-
锈色粒肩天牛Apriona swainsoni隶属于鞘翅目Coleoptera天牛科Cerambycidae沟颈天牛亚科Lamiinae白条天牛族Batocerini粒肩天牛属Apriona[1-2],可危害国槐Sophora japonica、黄檀Dalbergia hupehana、三叉蕨Tectaria subtriphylla、悬铃木Plantanus acerifoli等多种植物[3-6],而其寄生于云实Caesalpinia decapetala树茎的幼虫,异名云实蛀虫、云实蠹虫、黄牛刺虫、黄寮刺虫,在民间一直都有食服的习惯,因“一斗米换一条虫”而得名“斗米虫”[7-9]。近年来,斗米虫因其良好的药食功效[7-8, 10-11]日益获得认同。斗米虫野外采集困难,人工规模化生产尚未成熟,其市场价格因此逐步走高。受经济利益驱使,目前市场上时常发生其他天牛幼虫伪充斗米虫的现象,导致误食中毒等事件,扰乱药食昆虫市场的秩序。天牛科昆虫分类主要依据其成虫形态特征,幼虫形态描述信息缺乏,幼虫分类存在一定难度[12]。在实际工作中也发现,斗米虫与其他天牛幼虫,例如桑天牛A. germari幼虫,在形态方面具有一定的相似性,仅靠幼虫形态特征很容易鉴别错误。此外,因加工炮制等处理导致幼虫形态被破坏,也提高了鉴别难度。分子生物学手段的应用在很大程度上弥补了物种形态学鉴定的不足[13-16]。线粒体细胞色素氧化酶C亚基Ⅰ(Cytochrome C oxidase subunitⅠ,COⅠ)[17-20]、COⅡ[21]、16S核糖体RNA(16S rRNA,16S)[22]、ND基因(NADH dehydrogenase subunit,ND)[23]和细胞色素B(Cytochrome b,Cytb)[24-25]等,以及细胞核28S核糖体RNA(28S rRNA,28S)[23]和核糖体DNA的内转录间隔区(Internal transcribed spacer,ITS)[26-28]等多种基因已广泛应用于多种昆虫的分类鉴别。目前尚未发现锈色粒肩天牛幼虫线粒体基因或核基因序列鉴定的相关报道,因此,本研究对锈色粒肩天牛幼虫的3种线粒体编码基因(COⅠ、COⅡ和Cytb),以及1种核糖体大亚基编码基因(28S)共4种序列展开了研究。此次也对来自3个地区的桑天牛A. germari幼虫样品进行了测序,以补充桑天牛幼虫上述4种基因的序列信息,提高天牛幼虫进化树拓扑结构的准确性。本研究通过多种基因片段的序列分析,为锈色粒肩天牛幼虫的准确鉴定提供依据,也为该虫的食药用安全提供保障。
HTML
-
用于测序的4份锈色粒肩天牛幼虫样品分别来自于浙江金华(S1)、江西上饶(S2)、浙江开化(S3)、浙江淳安(S4),3份桑天牛幼虫样品分别来自于浙江金华(G1)、江西上饶(G2)、浙江开化(G3),幼虫活体置于-4 ℃冰箱保存备用。
-
TIANamp Genomic DNA Kit,天根生物科技(北京)有限公司;DNA凝胶回收试剂盒(GE0101-50)、感受态细胞(Escherichia coli DH5α)、TS-GelRed核酸染料(10 000×水溶液),北京擎科新业生物技术有限公司;pGEM-T Easy载体,美国Promega公司;Regular Agarose,西班牙Biowest公司;50×TAE Tris-乙酸电泳缓冲液,福州Phygene生物科技有限公司。
-
DK-S26电热恒温水浴锅,上海森信实验仪器有限公司;FRESCO 21微量冷冻离心机、NanoDrop 2000超微量分光光度计,美国Thermo Scientific公司;Life ECO扩增仪,杭州博日科技有限公司;DYY-12C型电泳仪,北京六一生物科技有限公司;Bio-Rad凝胶成像系统,美国伯乐公司。
-
所采用的COⅠ、COⅡ、Cytb和28S基因片段扩增引物信息详见表 1,供试引物委托北京擎科新业生物技术有限公司合成。
扩增基因片段 引物序列(5'→3') 退火温度Tm/℃ 参考文献 COⅠ 2195:TTGATTTTTTGGTCATCCAGAAGT 54 [22, 29] 3014:TCCAATGCACTAATCTGCCATATTA COⅡ COⅡSz-F:TGCTTCAAGATAGAGCCTCTCC 54 [21, 28] COⅡSz-R:GGTTTGCTCCACAGATTTCAG Cytb CytbF:TATGTACTACCATGAGGACAAATAT 58 [25, 30] CytbR:ATTACACCTCCTAATTTATTAGGAAT 28S D2-3665F:AGAGAGAGTTCAAGAGTACGTG 58 [22, 31] D5-4749R:GTTACACACTCCTTAGCGGA Table 1. Information of tested primers
-
取待提取幼虫,用无菌水清洗数遍,滤纸吸干水分,加液氮彻底磨碎。基因组DNA的提取参照天根TIANamp Genomic DNA Kit试剂盒操作说明书。利用NanoDrop 2000超微量分光光度计对样品基因组DNA进行核酸纯度及浓度的检测,于-20 ℃冰箱储藏备用。
-
PCR扩增反应体系为(总体积20 μL):2×TSINGKE Master Mix 10 μL,10 umol·L-1引物对各1.5 uL,模板DNA 3 uL(20 mg·L-1),ddH2O补足至20 μL。扩增程序为:94 ℃预变性5 min后;94 ℃变性30 s,退火45 s(不同引物对的退火温度Tm详见表 1),72 ℃延伸2 min,共35个循环;最后于72 ℃补平7 min,终止温度为4 ℃。PCR扩增反应在LifeECO基因扩增仪(杭州博日)上进行。取PCR扩增产物3 μL,点样于质量分数为1.5%的琼脂糖凝胶上,TS-GelRed核酸染料染色,1×TAE缓冲液中电泳30~40 min(120 V),扩增图谱摄取采用Bio-Rad凝胶成像分析仪。PCR扩增产物经切胶回收、纯化、pGEM-Teasy载体连接、感受态细胞转化(E. coli DH5α)和蓝白斑筛选后,随机挑选5个阳性克隆由北京擎科新业生物技术有限公司进行双向测序。
-
测序结果利用Contig Express软件进行拼接,得到目的基因片段在美国国家生物信息中心(NCBI)数据库中进行BLAST比对和相似性检索,以确定所获片段是否为目的基因并获得同源序列。利用MEGA 6.0软件将本研究获得的基因片段与GenBank检索获得的同源序列进行多重比对,两端对齐,进行序列信息分析,包括碱基组成、变异位点、简约信息位点等。同时运用邻接法(Neighbor-joining,NJ)构建进化树,模型为K2P(Kimura 2 parameter),Bootstrap置信值估算重复数设定为1 000次。
1.1. 材料
1.1.1. 供试昆虫
1.1.2. 供试试剂
1.1.3. 供试仪器
1.1.4. 供试引物
1.2. 方法
1.2.1. 基因组DNA提取及浓度测定
1.2.2. PCR扩增、电泳及测序
1.2.3. 数据分析
-
由表 1所示:引物成功扩增获得2种天牛幼虫的4种基因片段。经测序、拼接并去除上下游引物及两端不稳定序列,锈色粒肩天牛幼虫最终的COⅠ、COⅡ、Cytb和28S基因片段大小分别为817、545、434和1 088 bp,桑天牛幼虫基因片段大小则分别为783、545、435和1 096 bp。将各基因片段在GenBank进行BLAST,比对结果显示所获基因片段与其他天牛等昆虫的目标基因片段的相似性达80%以上,说明本研究成功获得2种天牛幼虫的4种不同基因片段。
-
将锈色粒肩天牛幼虫的4种基因序列与桑天牛幼虫序列以及GenBank中比对获得的其他天牛同源序列进行多重比较并分析各基因序列信息(表 2~4)。其中,COⅠ基因同源序列来自天牛科7属16种,COⅡ基因同源序列来自天牛科12属15种,Cytb基因同源序列来自天牛科3属4种,28S基因同源序列来自天牛科7属7种。
基因名称 保守位点 变异性位点 简约信息位点
数量/个单突变位点数
量/个片段大小/bp 数量/个 百分比/% 数量/个 百分比/% COⅠ 473 63.57 271 36.42 218 53 744 COⅡ 288 54.86 237 45.14 190 47 525 Cytb 303 66.45 153 33.55 130 23 456 28 480 73.28 166 25.34 92 74 655 Table 2. Distribution of specific sites of COⅠ, COⅡ, Cytb and 28S gene homologous sequences
基因
名称碱基含量/% A+T含量/% G+C含量/% T C A G COⅠ 38.75 15.37 30.97 14.90 69.72 30.27 COⅡ 40.90 15.96 32.91 10.23 73.81 26.19 Cytb 39.70 17.76 31.63 10.91 72.53 27.47 28 21.30 28.78 17.42 32.50 38.72 61.28 Table 3. Base composition characteristic of COⅠ, COⅡ, Cytb and 28S homologous sequences
基因
名称位点 转换 颠换 R 碱基替换频次 TT TC TA TG CC CA CG AA AG GG COⅠ 第1 27 46 0.57 80 23 39 1 5 6 0 89 4 0 第2 14 6 2.46 64 10 3 1 34 1 0 62 5 67 第3 3 3 1.12 104 2 1 0 54 0 1 48 1 36 总计 44 55 0.80 248 35 43 2 93 7 1 199 10 103 COⅡ 第1 14 10 1.36 46 8 5 2 27 3 0 47 6 32 第2 3 3 1.07 72 3 2 0 36 1 0 45 0 16 第3 20 34 0.58 64 18 30 0 3 4 0 55 2 0 总计 37 47 0.79 182 29 37 2 66 8 0 147 8 48 Cytb 第1 18 27 0.68 50 17 22 0 7 5 0 48 2 1 第2 10 7 1.32 40 7 5 1 23 1 1 42 3 29 第3 2 1 2.13 63 2 1 0 34 0 0 35 0 16 总计 30 35 0.86 153 26 28 1 64 6 1 125 5 46 28 第1 7 6 1.24 37 4 2 2 48 1 2 32 3 73 第2 12 8 1.58 44 10 3 2 53 1 2 36 3 51 第3 11 6 1.92 30 8 1 1 59 2 1 29 3 67 总计 30 20 1.50 111 22 6 5 160 4 5 97 9 191 Table 4. Base substitution mode of COⅠ, COⅡ, Cytb and 28S gene homologous sequences
所有序列多重比较,两端对齐去除冗余位点后,最终得到不同长度的各基因片段(456~744 bp),并对所有样品4种基因序列中含有保守位点、变异性位点、简约信息位点和单突变位点的数量及百分比进行了分析统计。表 2显示:28S的保守位点所占比例最高(73.28%),其后依次为Cytb(66.45%)、COⅠ(63.57%)和COⅡ(54.86%);4种基因的变异率则恰好相反,COⅡ序列的变异率最高(45.14%),其后依次为COⅠ(36.42%)和Cytb(33.55%),28S变异率最低(25.34%)。
COⅠ、COⅡ、Cytb和28S各基因所比对的同源序列碱基组成特点如表 3所示。其中,COⅠ、COⅡ和Cytb基因的T碱基含量最高,其次是A碱基,A+T含量(69.72%、73.81%、72.53%)均明显高于G+C含量(30.27%、26.19%、27.47%),而28S基因的G碱基含量最高,其次是C碱基,A+T含量(38.72%)则明显低于G+C含量(61.28 %)。
COⅠ、COⅡ、Cytb和28S各基因所比对的同源序列碱基替换信息见表 4。从表 4可知:COⅠ、COⅡ和Cytb基因序列的颠换均高于转换,转换与颠换比(R)平均值约0.8,颠换以T和A之间为主;而28S基因序列R平均值为1.5,即转换高于颠换,转换以T和C之间为主。
-
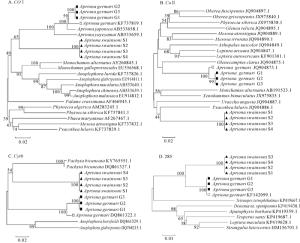
分别基于COⅠ、COⅡ、Cytb和28S基因片段,运用邻接法构建锈色粒肩天牛与其他天牛的分子进化树(图 1A~D)。可知,基于COⅠ和28S基因序列的分子进化树中,同属的昆虫均聚集为一类,其中粒肩天牛属的锈色粒肩天牛或桑天牛同种不同样品亦分别聚成一小分支,这与传统的分类方法相一致(图 1A,D)。基于COⅡ和Cytb基因序列的分子进化树中,锈色粒肩天牛和桑天牛各自不同样品也以较高的置信值分别聚成一支,但这2种同属的昆虫并未如COⅠ基因序列的进化树聚为一大类。COⅡ基因进化树中,锈色粒肩天牛与星斑天牛属Psacothea的黄星天牛Psacothea hilaris聚集,桑天牛则与白天牛属Olenecamptus的六星天牛Olenecamptus clarus聚在一起。Cytb基因进化树中,锈色粒肩天牛先与厚花天牛属Pachyta的双斑厚花天牛Pachyta bicuneata聚集,再与桑天牛聚类(图 1B~C)。
2.1. 基因序列的克隆与鉴定
2.2. 基因序列信息分析
2.3. 基因序列的进化树构建
-
斗米虫等昆虫天然保健产品因其特有的功效,逐渐被大众熟知和接受,鉴于天牛幼虫形态学鉴别存在的不足之处,需要依靠分子生物学手段完善以提高天牛幼虫鉴别的准确性。相对于其他分子标记法,DNA测序在种及以上高级分类阶元的分析结果更为准确直接,可作为一种有用的“分类”辅助工具[15]。至今为止,国内外已将多种线粒体基因和核基因广泛运用于昆虫的系统发育、遗传进化和物种分类等研究中[32-37]。线粒体基因严格遵守母系遗传方式,进化速度适中,相对保守的同时又有足够的变异,特别适用于解决较低阶元的系统发育关系[24, 38-39],28S基因则是真核生物染色体上编码核糖体大亚基的基因,在生物体内含量较大,在进化过程中较线粒体基因保守,尤其在高级分类阶元系统发育关系研究中应用广泛[40-41]。
本研究的3种线粒体蛋白编码基因(COⅠ、COⅡ和Cytb)和1种核基因(28S)为昆虫分类的常用基因。锈色粒肩天牛的上述4种基因片段及其同源序列的序列信息分析,结果表明线粒体基因(COⅠ、COⅡ和Cytb)和核基因(28S)的组成结构等存在差异。3种线粒体基因同源序列的变异率比28S基因片段高,这与核基因较线粒体基因更为保守这一观点相符合[40]。此外,3种线粒体基因同源序列表现出A和T碱基偏倚,颠换高于转换,而28S同源序列则相反(G和C碱基偏倚且转换高于颠换)。此次所选择分析的锈色粒肩天牛28S基因及其同源序列的碱基偏倚性与其他研究报道的天牛科28S基因相同,其碱基组成特点也与28S rDNA相符合[40]。
不同物种,其分子分类适用的基因也存在差异,根据不同物种选择合适的基因十分关键。CHEN等[39]以大蚜亚科Lachninae为研究对象,评估了3种线粒体基因(COⅠ、COⅡ和Cytb)作为DNA条形码的有效性,发现COⅡ基因较其他两者更合适于不同大蚜的鉴定,Cytb因其序列多样性高,是蚜虫种群遗传学研究的有效标记。杨晋英[38]研究表明:COⅠ、COⅡ和Cytb基因适用于五倍子蚜分类,其分子聚类关系与形态学分类相一致。李成等[42]研究表明:28S非常适合于旋毛虫Trichinella spiralis的分类鉴定及其分子进化关系的研究。本研究显示:利用COⅠ、COⅡ、Cytb和28S基因的通用性引物均能有效地扩增锈色粒肩天牛和桑天牛幼虫的目的片段。基于这4种基因序列构建的进化树亦都能以较高的置信值将锈色粒肩天牛幼虫单独聚成一支,与其他种的天牛区分开来,表明上述4种基因均可作为快速鉴别锈色粒肩天牛幼虫与桑天牛等其他天牛幼虫的分子参考依据。
总体而言,COⅠ、COⅡ、Cytb和28S这4种基因的进化树拓扑结构与传统分类结果基本相一致,但COⅡ和Cytb基因进化树其属级别的聚类稍有差别,其原因可能是本研究涉及的COⅡ和Cytb序列仅是该基因的部分序列,其所覆盖的信息还不够充足,可通过增加昆虫的目标基因序列数量或长度,以获得更为理想的系统进化树。










 DownLoad:
DownLoad: