-
梅花Prunus mume是中国十大传统名花,在中国有3 000多年栽培历史,江南地区花期为2月左右[1]。梅花适应性较强,耐高温、较耐低温和干旱。在年均气温为16~23 ℃的地区生长最好,根系不耐−8 ℃以下的低温[2-3]。梅花适栽范围介于自然分布区和历史分布区之间,北界为西藏经四川至甘肃天水,陕西宝鸡、西安,河南洛阳,最后到山东烟台,通过海岸线与自然分布区相接,以长江流域为集中赏梅地带[2, 4]。在华北、东北和西北等北方地区不能露地越冬,在江南地区,春季的极端低温(低于−3 ℃)则会对花(蕾)造成伤害,极大地影响观赏价值。因此,抗寒育种一直是梅花育种的重要方向[4-6]。WRKY转录因子是一类主要存在于植物中的锌指型转录因子,在植物响应生物胁迫与非生物胁迫的过程中起着重要作用。根据WRKY结构域数量和锌指结构特征,WRKY家族可分为3类:I类包含2个WRKY结构域和1个CX4−5CX22−23 HXH (C2H2)型锌指结构;Ⅱ类包含1个WRKY结构域和1个CX4−5CX22−23HXH (C2H2)型锌指结构;Ⅲ类包含1个WRKY结构域和1个CX7CX23HXC (C2HC)型锌指结构[7]。WRKY家族参与了广泛的生物过程,包括种子萌发、植物发育和植物激素信号传递等[8]。研究表明:WRKYs最重要的功能之一是参与防御非生物胁迫[9],可显著提高小麦Triticicum aestivum[10]、水稻Oryza sativa[11]等的耐寒性和香蕉Musa acuminate[12]、棉花Gossypium hirsutum[13]等的抗旱性。近年来,随着梅花基因组正式公布[14],关于梅花抗寒和抗旱基因的挖掘和研究逐渐展开,这为深入研究梅花抗寒、抗旱等机制提供了重要的基础。梅花中共鉴定出58个WRKY成员,PmWRKYs在梅花的不同组织(根、茎、叶、花和果实)中有不同程度的表达,其中17个PmWRKYs可能是调控梅花抗寒性的潜在转录因子[15]。本研究以梅花品种‘骨红朱砂’‘Guhong Zhusha’为材料,采用反转录PCR(RT-PCR)技术克隆获得了2个PmWRKY2转录因子,通过生物信息学分析和同源基因序列比对,检测PmWRKY2基因在不同非生物胁迫下的表达模式,以期为后续开展WRKY转录因子在梅花抗寒和抗旱方面的作用机制研究奠定基础。
HTML
-
梅花‘骨红朱砂’来自浙江农林大学梅花种质资源圃。采集生长势一致,无病虫害的1年生枝条,插于加入清水的培养瓶中,参照PENG等方法[16]处理,湿度为50%,光周期为16 h/8 h。低温(2 ℃)处理组取样时间为0(ck)、1、2、4、6、12、24、48、72 h。采用200 mmol·L−1的甘露醇溶液模拟干旱处理,取样时间为0(ck)、3、6、12、24、36、48 h。采用100 μmol·L−1脱落酸(ABA)处理,取样时间为0(ck)、3、6、12、24、36、48 h。所有新鲜样品采集后立即用液氮速冻,保存于−80 ℃,3次生物学重复。
-
RNA的提取采用购自天津诺禾致源公司的UltraClean Polysaccharide and Phenol Plant RNA Purification Kit,方法参照试剂盒的提取说明书。cDNA的合成根据TAKARA PrimeScript™ RT Master Mix (Perfect Real Time)说明书在冰上进行。
-
通过梅花基因组和表达谱数据获得PmWRKY2-1和PmWRKY2-2序列,利用Prime 5.0设计特异性引物(表1),以‘骨红朱砂’叶片cDNA为模板,利用r-Taq DNA聚合酶进行PCR扩增。扩增条件为:95 ℃预变性5 min;95 ℃变性30 s,54 ℃退火30 s,72 ℃延伸3 min,35个循环;72 ℃延伸10 min。PCR扩增产物经切胶回收试剂盒回收后连接到pMD18-T载体(Takara公司,大连)中,转化大肠埃希菌Escherichia coli DH5α感受态细胞后挑取阳性克隆,经PCR验证后送往杭州有康科技有限公司测序。
用途 引物名称 序列(5′→3′) 基因克隆 PmWRKY2-1F ATGGCTGGCATCGATGA PmWRKY2-1R CTACATCTGTGGTCCAAG PmWRKY2-2F ATGGGATTTTTAAGAACC PmWRKY2-2R CTAGTACGATTGATGACTGCTTC 实时荧光定量PCR QPmWRKY2-1F GTCCCCTTATCTGACAATACCTC QPmWRKY2-1R AAAGCGAATGAAGTATTTATGTCCT QPmWRKY2-2F TCCGTTGCTTCCTCCCAATGATGAC QPmWRKY2-2R CAAAATCTATTGGTTGTTGCTCC QPmEF1αS CGGATTCAATGTTAAGAATGTTGC QPmEF1αA AGAACTGGAGCATATCCGTTACC Table 1. Primers used in Gene clone and Quantitative real-time PCR
-
采用在线软件BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) 进行基因序列比对分析,用ORF finder在线分析开放阅读框,运用ProtParam 在线软件(http://web.expasy.org/protparam/)预测编码蛋白质的分子量、理论等电点;利用 SOPMA 在线工具分析PmWRKY2蛋白质的二级结构组成;利用WOLFPSORT在线软件预测基因的亚细胞定位;利用 DNAMAN 9.0软件对梅花PmWRKY2蛋白质与其他物种WRKY蛋白质进行比对分析;使用ClutsalX-v1.83程序进行多序列比对,然后将比对结果输入到MEGA 6.0软件中,利用邻接法(neighbor-joining, NJ)构建系统发育树,Bootstrap值取1 000次。
-
以不同处理的叶片为模板,反转录为cDNA,并进行实时荧光定量PCR。利用Prime 5.0设计PmWRKY2-1和PmWRKY2-2的特异性引物,以梅花PmEF1α为内参基因。反应体系为SYBR Premix Ex Taq 酶(Takara,大连)10.0 μL,cDNA 2.0 μL,上下游引物(10 μm·L−1)各0.8 μL,双蒸水 6.4 μL,每个样品设置3次重复。反应程序为两步法:95 ℃预变性30 s,95 ℃变性5 s,60 ℃复性30 s,共40个循环;然后以95 ℃持续5 s,60 ℃持续1 min,95 ℃持续15 s作为溶解曲线分析程序,最后根据
$ {2^{ - \Delta \Delta {C_{\rm{t}}}}} $ 法计算目的基因的相对表达量。
1.1. 植物材料与处理
1.2. 方法
1.2.1. RNA提取及cDNA合成
1.2.2. 基因的克隆
1.2.3. 序列的生物信息学分析
1.2.4. 基因表达分析
-
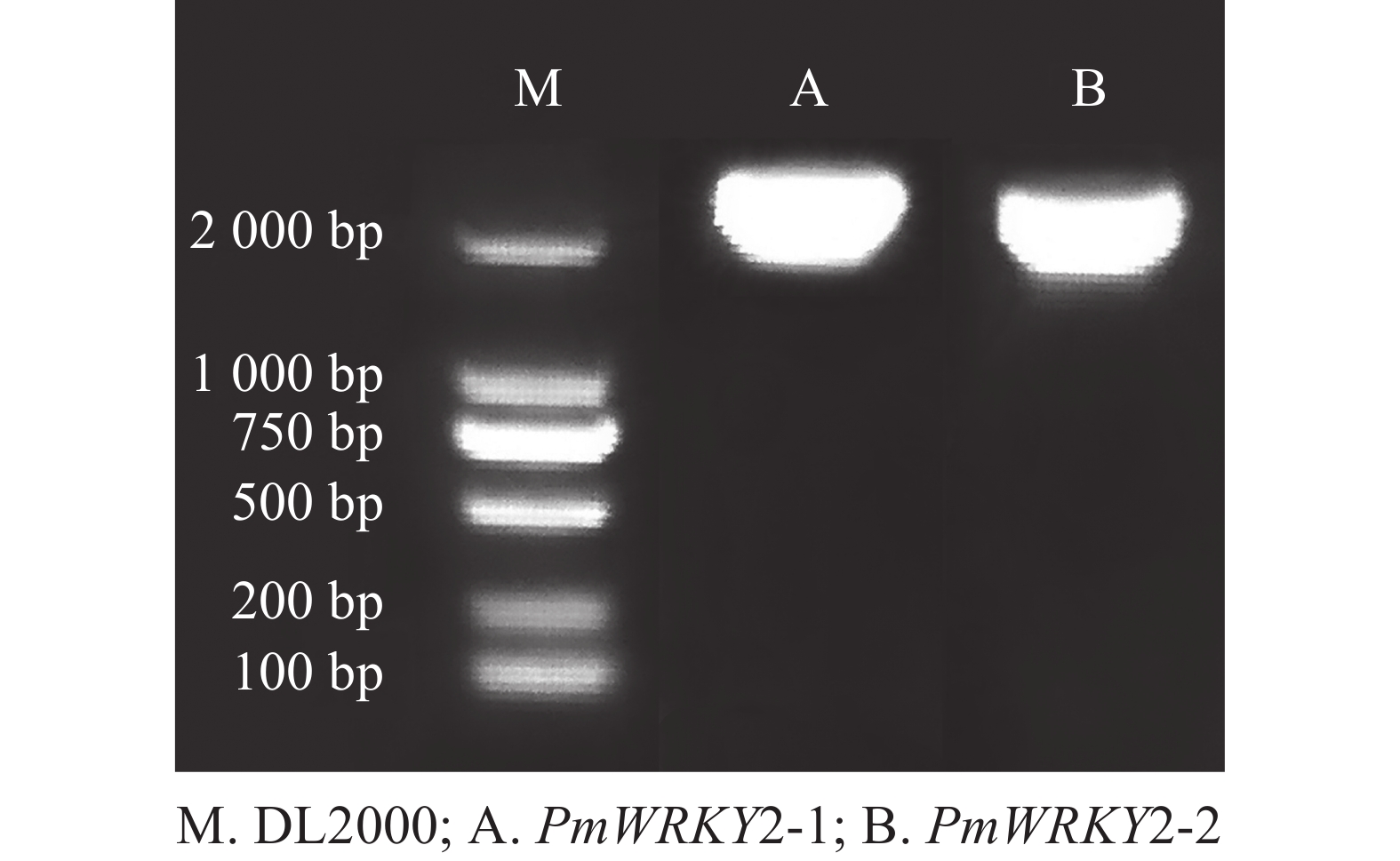
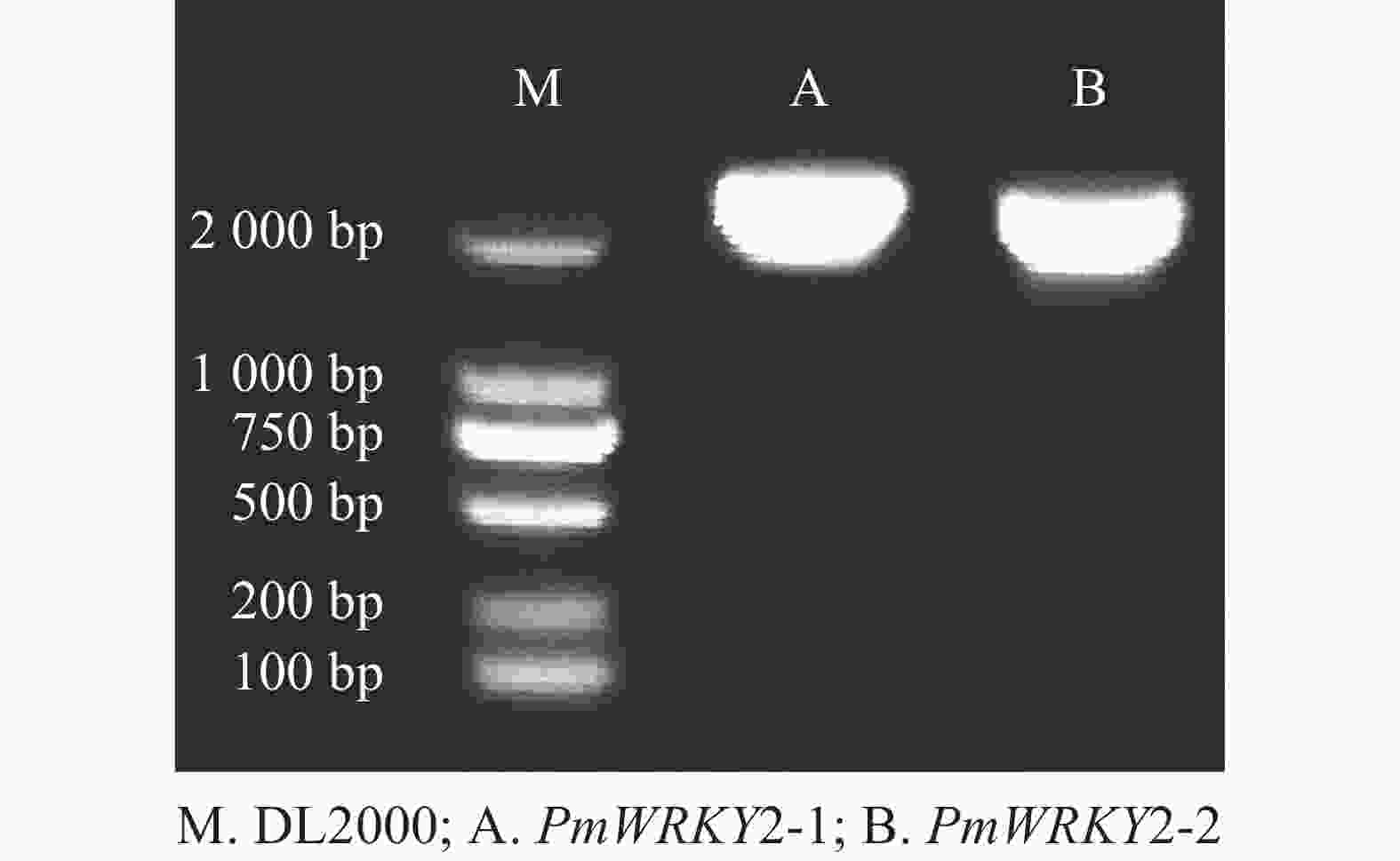
利用特异性引物进行PCR扩增,经过连接、转化、测序后获得编码序列(CDS)。测序结果显示:PmWRKY2-1和PmWRKY2-2的CDS长度分别为2 223和2 220 bp(图1),编码的氨基酸数目分别为740和739个,蛋白质分子量分别为79.94和80.98 kD,理论等电点分别为5.65和5.82。不稳定系数分别为53.93和53.82,脂肪指数分别为54.18和59.53,预测它们为不稳定蛋白质。总平均亲水系数(GRAVY)分别为−0.774和−0.743,属于亲水性蛋白质。亚细胞定位预测结果显示:PmWRKY2-1和PmWRKY2-2均位于细胞核。
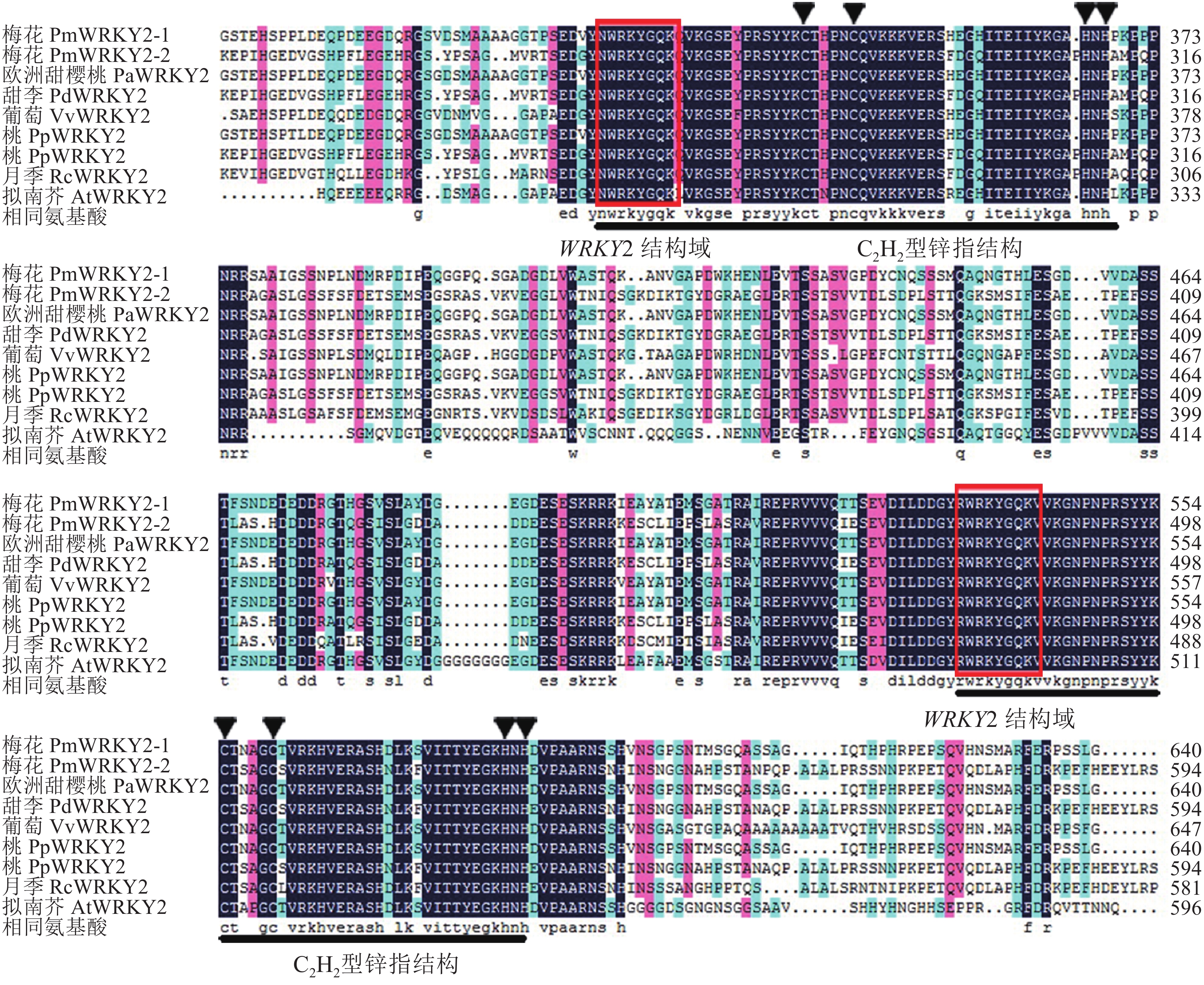
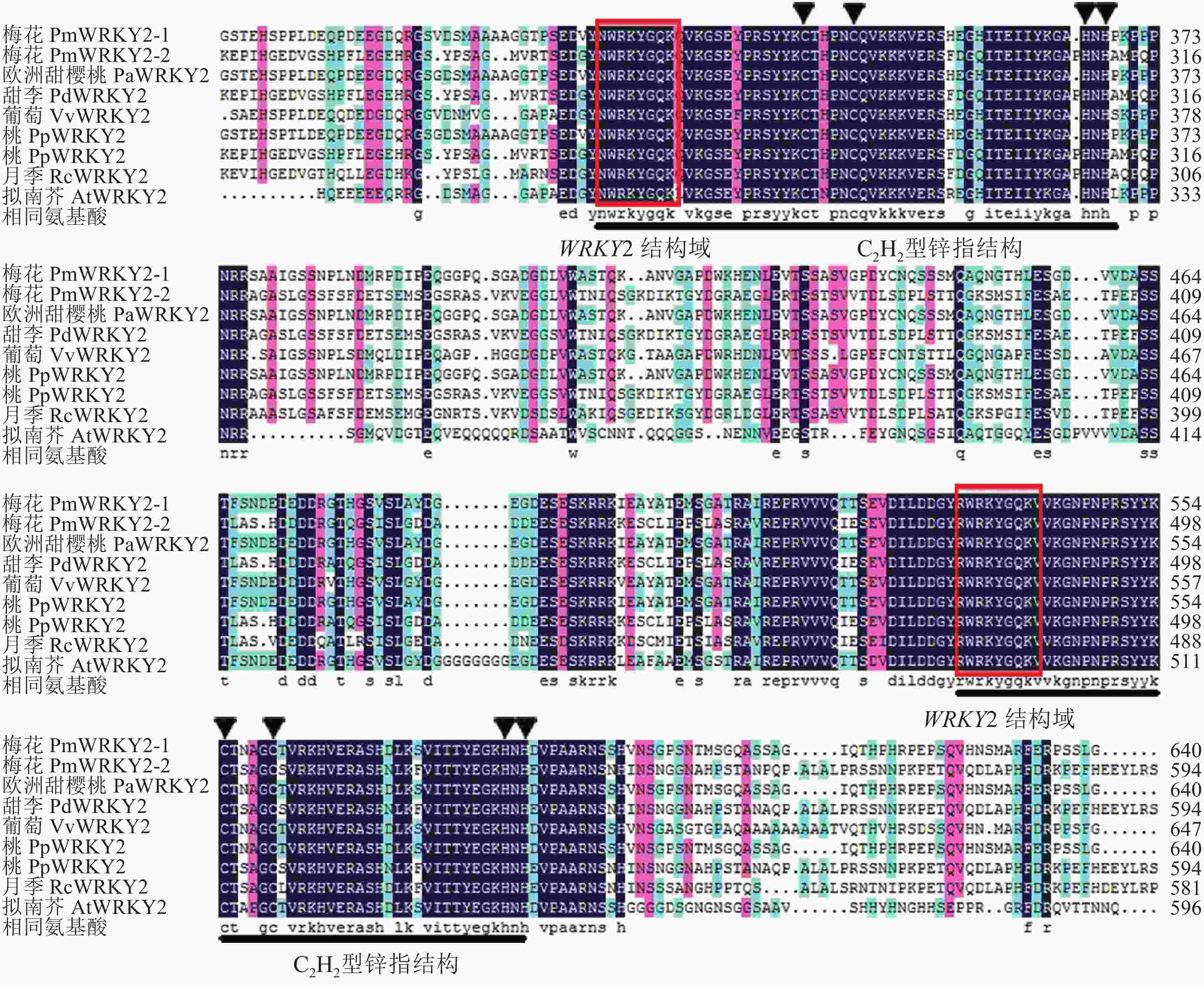
氨基酸序列比对结果显示(图2):梅花PmWRKY2-1和PmWRKY2-2的同源性仅为45.87%,与拟南芥Arabidopsis thaliana的AtWRKY2相似性分别为51.26%和32.07%;其中PmWRKY2-1与欧洲甜樱桃P. avium(XP_021826759.1)、桃P. persica(XP_007206427.1)的WRKY2同源性分别为98.65%,98.78%;PmWRKY2-2与甜李P. dulcis(XP_034218428.1)、桃(XP_007207009.2)的WRKY2同源性分别为98.51%,98.11%,与月季Rosa chinensis(XP_024188041.1)WRKY2同源性为77.97%。进一步分析发现:梅花PmWRKY2-1和PmWRKY2-2氨基酸序列与其他植物氨基酸序列一样,均包含2个WRKY结构域和1个CX4−5CX22−23 HXH(C2H2)型锌指结构,属于Group Ⅰ (图3)。
-
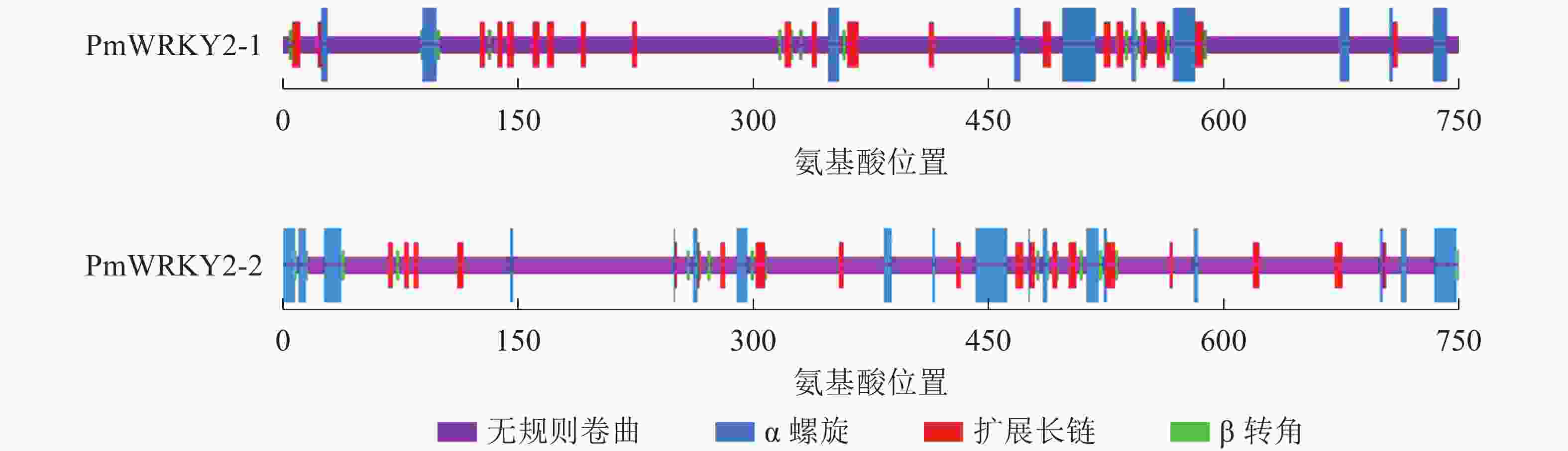
蛋白质二级结构预测结果显示(图4):PmWRKY2-1蛋白质的二级结构中包含75.68%的无规则卷曲、10.81%的α螺旋、10.41%的扩展长链和3.11%的β转角结构;PmWRKY2-2蛋白质的二级结构中包含74.02%的无规则卷曲、13.80%的α螺旋、8.80%的扩展长链和3.38%的β转角结构。
-
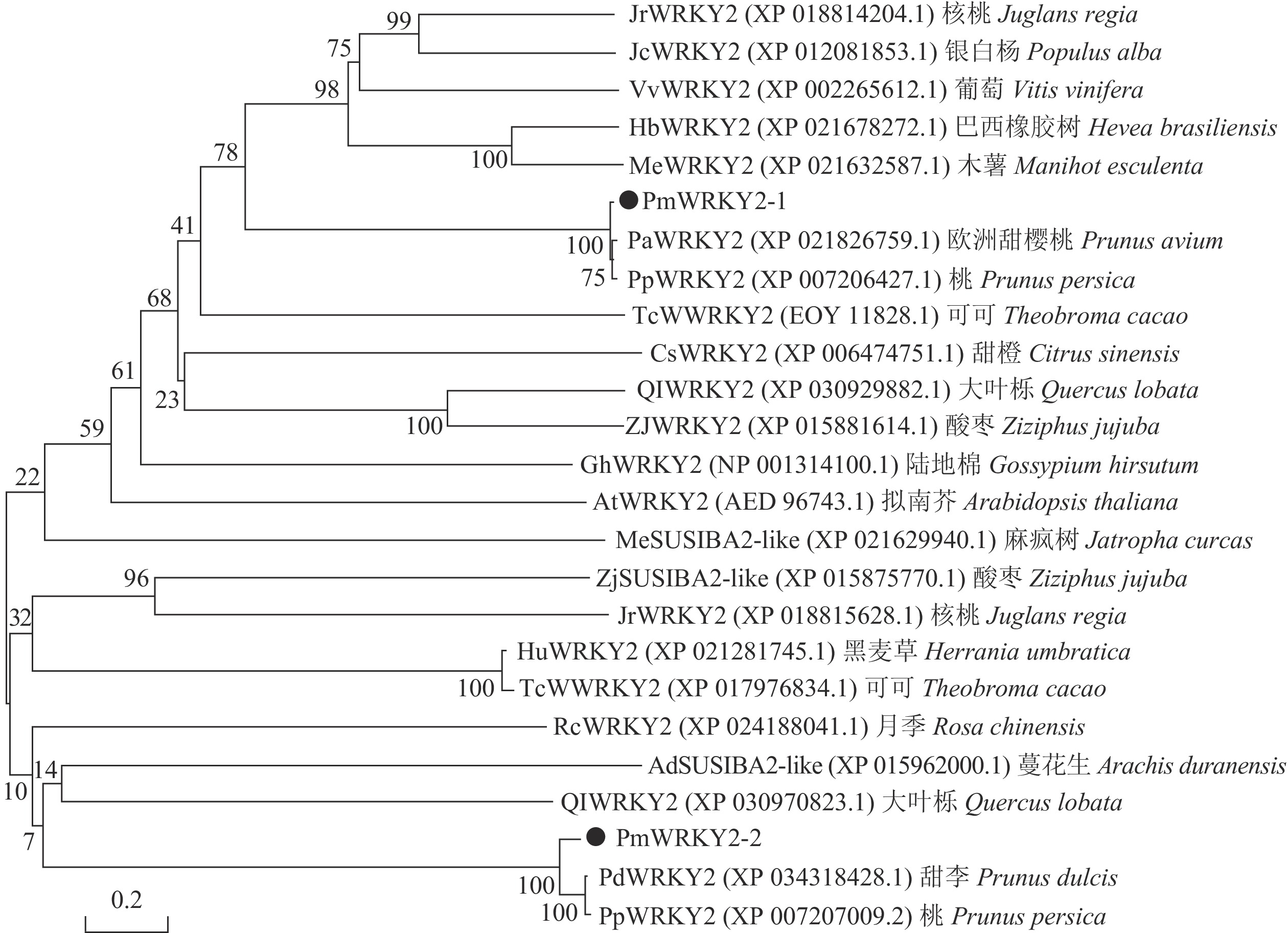
利用MEGA 6.0软件构建梅花PmWRKY2-1和PmWRKY2-2氨基酸序列的系统进化树(图5)。结果显示:梅花PmWRKY2-1与PmWRKY2-2的相似性较低,但与一些蔷薇科Rosaceae植物的亲缘关系都较近;其中PmWRKY2-1与桃(XP_007206427.1)、欧洲甜樱桃(XP_021826759.1)的WRKY亲缘关系较近,PmWRKY2-2与甜李(XP_034218428.1)、桃(XP_007207009.2)的WRKY氨基酸聚为一类,与麻疯树Jatropha curcas (XP_021629940.1)、酸枣Ziziphus jujube(XP_015875770.1)、蔓花生Arachis duranensis(XP_015962000.1)等SUSIBA2-Like(sugar signaling in barley)基因的氨基酸序列也有一定的相似性。
-
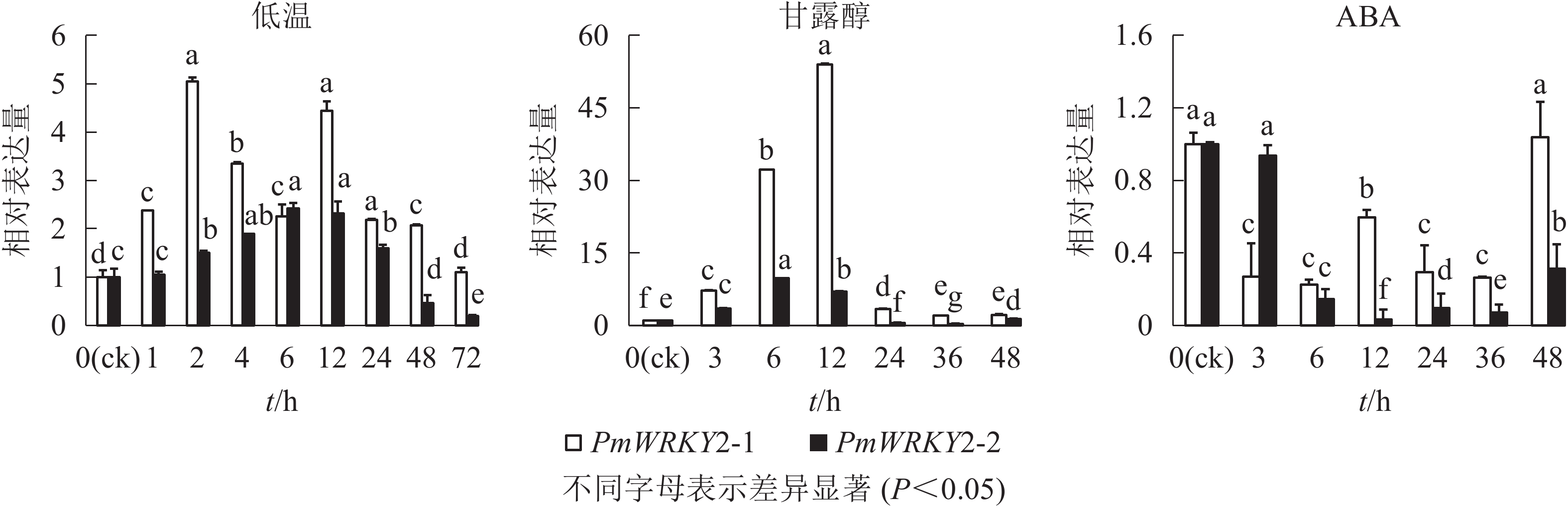
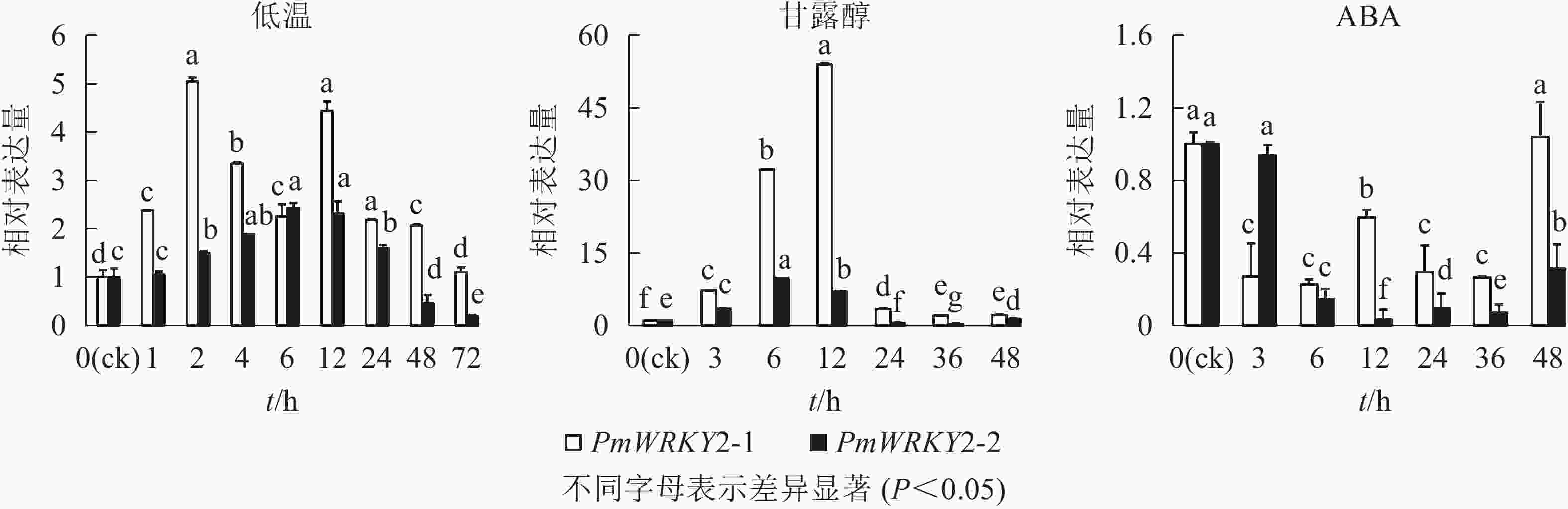
低温和干旱(甘露醇)处理后,PmWRKY2-1和PmWRKY2-2表达均发生显著变化(图6)。在低温处理下,PmWRKY2-1在2 和12 h时表达量最高,分别是对照的5.0和4.4倍,之后呈现下降的趋势;PmWRKY2-2的表达量呈现先上升后下降的趋势,在6 h时达到最大值,是对照的2.4倍。在干旱处理下,PmWRKY2-1和PmWRKY2-2的表达模式均为先上升后下降的趋势,PmWRKY2-1的表达量在12 h达到最大且为对照的53.9倍,之后便呈现下降的趋势;PmWRKY2-2的表达量在6 h处达到最大,为对照的9.7倍,之后呈现下降趋势。在脱落酸(ABA)处理下,处理48 h前,PmWRKY2-1和PmWRKY2-2均显著下调(P<0.05),说明其表达可被ABA抑制。
2.1. 梅花PmWRKY2-1和PmWRKY2-2基因的克隆及生物信息学分析
2.2. 梅花PmWRKY2-1和PmWRKY2-2蛋白的二级结构预测
2.3. 系统进化分析
2.4. PmWRKY2-1和PmWRKY2-2在非生物胁迫和脱落酸处理下的表达
-
植物在生长发育的过程中会受到多种因素的影响,而低温与干旱是常见的影响植物生长发育、果实品质以及地理分布的非生物胁迫因素,严重时可能会导致植物死亡。植物在长期适应进化的过程中逐渐形成了复杂而高效的应答机制,从分子、生理、细胞和生化等多方面做出适应性调整,以抵御和适应低温、干旱等胁迫。在植物响应低温、干旱胁迫过程中,普遍存在于植物中的WRKY转录因子发挥了重要作用[17],目前已在拟南芥[7]、番茄Solanum lycopersicum[18]、玉米Zea mays [19]、苹果Malus domestica[20]、水稻[21]等大多数物种中均有报道。
本研究克隆获得的PmWRKY2-1和PmWRKY2-2基因都含有2个WRKY结构域,C端都为C2H2型锌指结构;但2个蛋白质序列的差异较大,相似性仅为45.87%;与一些蔷薇科植物的亲缘关系较近;PmWRKY2-1与拟南芥AtWRKY2的相似性为51.26%,PmWRKY2-2为32.07%。值得一提的是,PmWRKY2-2与麻风树(XP_021629940.1)、酸枣(XP_015875770.1)、蔓花生 (XP_015962000.1)等植物的SUSIBA2-Like氨基酸序列具有一定的相似性,有研究[22]报道SUSIBA2属于WRKY转录因子超家族并参与碳水化合物合成代谢。
罗昌国等[23]发现:低温处理下湖北海棠Malus hupehensis MhWRKY40b基因表达量呈现先上升后下降的趋势;低温处理下黄瓜Cucumis sativus CsWRKY46[24]和水稻OsWRKY76[11]也呈现先上升后下降的表达趋势,与本研究中梅花PmWRKY2-1和PmWRKY2-2基因对低温的响应趋势一致。干旱处理下PmWRKY2-1和PmWRKY2-2的表达量先显著上升后下降,最高表达量分别上调了约50倍和10倍。ZHU等[25]发现:拟南芥中过表达甘薯Ipomoea batatas IbWRKY2和苦荞[26]Fagopyrum tataricum FtWRKY10能提高转基因植株的抗旱性;ZHANG等[27]发现:吲哚-3-乙酸(indole-3-acetic acid)处理白车轴草Trifolium repens,其WRKY2作为干旱响应基因可以提高白车轴草的耐旱性。JIANG等[28]发现:ABI5、ABI3、ABA2和ABA3等ABA途径基因诱导拟南芥2个WRKY2的表达从而介导种子萌发和萌发后的发育停滞。本研究中,ABA处理下,梅花2个PmWRKY2基因的表达都被抑制,预测启动子序列中的PmWRKY2-1和PmWRKY2-2分别含有1个和7个ABA响应元件ABRE,推测这2个基因可能通过ABA调控低温相关基因的表达进而调控梅花的耐寒性,但这些推论还需进一步验证。











 DownLoad:
DownLoad: