-
黑果枸杞Lycium ruthenicum是茄科Solanaceae枸杞属Lycium植物,主要分布在中国西北地区[1],野生种质资源非常丰富,耐干旱耐盐碱;其果实富含类黄酮、花青素、多糖、酚酸和脂肪酸等生物活性物质,具有预防多种疾病的功效[2],深受消费者喜爱。目前,对黑果枸杞果实活性物质的研究较多[2-3]。也有研究利用扩增片段长度多态性(AFLP)[4],简单重复序列间扩增多态性(ISSR)[5]和相关序列扩增多态性(SRAP)[6]标记进行遗传多样性评价,但鲜有利用简单重复序列(SSR)标记的报道。SSR标记是基于生物基因组中由1~6个核苷酸组成的串联重复序列特性开发的分子标记,具有多态性丰富、高通量检测、共显性遗传等优点,已广泛应用遗传图谱构建、分子标记辅助育种、种质资源鉴定和遗传多样性分析等[7-10]领域。由于缺乏基因组信息,基于基因组测序开发黑果枸杞SSR标记成本高,步骤繁琐[11-12],SSR标记在黑果枸杞种质资源研究中的应用受到限制。同时,美国生物技术信息中心(NCBI)中公布的黑果枸杞表达序列标签(EST)极少,除了CHEN等[16]研究了基于盐胁迫下黑果枸杞转录组表达序列标签-简单重复序列(EST-SSR)标记开发之外,基于转录组测序黑果枸杞EST-SSR标记开发尚未见报道。EST-SSR来源于表达的基因组区域,可直接反映相关基因多样性,在不同物种间具有良好的通用性;随着高通量测序技术和生物信息学方法快速发展,基于转录组测序已开发刺梨Rosa roxburghii,中国樱桃Cerasus pseudocerasus,马铃薯Solanum tuberosum等[13-15]多种植物的SSR标记。本研究以前期黑果枸杞抗旱转录组测序获得的数据为基础,利用生物信息学方法批量开发EST-SSR标记,并分析其分布特点、组成特征,以期为黑果枸杞种质资源遗传多样性分析研究提供参考。
HTML
-
取1年生黑果枸杞持续干旱胁迫下的叶和根,提取RNA后进行转绿组测序(无参,HiSeqTM2500,北京诺禾致源生物信息科技有限公司),共12个样本,计80 G数据。利用Trinnity等软件[17]对测序的数据进行拼接和组装,获得213 058条单基因簇(unigene)作为背景数据进行分析。
-
用于SSR引物筛选及评价的24份枸杞种质资源(表 1)均来自宁夏农林科学院国家枸杞工程技术研究中心枸杞种质资源圃(38°38′49″N,106°9′10″E)。利用天根试剂盒(DP5-02,天根)提取基因组DNA。
序号 种质名称 果实颜色 1 ‘宁杞1号’ Lycium bararum ‘Ningqi 1’ 红色 2 ‘宁杞2号’ L. bararum ‘Ningqi 2’ 红色 3 ‘宁杞5号’ L. bararum ‘Ningqi 5’ 红色 4 ‘宁杞7号’ L. bararum ‘Ningqi 7’ 红色 5 ‘宁杞菜1号’ L. bararum ‘Ningqicai 1’ 红色 6 ‘宁农杞1号’ L. bararum ‘Ningnongqi 1’ 红色 7 ‘宁农杞2号’ L. bararum ‘Ningnongqi 2’ 红色 8 ‘宁农杞3号’ L. bararum ‘Ningnongqi 3’ 红色 9 ‘宁农杞9号’ L. bararum ‘Ningnongqi 9’ 红色 10 ‘蒙杞1号’ L. bararum ‘Mengqi 1’ 红色 11 ‘蒙杞2号’ L. bararum ‘Mengqi 2’ 红色 12 ‘天精3号’ (拉丁名未知) 红色 13 黑果枸杞L. ruthenicum 黑色 14 中国枸杞L. chincnse 红色 15 昌吉枸杞L. changjicum 褐色 16 新疆枸杞L. daystemum 红色 17 ‘HGB’ L.bararum ‘HGB’ 黄色 18 SC-09(拉丁名未知) 红色 19 HB-09(拉丁名未知) 红色 20 DHG(拉丁名未知) 黄色 21 AN-YN-01(拉丁名未知) 红色 22 W-12-30(拉丁名未知) 黄色 23 W-12-27(拉丁名未知) 黄色 24 W-11-15(拉丁名未知) 黄色 说明:1-12为栽培品种,13-24为野生资源 Table 1. Information of twenty-four materials used in this study
-
使用MISA软件(http://pgrc.ipk-gatersleben.de/misa/misa.html)搜索Unigene序列的SSR位点。搜索标准为:重复单元长度1~6 bp,单核苷酸重复次数≥10次,二核苷酸重复次数≥6次,三、四、五、六核苷酸重复次数≥5次,复合型SSR位点碱基间隔≤100 bp。
采用BatchPrimer 3软件()对获得的SSR位点批量设计引物。引物设计参数为:引物长度18~27 bp(最佳22 bp),GC为40%~70%(最适50%),退火温度为50~60 ℃(最适55 ℃),PCR扩增产物长度为100~300 bp,其他参数为默认设置。
-
随机挑选128对引物,在上游引物5′端添加18 bp的M13通用引物序列(TGTAAAACGACGGCCAGT),3′端保持不变,用FAM荧光集团修饰后,由英潍捷基贸易有限公司合成。
PCR扩增体系(15.0 μL):DNA模板1.0 μL,M13通用荧光引物0.1 μL,上游引物和下游引物各0.4 μL,2×Taq PCR Master Mix 7.5 μL,双蒸水5.6 μL。PCR扩增在ABI-2720(应用生物系统公司,美国)上进行。扩增程序为:95 ℃ 5 min;95 ℃ 30 s,60~45 ℃ 30 s,72 ℃ 30 s,15个循环;95 ℃ 30 s,50 ℃ 30 s,72 ℃ 30 s,20个循环;72 ℃ 7 min。扩增产物经过ABI3730XL DNA(应用生物系统公司,美国)检测,用LIZ500作为分子量内标。
-
以SSR出现频率和SSR平均分布距离描述EST-SSR。公式如下:①SSR出现频率fc=c/n×100%,其中c为搜索到的SSR数量(个),n为无余EST数量(个)。②SSR平均分布距离fN=N/c,其中N为无余EST数量的总碱基数(个)。由GeneMapper 4.0软件获得扩增产物片段大小,用DataFormater 2.7软件[18]将数据转化为PowerMarker v3.25软件[19]的输入格式,计算等位基因数(number of alleles,NA)、主效等位基因频率(major allele frequency, fMA)、期望杂合度(expected heterozygosity,HE)、观察杂合度(observed heterozygosity,HO)和多态信息量(polymorphism information content,CPI)。
1.1. 转录组数据来源
1.2. 植物材料及DNA提取
1.3. 转录组SSR位点鉴别及引物设计
1.4. EST-SSR引物筛选及验证
1.5. 数据分析
-
利用MISA软件对黑果枸杞转录组测序获得的213 058条Unigene(序列总长度为262 643 598 bp)序列进行搜索,发现其中56 170条Unigene序列中含有73 896个SSR位点,其中13 611条Unigene含有2个或2个以上EST-SSR位点。总体上,SSR发生频率为26.36%,平均3.55 kb出现1个SSR位点。SSR类型丰富,单核苷酸至六核苷酸重复类型均存在;单核苷酸、二核苷酸和三核苷酸重复出现频率占优势,分别占总SSR的74.33%,13.30%和11.81%;四核苷酸、五核苷酸和六核苷酸重复类型数量较少,分别占总数的0.49%,0.03%和0.04%(表 2)。
重复类型 不同重复次数下SSR数量/条 总数/条 百分比/% 5 6 7 8 9 10 > 10次 单核苷酸 20 340 34 587 54 927 74.33 二核苷酸 3 661 2 116 1 502 1 282 970 298 9 829 13.30 三核苷酸 5 285 2 414 982 39 1 1 4 8 726 11.81 四核苷酸 317 32 5 2 1 3 360 0.49 五核苷酸 13 1 5 1 1 21 0.03 六核苷酸 16 9 2 2 4 33 0.04 总数 5 631 6 117 3 110 1 546 1 284 21 311 34 897 73 896 百分比/R 7.62 8.28 4.21 2.09 1.74 28.84 47.22 100.00 Table 2. Frequencies of the different SSR repeat motif types observed in the Lyciurn ruthenicum transcriptome
-
从黑果枸杞转录组SSR核苷酸基序重复类型来看,73 896个SSR位点共有84种重复基元,单核苷酸至六核苷酸重复分别有2,4,10,26,18和24种。从出现的频率来看(表 3):占优势的前3种重复单元类型是单核苷酸、二核苷酸和三核苷酸重复基元;单核苷酸重复基元以A/T为主,占该类型SSR位点总数的95.64%;二核苷酸主要以AG/CT为主,占二核苷酸总数的40.42%,其次是AT/AT,AC/GT和CG/CG,分别所占比例为35.37%,23.74%和0.46%;三核苷酸重复单元以AAC/GTT,AAG/CTT,AAT/ATT,ATC/ATG和ACC/GGT为主,占所有三核苷酸重复单元的75.88%;四核苷酸、五核苷酸和六核苷酸重复单元类型较为分散,出现频率相对较低,仅为0.56%。
重复基序类型 数量/条 占该类型SSR比例/% 占总SSR比例/% A/T 52 531 95.64 71.09 C/G 2 396 4.36 3.24 AC/GT 2 333 23.74 3.16 AG/CT 3 973 40.42 5.38 AT/AT 3 477 35.37 4.71 CG/CG 46 0.47 0.06 AAC/GTT 2 108 24.16 2.85 AAG/CTT 1 865 21.37 2.52 AAT/ATT 1 035 11.86 1.40 ACC/GGT 635 7.28 0.86 ACG/CGT 174 1.99 0.24 ACT/AGT 319 3.66 0.43 AGC/CTG 597 6.84 0.81 AGG/CCT 424 4.86 0.57 ATC/ATG 978 11.21 1.32 CCG/CGG 591 6.77 0.80 其他类型 414 0.56 Table 3. Number and frequencies of repeat motif types in Lycium ruthenicum
-
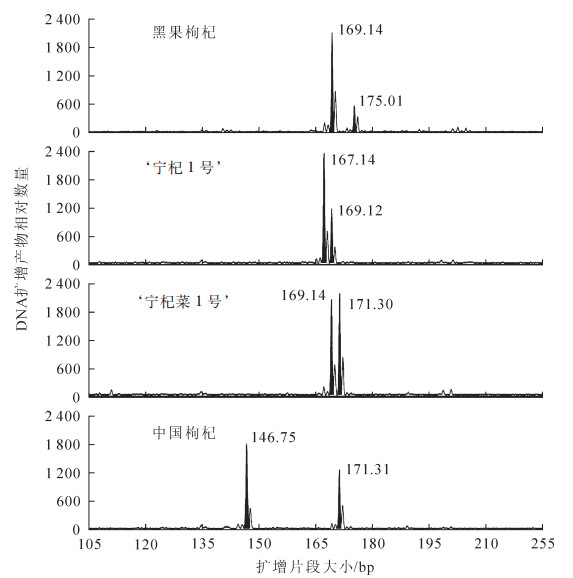
为检测所开发的EST-SSR标记的可用性,对56 170条Unigene序列的73 896个EST-SSR位点设计引物,共得到引物12 674对。随机挑选128对(包括二核苷酸、三核苷酸、四核苷酸、五核苷酸和六核苷酸),以表型性状差异较大的‘黑果枸杞’‘宁杞1号’‘宁杞菜1号’和‘中国枸杞’的DNA为模板进行PCR扩增,其中74对引物能扩增出清晰的扩增产物,引物扩增有效率为57.8%;其中28对引物具有多态性,多态性引物占有效引物的37.8%。图 1为引物LM-02对4份种质的基因型图。
-
以筛选的28对多态性引物对24份枸杞种质作遗传多样性分析。共检测到等位基因256个,各引物检测到的等位基因为4~19个,平均为9.1个;共检测到基因型数303个;主效等位基因频率变化范围为0.188~0.729,平均值为0.432;观察杂合度为0.167~0.833,平均值为0.439;期望杂合度为0.433~0.904,平均值为0.712,多态信息量为0.395~0.897,平均值为0.678。由Botstein理论[20]判定(表 4),在开发出的28个多态位点中有24个高度多态位点(CPI≥0.5),4个中度多态位点(0.25≤CPI < 0.5)。
引物名称 等位基因数 主效等位基因频中 期望杂合度 观察杂合度 多态信息量 LM-02 6 0.458 0.6831 0.458 0.634 LM-04 8 0.375 0.773 0.250 0.743 LM-05 10 0.438 0.747 0.292 0.722 LM-08 8 0.5CX) 0.682 0.458 0.647 LM-20 9 0.250 0.805 0.292 0.777 LM-22 10 0.5CX) 0.697 0.542 0.669 LM-28 9 0.250 0.835 0.417 0.815 LM-30 15 0.250 0.882 0.542 0.873 LM-33 8 0.542 0.655 0.500 0.622 LM-42 4 0.729 0.433 0.167 0.395 LM-43 4 0.646 0.513 0.583 0.454 LM-50 4 0.646 0.484 0.417 0.402 LM-51 7 0.583 0.615 0.542 0.584 LM-56 15 0.229 0.872 0.542 0.861 LM-61 10 0.479 0.722 0.167 0.699 LM-64 11 0.438 0.769 0.667 0.753 LM-68 7 0.354 0.724 0.333 0.680 LM-71 9 0.3% 0.760 0.458 0.730 LM-75 7 0.521 0.586 0.250 0.508 LM-86 6 0.583 0.602 0.292 0.560 LM-96 7 0.417 0.745 0.667 0.712 LM-99 11 0.333 0.819 0.375 0.800 LM-101 5 0.667 0.481 0.458 0.414 LM-110 15 0.250 0.876 0.708 0.865 LM-111 19 0.188 0.904 0.542 0.897 LM-112 7 0.500 0.591 0.250 0.511 LM-114 12 0.313 0.832 0.292 0.814 LM-125 13 0.271 0.854 0.833 0.840 平均值 9.1 0.432 0.712 0.439 0.678 Table 4. Genetic diversity of 24 wolfberry germplasm revealed by 28 EST-SSR markers
2.1. 黑果枸杞转录组中SSR位点的分布特点
2.2. 转录组SSR基序重复类型和频率特征
2.3. SSR引物有效性及多态性检测
2.4. SSR位点遗传多样性分析
-
本研究搜索了黑果枸杞的213 058条Unigene序列,发现其中的56 170条中含有73 896个SSR位点,发生频率为26.36%;高于中国樱桃Cerasus pseudocerasus(15.62%)[14],马铃薯Solanum tuberosum(3.43%)[15],夏蜡梅Sinocalycanthus chinensis(21.25%)[16],马尾松Pinus massoniana(4.69%)[23]和中间锦鸡儿Caragana intermedia(14.78%)[24],低于蓝靛果忍冬Lonicera caerulea var. edulis(32.51%)[25],山桐子Idesia polycarpa(35.00%)[26],普通油茶Camellia oleifera(39.67%)[27]和芙蓉李Prunus salicina(54.51%)[28]。可见,不同物种转录组SSR位点的出现频率不同,出现差异的原因除了物种本身差异,还可能与分析数据库大小、SSR挖掘工具及搜索条件有关[13]。
从重复单元频率来看,黑果枸杞转录组的SSR主要类型为单核苷酸重复基序(74.33%),其次为二核苷酸重复(13.30%)和三核苷酸重复(11.81%)。研究发现:多数植物中EST-SSR以二核苷酸和三核苷酸为主,但主要的重复基序类型不同[29]。推测原因可能与SSR搜索参数设置有关,多数植物在进行SSR位点搜索时并未将单核苷酸重复设置为搜索对象[13-14]。
本研究从213 058条Unigene序列中鉴定出73 896个SSR位点,丰富了黑果枸杞EST-SSR标记的数量。为了进一步评估这些EST-SSR引物的质量,从设计到的12 674对EST-SSR引物中随机挑选出128对引物,54对引物未能扩增出产物,原因可能是扩增产物中存在大量内含子或引物设计不合理[30-31];有74对引物成功扩增出产物,引物扩增有效性达57.8%,表明开发的引物质量较高,对后续黑果枸杞种质资源遗传多样性分析、品种鉴定、遗传图谱构建和分子标记辅助育种等均有应用价值。










 DownLoad:
DownLoad: