-
简单重复序列(simple sequence repeat,SSR)是一种由1~6个核苷酸为单位多次串联重复组成的核苷酸序列,广泛分布于生物基因组中[1-2]。SSR分子标记技术具有共显性、多态性高、遗传信息量大、稳定性高、重复性好等优点[3-5],已被广泛应用于遗传多样性分析、种质鉴定、基因定位、功能基因挖掘等领域[1, 6-7]。传统的基因组SSR标记技术(genomic-SSR)需要构建DNA文库,耗时耗力[6, 8]。表达序列标签SSR(expressed sequence tag SSR,EST-SSR),来源于基因的转录组区,相对genomic-SSR更简便,经济且信息量大、通用性好[6, 9]。宋跃朋等[10]利用杨树Populus对genomic-SSR和EST-SSR这2种标记的遗传差异做了比较研究,发现在鉴定基因型方面EST-SSR相对更精确。目前,基于转录组序列的SSR标记在很多植物上得到应用[11-16]。夏蜡梅Sinocalycanthus chinensis隶属于蜡梅科Calycanthaceae夏蜡梅属Sinocalycanthus,为第三纪孑遗植物[17],其系统地位特殊,且具有较高的观赏价值。由于自然种群规模小,生境片段化使夏蜡梅不同种群间存在不同距离的隔离[18],种群间基因交流少,内部近交率增加,导致种群内遗传多样性低,种群间遗传分化增加[19-24]。尽管已有多种标记[同工酶[19],随机扩增多态DNA(RAPD)[20, 22, 24],ISSR[21, 24]和扩增片段长度多态性分子标记(AFLP)[25]]用于夏蜡梅的遗传多样性和交配系统分析,但之前标记均是显性标记,且所揭示的多态性信息普遍偏低。因此,开发共显性的SSR标记用于夏蜡梅的遗传多样性相关研究势在必行。本研究以之前获得的夏蜡梅转录组数据,分析SSR位点特征,设计相应引物,建立最优的SSR-PCR体系,从不同水平(种群间和种群内)筛选高效、多态的引物。
HTML
-
夏蜡梅不同野生种群材料:选择浙江天台的经过坪(JGP)和捣臼孔(DJK),浙江临安的龙塘山(LTS),西坑(XK),前坑(QK)和安徽绩溪的龙须山(LXS)等6个野生种群,采集新鲜叶片,硅胶干燥低温保存,随机选取个体3个·种群-1。
同一野生种群材料:选择经过坪(JGP)种群,按比例估算采样植株的间距和大小,采集24个个体叶片样本(间隔距离视种群大小适当调整),各年龄级采集数量大致相等,硅胶干燥低温保存。
-
实验采用十六烷基三甲基溴化铵(CTAB)法提取基因组DNA[26]。
-
基于夏蜡梅转录组数据库组装的125 014条unigene序列,运用MISA进行SSR位点搜索[27]:重复单元长度为1~6个核苷酸,最少重复次数为12,6,5,5,4,4次;SSR位点侧翼序列的长度≥150 bp。据检测到的位点,用Primer 3.0引物批量设计程序设计引物[4]:引物序列中没有SSR;序列在DNA保守序列区内;长度为15~30 bp;上下游引物不存在互补序列;引物自身不存在互补序列;引物退火温度(Tm)为53~63 ℃;上下游引物的Tm值相差≤5 ℃;鸟嘌呤和胞嘧啶所占的比率(GC含量)为40%~60%。随机挑选2~6核苷酸重复基序的引物220对(编号:XLM001~XLM220),由上海生工生物工程股份有限公司合成,用于后续SSR引物筛选。
-
以夏蜡梅叶片DNA为模板,SSR-PCR扩增程序参考WU等[28]:94 ℃预变性3 min;30个循环(94 ℃变性30 s,最佳Tm退火30 s,72 ℃ 1 min),72 ℃延伸10 min。
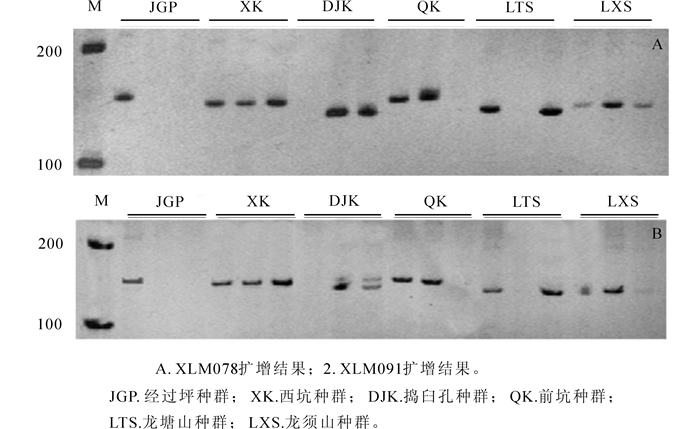
SSR-PCR扩增采用20.0 μL体系,利用五因素四水平正交试验设计L16(45)(表 1)筛选Taq酶,镁离子(Mg2+),DNA,dNTP和引物5种因素的浓度,建立夏蜡梅SSR-PCR扩增体系。选择编号XLM78和XLM91的2对引物用于优化PCR扩增体系。
编号 Taq酶/(×16.67 nkat) 镁离子/(mmol • L-1) DNA/ng dNTP/(mmol • L-1) 引物/(μmol • L-1) 1 0.1 1.5 25 0.10 0.10 2 0.1 2.0 50 0.15 0.20 3 0.1 2.5 75 0.20 0.30 4 0.1 3.0 100 0.25 0.40 5 0.2 1.5 50 0.20 0.40 6 0.2 2.0 25 0.25 0.30 7 0.2 2.5 100 0.10 0.20 8 0.2 3.0 75 0.15 0.10 9 0.3 1.5 75 0.25 0.20 10 0.3 2.0 100 0.20 0.10 11 0.3 2.5 25 0.15 0.40 12 0.3 3.0 50 0.10 0.30 13 0.4 1.5 100 0.15 0.30 14 0.4 2.0 75 0.10 0.40 15 0.4 2.5 50 0.25 0.10 16 0.4 3.0 25 0.20 0.20 Table 1. Orthogonal experimental design of SSR-PCR technique system for Sinocalycanthus chinensis
PCR扩增产物用体积分数为8%非变性聚丙烯酰胺凝胶电泳检测,160 V电压,90 min,银染显色。
-
利用优化的PCR扩增体系和6个种群(个体3个·种群-1)对合成的220对引物进行多态性筛选[29]:根据条带扩增情况统计能够稳定扩增出清晰目的条带的引物;根据条带迁移情况统计目标范围内出现相异条带的引物;根据条带大小的不同分类标记带型“A,B,C,D,…”,纯合子条带可记为“AA,BB,CC,DD,…”,杂合子条带根据带型表示为“AB,BD,CD,BD,…”,带型数据可用于后续遗传多样性等遗传分析。
利用同一种群(JGP)的24个个体对筛选的种群间多态性引物进行PCR扩增,统计目标范围内出现相异条带的引物。读带方法同上。
1.1. 试验材料
1.2. 基因组DNA的提取
1.3. SSR位点开发与引物设计
1.4. 聚合酶链式反应(PCR)扩增体系的优化
1.5. SSR引物筛选和验证
-
从夏蜡梅125 014条unigene序列(总覆盖长度为137.52 Mb)中检测到22 113条序列上包含了共26 564个SSR位点(21.25%),平均密度为5.18 kb。其中含有2个或2个以上SSR位点的序列有3 664条(16.57%),复合SSRs有1 066条(4.01%)。
夏蜡梅不同重复类型的EST-SSRs分布频率不同(表 2)。最丰富的类型是二碱基重复型,占42.43%;其次是单碱基重复型和三碱基重复型,分别占了29.50%和23.25%;四碱基重复型、五碱基重复型、六碱基重复型较少,占4.82%。从重复次数看(表 2),二碱基及二碱基以上重复型中,以5~7次重复为主,其次是8~10次重复:二碱基重复型集中在6~10次;三碱基重复型、四碱基重复型集中在5~6次;五碱基重复型、六碱基重复型集中在4次。从SSR重复基元出现的频率看(表 3):单碱基重复型以(A/T)n形式为主,占99.07%;(C/G)n极少,仅占0.93%。二碱基重复型以(AG/CT)n形式为主,占85.33%,占SSR总数的36.21%;(CG/CG)n最少仅占本类基序比例的0.21%,占SSR总数的0.09%。碱基重复型以(AAG/CTT)n形式最多,占40.8%,其次是(ATC/ATG)n和(AGC/CTG)n,分别占18.26%和11.69%,(ACT/AGT)n最少,占1.12%。四碱基重复型、五碱基重复型、六碱基重复型的重复基元种类较多,占SSR总数比例较少(4.82%)。
重复基元 不同重复次数的EST-SSRs 总计 分布频率/% 4 5 6 7 8 9 10 11 ≥12 单碱基重复 7 837 7 837 29.50 二碱基重复 3 360 2 017 1 665 1 992 1 672 552 14 11 272 42.43 三碱基重复 3 645 1 603 818 110 6 176 23.25 四碱基重复 226 55 281 1.06 五碱基重复 416 48 464 1.75 六碱基重复 534 534 2.01 总计 950 3 919 5 018 2 835 1 775 1 992 1 672 552 7 851 分布频率/% 3.58 14.75 18.89 10.67 6.68 7.5 6.29 2.08 29.56 Table 2. Distribution of the SSR motifs in Sinocalycanthus chinensis
主要SSR基序 数量 占本类SSR重复基序比例/% 占总SSR重复基序比例/% A/T 7 764 99.07 29.23 C/G 73 0.93 0.27 AC/GT 1 044 9.26 3.93 AG/CT 9 618 85.33 36.21 AT/AT 586 5.20 2.21 CG/CG 24 0.21 0.09 AAC/GTT 240 3.89 0.90 AAG/CTT 2 520 40.80 9.49 AAT/ATT 474 7.67 1.78 ACC/GGT 363 5.88 1.37 ACG/CGT 96 1.55 0.36 ACT/AGT 69 1.12 0.26 AGC/CTG 722 11.69 2.72 AGG/CCT 439 7.11 1.65 ATC/ATG 1 128 18.26 4.25 CCG/CGG 125 2.02 0.47 Table 3. Distribution of types for EST-SSR in Sinocalycanthus chinensis
-
正交实验结果(图 1)显示:5,9,10,13号组合均可扩增出比较清晰、稳定的带且杂带较少(XLM078扩增结果),重复试验(XLM091扩增结果),结果一致。
选用6个种群的18个样本(随机选择样本3个·种群-1)验证以上4种体系。结果发现:仅9号组合可重复稳定地扩增出清晰的条带(图 2)。为此选择9号体系作为夏蜡梅SSR-PCR最优体系:20.0 μL体系含Taq酶0.6 U(1 U=16.67 nkat),镁离子1.50 mmol·L-1,dNTP 0.25 mmol·L-1,引物0.20 μmol·L-1,DNA 75.0 ng。
-
试验选取6个种群的18个样本(随机选择样本3个·种群-1)对随机选择合成的220对引物(编号:XLM001~XLM220)进行PCR扩增,结果表明(表 4):总体而言,共120对引物扩增出特异性产物,有效扩增率为54.54%,其中14对引物扩增出多态性条带,占引物总数的6.36%。220随机引物对应的SSRs中,最多的为二碱基重复型、三碱基重复型,最少的为四碱基重复型。各自类型的引物中,可成功扩增出产物的引物比例与总体(54.54%)相差不大,其中最高的是四碱基重复型(64.29%),最低的为三碱基重复型(48.57%);就多态性引物所占各自类型引物总量的比例而言,二碱基重复型SSRs对应的引物最高(7.79%),其次是四碱基重复型(7.14%),最低的为五碱基重复型(4.35%)。14对多态性引物共扩增出37个等位基因,平均扩增为2.64,其中五碱基重复型SSRs对应的引物扩增的等位基因数最高(平均为4),其次是四碱基重复型(平均为3),最低的为六碱基重复型(平均为2)。
碱基重复类型 不同重复次数的引物/对 总数/对 成功扩增/对 比例/% 多态引物/对 比例/% 平均位点数 4 5 6 7 8 9 10 11 二 24 16 14 14 5 4 77 45 58.44 6 7.79 2.50 三 23 25 15 7 70 34 48.57 4 5.71 2.85 四 10 4 14 9 64.29 1 7.14 3.00 五 23 23 13 56.52 1 4.35 4.00 六 36 36 19 52.78 2 5.56 2.00 总体 59 33 53 31 21 14 5 4 220 120 54.55 14 6.36 2.64 Table 4. SSR types and amplification results of 220 primers
选取经过坪(JGP)种群24个样本,对上述具有种群间多态性的14对引物进行SSR-PCR扩增。结果发现:7对引物(3.18%)扩增出多态性条带:即XLM003,XLM007,XLM021,XLM035,XLM078,XLM139和XLM146,其中XLM007和XLM139扩增产物条带比较丰富,等位变异较多,XLM007在经过坪(JGP)种群的个体中扩增出4个(图 3A),XLM139扩增出5个(图 3B)。
2.1. 夏蜡梅转录组SSR位点的分布频率与基序特征
2.2. 夏蜡梅SSR-PCR体系优化结果
2.3. SSR引物筛选和验证
-
夏蜡梅转录组序列中的SSRs较为丰富,从125 014条unigene序列中共检测到26 564个SSR位点,发生频率为21.25%,平均5.18 kb就有1个SSR位点,其中含有2个或2个以上SSR位点的序列有3 664条(16.57%),复合SSRs有1 066条(4.01%)。就SSR位点发生频率而言,夏蜡梅比蜡梅Chimonanthus praecox(12.35%)[3]高,相较其他观赏植物,如百合Lilium(5.98%)[31],蝴蝶兰Phalaenopsis(3.19%)[32],辣椒Capsicum annuum(7.83%)[4],南方红豆杉Taxus chinensis var. mairei(2.24%)[5]和红掌Anthurium andraeanum(12.70%)[16]等也是如此;就SSR位点平均密度而言,夏蜡梅(1/5.18 kb)与蜡梅(1/5.00 kb)[30]相差无几,显著高于上述几种观赏植物。SSRs发生频率和平均密度除了因物种的不同而差异较大外,还可能与搜索标准,数据库的大小有关[4]。
研究表明:多数物种的EST-SSRs中以三碱基重复型出现的频率较高[33]。夏蜡梅不同重复类型的EST-SSRs最丰富的类型是二碱基重复型,占42.43%,其中以(AG/CT)n形式的二碱基重复型最多,占总SSRs的36.21%,这与蜡梅[44.81%,(AG/CT)n占总SSRs的39.85%][30]一致;三碱基重复型中都以(AAG/CTT)n为主要的重复基元[30],两者四、五、六碱基重复型所占比例都相对较少;就重复次数而言,除单核苷重复外,两者都以4~10次重复为主[30]。有所不同的是,夏蜡梅转录组中单碱基重复型[基本为(A/T)n]所占数量仅次于二碱基重复型,蜡梅转录组中三碱基重复型[比例最大的为(AAG/CTT)n]要多于单碱基重复型[30]。形成这种差异的原因除了基于植物本身的EST-SSR特点不同外,与转录组数据来源和数量也有关系[4]。
-
根据22 113条EST序列中检测到的26 564个SSR-ESTs,共设计出15 585对引物。在随机选择220对引物中,有120对引物(54.55%)成功扩增出产物,14对(6.36%)在不同种群个体中扩增出多态性条带,平均位点数为2.64;7对(3.18%)在经过坪种群内不同个体间扩增出多态性条带,说明夏蜡梅EST-SSR位点多,但多态性较低,原因是基因转录区的具有较高的保守性,使得EST-SSR引物具有较强的种属间通用性,可以用于亲缘关系较远的物种[30, 34]。胥猛等[35]基于鹅掌楸Liriodendron的6 520条EST设计了176对SSR引物,66对扩增出多态性条带,这66对中85%在鹅掌楸中有扩增,54%在白玉兰Michelia alba中有扩增。宋跃朋等[10]在比较杨树Populus基因组SSR和EST-SSR这2种标记的遗传差异性中发现其EST-SSR多态性明显低于基因组SSR,保守性很强,通用性高。夏蜡梅EST-SSR引物的种属间通用性还有待验证。
夏蜡梅EST-SSR引物中有14对(6.36%)在6个种群间扩增出多态性条带,而仅7对(3.18%)在同一种群(JGP)中有多态性条带,可能源于其种群自然分布的特点:种群规模小,片段化分布,不同种群间相互隔离,基因流阻断,种群内部近交率增加,种群间遗传分化增加,导致种群内遗传多样性低,大部分遗传多样性来自种群间[19-24]。
本研究开发了基于夏蜡梅转录组的SSR标记,为夏蜡梅种群的遗传多样性分析、交配系统估算、遗传图谱构建等提供了新的途径,同时夏蜡梅EST序列的开发为挖掘新的SSR提供了丰富的资源。













 DownLoad:
DownLoad: