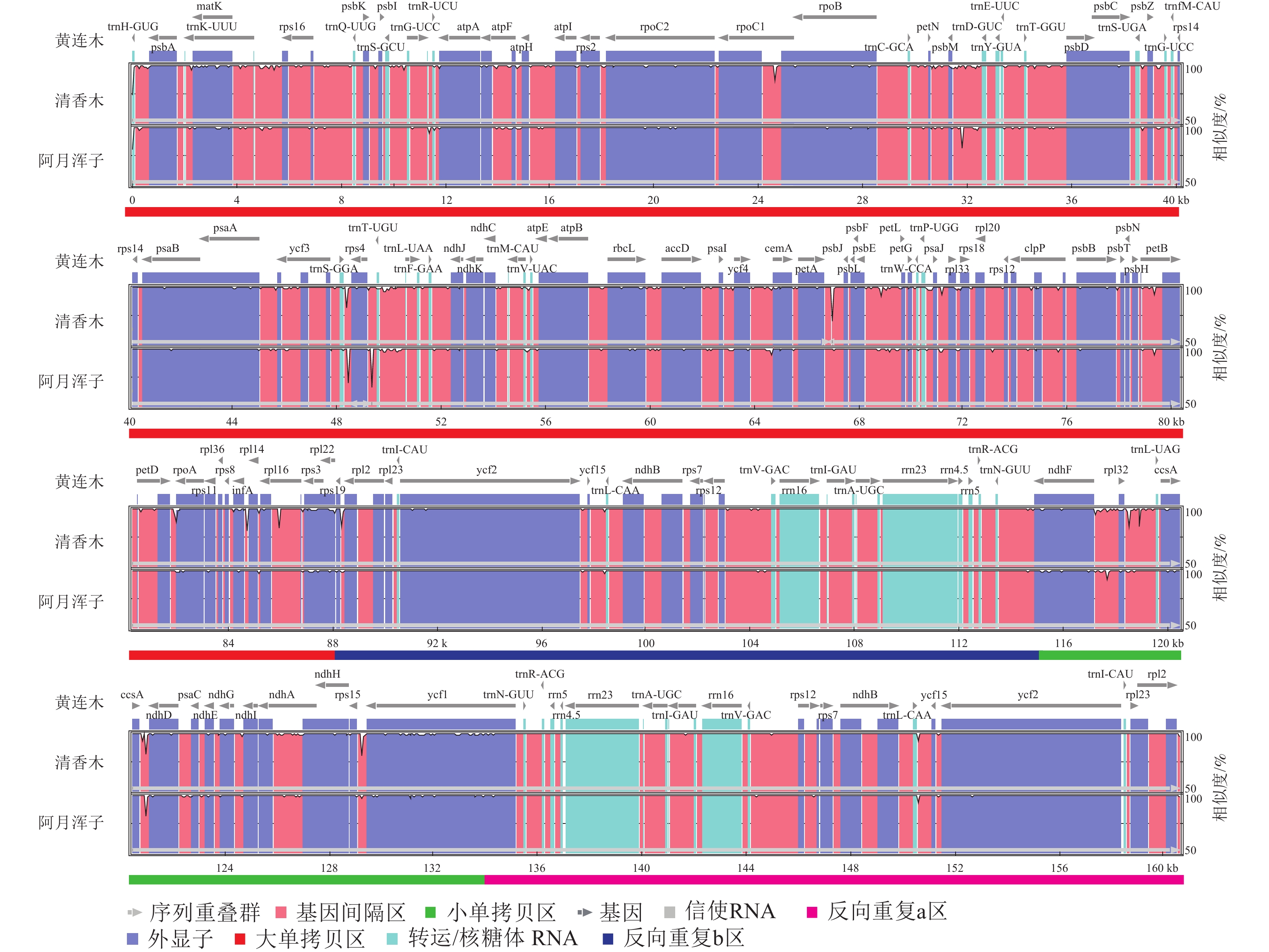
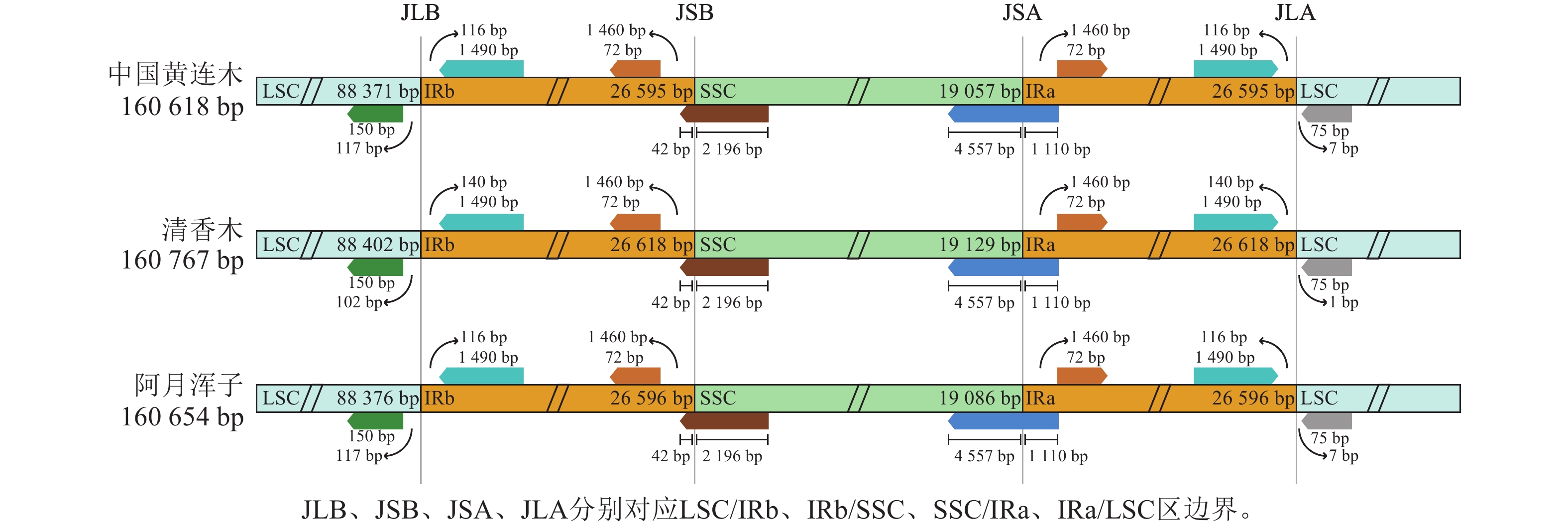
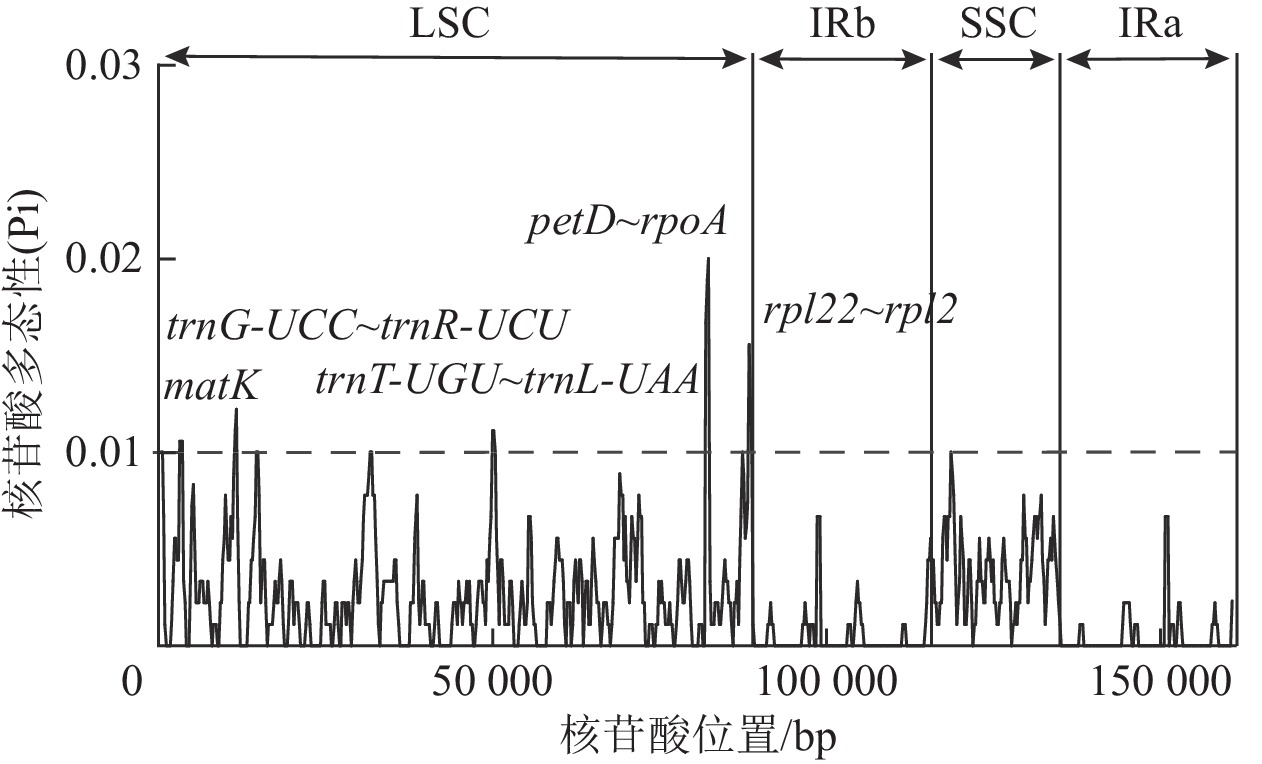
| [1] |
ALZAHRANI D A. Complete chloroplast genome of
Abutilon fruticosum: genome structure, comparative and phylogenetic analysis [J].
Plants, 2021,
10(2): 270. DOI: 10.3390/plants10020270. |
| [2] |
柳婷婷, 许丽爱, 胡紫蔚, 等. 菜心叶绿体基因组特征及系统发育分析[J/OL]. 浙江农林大学学报, 2025-09-29. https://kns.cnki.net/kcms/detail/33.1370.S.20250928.1915.006.html. LIU Tingting, XU Liai, HU Ziwei, et al. Characteristics of the chloroplast genome and phylogenetic analysis of flowering Chinese cabbage[J/OL]. Journal of Zhejiang A&F University, 2025-09-29 . https://kns.cnki.net/kcms/detail/33.1370.S.20250928.1915.006.html. |
LIU Tingting, XU Liai, HU Ziwei, et al. Characteristics of the chloroplast genome and phylogenetic analysis of flowering Chinese cabbage[J/OL]. Journal of Zhejiang A&F University, 2025-09-29 . https://kns.cnki.net/kcms/detail/33.1370.S.20250928.1915.006.html. |
| [3] |
PALMER J. Comparative organization of chloroplast genomes [J].
Annual Review of Genetics, 1985,
19: 325−354. DOI: 10.1146/annurev.genet.19.1.325. |
| [4] |
TAN Wei, GAO Han, JIANG Weiling,
et al. The complete chloroplast genome of
Gleditsia sinensis and
Gleditsia japonica: genome organization, comparative analysis, and development of taxon specific DNA mini-barcodes [J].
Scientific Reports, 2020,
10: 16309. DOI: 10.1038/s41598-020-73392-7. |
| [5] |
朱梦飞, 胡迎峰, 师雪芹. 濒危植物新绒苔叶绿体基因组特征及系统发育位置分析[J]. 浙江农林大学学报, 2025,
42(1): 55−63. ZHU Mengfei, HU Yingfeng, SHI Xueqin. Characterization and phylogenetic location analysis of chloroplast of the endangered plant
Neotrichocolea bissetii [J].
Journal of Zhejiang A&F University, 2025,
42(1): 55−63. DOI: 10.11833/j.issn.2095-0756.20240356. |
ZHU Mengfei, HU Yingfeng, SHI Xueqin. Characterization and phylogenetic location analysis of chloroplast of the endangered plant
Neotrichocolea bissetii [J]. Journal of Zhejiang A&F University, 2025, 42(1): 55−63. DOI:
10.11833/j.issn.2095-0756.20240356. |
| [6] |
江转转, 陈红, 鲍红艳, 等. 狼尾草属叶绿体基因组特征与分子标记开发[J]. 浙江农林大学学报, 2025,
42(2): 365−372. JIANG Zhuanzhuan, CHEN Hong, BAO Hongyan,
et al. Chloroplast genome characteristics and molecular marker development of
Pennisetum [J].
Journal of Zhejiang A&F University, 2025,
42(2): 365−372. DOI: 10.11833/j.issn.2095-0756.20240371. |
JIANG Zhuanzhuan, CHEN Hong, BAO Hongyan, et al. Chloroplast genome characteristics and molecular marker development of
Pennisetum [J]. Journal of Zhejiang A&F University, 2025, 42(2): 365−372. DOI:
10.11833/j.issn.2095-0756.20240371. |
| [7] |
MOLINA A, HERVÁS-STUBBS S, DANIELL H,
et al. High-yield expression of a viral peptide animal vaccine in transgenic tobacco chloroplasts [J].
Plant Biotechnology Journal, 2004,
2(2): 141−153. DOI: 10.1046/j.1467-7652.2004.00057.x. |
| [8] |
JIN Honglei, DUAN Sujuan, ZHANG Pengxiang,
et al. Dual roles for CND1 in maintenance of nuclear and chloroplast genome stability in plants [J].
Cell Reports, 2023,
42(3): 112268. DOI: 10.1016/j.celrep.2023.112268. |
| [9] |
沈植国, 宋宏伟, 韩健, 等. 世界黄连木属种质资源种类与分布综述[J]. 世界林业研究, 2012,
25(5): 29−34. SHEN Zhiguo, SONG Hongwei, HAN Jian,
et al. Species and distribution for world germplasm resources of
Pistacia L. [J].
World Forestry Research, 2012,
25(5): 29−34. DOI: 10.13348/j.cnki.sjlyyj.2012.05.010. |
SHEN Zhiguo, SONG Hongwei, HAN Jian, et al. Species and distribution for world germplasm resources of
Pistacia L. [J]. World Forestry Research, 2012, 25(5): 29−34. DOI:
10.13348/j.cnki.sjlyyj.2012.05.010. |
| [10] |
郑万钧. 中国树木志: 第4卷[M]. 北京: 中国林业出版社, 2004: 4227−4230. ZHENG Wanjun. Chinese Tree Chronicles: Volume 4[M]. Beijing: China Forestry Publishing House, 2004: 4227−4230. |
ZHENG Wanjun. Chinese Tree Chronicles: Volume 4[M]. Beijing: China Forestry Publishing House, 2004: 4227−4230. |
| [11] |
LIU Xianghua, XING Chao, RUAN Ying,
et al. Determination of fatty acid methyl esters in biodiesel produced from
Pistacia chinensis oil by GC [J].
Applied Mechanics and Materials, 2013,
291/294: 253−256. DOI: 10.4028/www.scientific.net/amm.291-294.253. |
| [12] |
TANG Mingli, ZHANG Pingping, ZHANG Liyun,
et al. A potential bioenergy tree:
Pistacia chinensis bunge [J].
Energy Procedia, 2012,
16: 737−746. DOI: 10.1016/j.egypro.2012.01.119. |
| [13] |
乔永锋, 彭永芳, 方云山, 等. 云南清香木绿叶和嫩红叶挥发性成分对比研究[J]. 安徽农业科学, 2013,
41(4): 1583−1584, 1587. QIAO Yongfeng, PENG Yongfang, FANG Yunshan,
et al. Study on the volatile component of green and red leave of the
Pistacia weinmannifolia [J].
Journal of Anhui Agricultural Sciences, 2013,
41(4): 1583−1584, 1587. DOI: 10.13989/j.cnki.0517-6611.2013.04.067. |
QIAO Yongfeng, PENG Yongfang, FANG Yunshan, et al. Study on the volatile component of green and red leave of the
Pistacia weinmannifolia [J]. Journal of Anhui Agricultural Sciences, 2013, 41(4): 1583−1584, 1587. DOI:
10.13989/j.cnki.0517-6611.2013.04.067. |
| [14] |
ZENG Lin, TU Xiaolong, DAI He,
et al. Whole genomes and transcriptomes reveal adaptation and domestication of pistachio [J].
Genome Biology, 2019,
20(1): 79. DOI: 10.1186/s13059-019-1686-3. |
| [15] |
ZHENG Shuyu, POCZAI P, HYVÖNEN J,
et al. Chloroplot: an online program for the versatile plotting of organelle genomes [J].
Frontiers in Genetics, 2020,
11: 576124. DOI: 10.3389/fgene.2020.576124. |
| [16] |
BEIER S, THIEL T, MÜNCH T,
et al. MISA-web: a web server for microsatellite prediction [J].
Bioinformatics, 2017,
33(16): 2583−2585. DOI: 10.1093/bioinformatics/btx198. |
| [17] |
KURTZ S, CHOUDHURI J V, OHLEBUSCH E,
et al. REPuter: the manifold applications of repeat analysis on a genomic scale [J].
Nucleic Acids Research, 2001,
29(22): 4633−4642. DOI: 10.1093/nar/29.22.4633. |
| [18] |
FRAZER K A, PACHTER L, POLIAKOV A,
et al. VISTA: computational tools for comparative genomics [J].
Nucleic Acids Research, 2004,
32: W273−W279. DOI: 10.1093/nar/gkh458. |
| [19] |
LI Huie, GUO Qiqiang, XU Lei,
et al. CPJSdraw: analysis and visualization of junction sites of chloroplast genomes [J].
PeerJ, 2023,
11: e15326. DOI: 10.7717/peerj.15326. |
| [20] |
ROZAS J, FERRER-MATA A, SÁNCHEZ-DELBARRIO J C,
et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets [J].
Molecular Biology and Evolution, 2017,
34(12): 3299−3302. DOI: 10.1093/molbev/msx248. |
| [21] |
KATOH K, MISAWA K, KUMA K I,
et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform [J].
Nucleic Acids Research, 2002,
30(14): 3059−3066. DOI: 10.1093/nar/gkf436. |
| [22] |
KUMAR S, NEI M, DUDLEY J,
et al. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences [J].
Briefings in Bioinformatics, 2008,
9(4): 299−306. DOI: 10.1093/bib/bbn017. |
| [23] |
刘潮, 叶秀传, 杨春会, 等. 万寿菊属物种叶绿体基因组特征及系统发育分析[J]. 曲靖师范学院学报, 2024,
43(3): 24−30. LIU Chao, YE Xiuchuan, YANG Chunhui,
et al. Chloroplast genomic characterization and phylogenetic analysis of
Tagetes species [J].
Journal of Qujing Normal University, 2024,
43(3): 24−30. DOI: 10.3969/j.issn.1009-8879.2024.03.005. |
LIU Chao, YE Xiuchuan, YANG Chunhui, et al. Chloroplast genomic characterization and phylogenetic analysis of
Tagetes species [J]. Journal of Qujing Normal University, 2024, 43(3): 24−30. DOI:
10.3969/j.issn.1009-8879.2024.03.005. |
| [24] |
LU Ruisen, LI Pan, QIU Yingxiong. The complete chloroplast genomes of three
Cardiocrinum (Liliaceae) species: comparative genomic and phylogenetic analyses [J].
Frontiers in Plant Science, 2016,
7: 2054. DOI: 10.3389/fpls.2016.02054. |
| [25] |
STRAUSS S H, PALMER J D, HOWE G T,
et al. Chloroplast genomes of two conifers lack a large inverted repeat and are extensively rearranged [J].
Proceedings of the National Academy of Sciences of the United States of America, 1988,
85(11): 3898−3902. DOI: 10.1073/pnas.85.11.3898. |
| [26] |
修志莹, 赵艳玲, 程永琴, 等. 牡荆属叶绿体基因组比较分析及系统发育分析[J]. 广西植物, 2024,
44(9): 1755−1771. XIU Zhiying, ZHAO Yanling, CHENG Yongqin,
et al. Comparative analysis of chloroplast genomes and phylogenetic analysis of
Vitex [J].
Guihaia, 2024,
44(9): 1755−1771. DOI: 10.11931/guihaia.gxzw202307027. |
XIU Zhiying, ZHAO Yanling, CHENG Yongqin, et al. Comparative analysis of chloroplast genomes and phylogenetic analysis of
Vitex [J]. Guihaia, 2024, 44(9): 1755−1771. DOI:
10.11931/guihaia.gxzw202307027. |
| [27] |
YANG Tiange, WU Zhihua, TIE Jun,
et al. A comprehensive analysis of chloroplast genome provides new insights into the evolution of the genus
Chrysosplenium [J].
International Journal of Molecular Sciences, 2023,
24(19): 14735. DOI: 10.3390/ijms241914735. |
| [28] |
SUN Jiahui, WANG Yiheng, LIU Yanlei,
et al. Evolutionary and phylogenetic aspects of the chloroplast genome of
Chaenomeles species [J].
Scientific Reports, 2020,
10: 11466. DOI: 10.1038/s41598-020-67943-1. |
| [29] |
DING Shixiong, LI Jiachen, HU Ke,
et al. Morphological characteristics and comparative chloroplast genome analyses between red and white flower phenotypes of
Pyracantha fortuneana (Maxim. ) Li (Rosaceae), with implications for taxonomy and phylogeny [J].
Genes, 2022,
13(12): 2404. DOI: 10.3390/genes13122404. |
| [30] |
BODIN S S, KIM J S, KIM J H. Complete chloroplast genome of
Chionographis japonica (Willd. ) Maxim. (Melanthiaceae): comparative genomics and evaluation of universal primers for Liliales [J].
Plant Molecular Biology Reporter, 2013,
31(6): 1407−1421. DOI: 10.1007/s11105-013-0616-x. |
| [31] |
ZHAO Yongbing, YIN Jinlong, GUO Haiyan,
et al. The complete chloroplast genome provides insight into the evolution and polymorphism of
Panax ginseng [J].
Frontiers in Plant Science, 2014,
5: 696. DOI: 10.3389/fpls.2014.00696. |
| [32] |
戴前莉, 朱恒星, 魏卓, 等. 2种楠属植物叶绿体基因组特征及系统发育[J]. 东北林业大学学报, 2024,
52(6): 58−63, 84. DAI Qianli, ZHU Hengxing, WEI Zhuo,
et al. Characteristics and phylogenetic of the chloroplast genomes of two
Phoebe plants [J].
Journal of Northeast Forestry University, 2024,
52(6): 58−63, 84. DOI: 10.13759/j.cnki.dlxb.2024.06.004. |
DAI Qianli, ZHU Hengxing, WEI Zhuo, et al. Characteristics and phylogenetic of the chloroplast genomes of two
Phoebe plants [J]. Journal of Northeast Forestry University, 2024, 52(6): 58−63, 84. DOI:
10.13759/j.cnki.dlxb.2024.06.004. |
| [33] |
付文佛, 肖涛, 张永洪, 等. 石蒜属植物叶绿体基因组结构及系统学研究[J]. 亚热带植物科学, 2023,
52(4): 271−286. FU Wenfo, XIAO Tao, ZHANG Yonghong,
et al. Chloroplast genome characteristics and phylogeny of the genus
Lycoris(Amaryllidaceae) [J].
Subtropical Plant Science, 2023,
52(4): 271−286. DOI: 10.3969/j.issn.1009-7791.2023.04.001. |
FU Wenfo, XIAO Tao, ZHANG Yonghong, et al. Chloroplast genome characteristics and phylogeny of the genus
Lycoris(Amaryllidaceae) [J]. Subtropical Plant Science, 2023, 52(4): 271−286. DOI:
10.3969/j.issn.1009-7791.2023.04.001. |
| [34] |
熊瑶, 童凌斐, 曹岚, 等. 四种忍冬属植物叶绿体基因组结构特征及系统发育分析[J]. 药学学报, 2024,
59(11): 3164−3171. XIONG Yao, TONG Lingfei, CAO Lan,
et al. Structural characteristics and phylogenetic analysis of chloroplast genomes of four species of
Lonicera [J].
Acta Pharmaceutica Sinica, 2024,
59(11): 3164−3171. DOI: 10.16438/j.0513-4870.2024-0306. |
XIONG Yao, TONG Lingfei, CAO Lan, et al. Structural characteristics and phylogenetic analysis of chloroplast genomes of four species of
Lonicera [J]. Acta Pharmaceutica Sinica, 2024, 59(11): 3164−3171. DOI:
10.16438/j.0513-4870.2024-0306. |
| [35] |
秃玉翔, 赵文植, 沈伟祥, 等. 紫麻属叶绿体全基因组特征及系统发育分析[J]. 种子, 2023,
42(12): 24−30, 37. TU Yuxiang, ZHAO Wenzhi, SHEN Weixiang,
et al. Characterization of complete chloroplast genome and phylogenetic analysis of
Oreocnide [J].
Seed, 2023,
42(12): 24−30, 37. DOI: 10.16590/j.cnki.1001-4705.2023.12.024. |
TU Yuxiang, ZHAO Wenzhi, SHEN Weixiang, et al. Characterization of complete chloroplast genome and phylogenetic analysis of
Oreocnide [J]. Seed, 2023, 42(12): 24−30, 37. DOI:
10.16590/j.cnki.1001-4705.2023.12.024. |
| [36] |
YAN Linjun, WANG Huanli, HUANG Xi,
et al. Chloroplast genomes of genus
Tilia: comparative genomics and molecular evolution [J].
Frontiers in Genetics, 2022,
13: 925726. DOI: 10.3389/fgene.2022.925726. |
| [37] |
ZHAO Fei, LI Bo, DREW B T,
et al. Leveraging plastomes for comparative analysis and phylogenomic inference within Scutellarioideae (Lamiaceae) [J].
PLoS One, 2020,
15(5): e0232602. DOI: 10.1371/journal.pone.0232602. |
| [38] |
XU Chao, DONG Wenpan, LI Wenqing,
et al. Comparative analysis of six
Lagerstroemia complete chloroplast genomes [J].
Frontiers in Plant Science, 2017,
8: 15. DOI: 10.3389/fpls.2017.00015. |
| [39] |
YANG Yanci, ZHOU Tao, DUAN Dong,
et al. Comparative analysis of the complete chloroplast genomes of five
Quercus species [J].
Frontiers in Plant Science, 2016,
7: 959. DOI: 10.3389/fpls.2016.00959. |
| [40] |
YU Xiaoyue, ZUO Lihui, LU Dandan,
et al. Comparative analysis of chloroplast genomes of five
Robinia species: genome comparative and evolution analysis [J].
Gene, 2019,
689: 141−151. DOI: 10.1016/j.gene.2018.12.023. |
| [41] |
PENJOR T, YAMAMOTO M, UEHARA M,
et al. Phylogenetic relationships of
Citrus and its relatives based on matK gene sequences [J].
PLoS One, 2013,
8(4): e62574. DOI: 10.1371/journal.pone.0062574. |
| [42] |
GAO Ting, SUN Zhiying, YAO Hui,
et al. Identification of Fabaceae plants using the DNA barcode matK [J].
Planta Medica, 2011,
77(1): 92−94. DOI: 10.1055/s-0030-1250050. |
| [43] |
郑梦迪, 张春, 马瑞龙, 等. 基于matK和ITS2及二级结构对药材香薷及其混伪品的鉴别研究[J]. 中国现代应用药学, 2022,
39(17): 2222−2228. ZHENG Mengdi, ZHANG Chun, MA Ruilong,
et al. Identification of moslae herba and its adulterants based on matK, ITS2 and its secondary structure [J].
Chinese Journal of Modern Applied Pharmacy, 2022,
39(17): 2222−2228. DOI: 10.13748/j.cnki.issn1007-7693.2022.17.008. |
ZHENG Mengdi, ZHANG Chun, MA Ruilong, et al. Identification of moslae herba and its adulterants based on matK, ITS2 and its secondary structure [J]. Chinese Journal of Modern Applied Pharmacy, 2022, 39(17): 2222−2228. DOI:
10.13748/j.cnki.issn1007-7693.2022.17.008. |
| [44] |
TAUFIQ PROBOJATI R, HADIYANTI N, HAPSARI L. Identification and genetic diversity of pineapple local accessions from kediri and closely related species (Bromeliaceae) based on
matK gene [J].
Biotropika: Journal of Tropical Biology, 2024,
12(3): 111−118. DOI: 10.21776/ub.biotropika.2024.012.03.01. |
| [45] |
YI Tingshuang, WEN Jun, GOLAN-GOLDHIRSH A,
et al. Phylogenetics and reticulate evolution in
Pistacia (Anacardiaceae) [J].
American Journal of Botany, 2008,
95(2): 241−251. DOI: 10.3732/ajb.95.2.241. |
| [46] |
LIU Li, WANG Zhen, SU Yingjuan,
et al. Population transcriptomic sequencing reveals allopatric divergence and local adaptation in
Pseudotaxus chienii (Taxaceae) [J].
BMC Genomics, 2021,
22(1): 388. DOI: 10.1186/s12864-021-07682-3. |









 DownLoad:
DownLoad: