-
山麦冬Liriope spicata为百合科Liliaceae多年生草本植物,在园林绿化中多栽培于林下或林缘半阴处,掩饰裸露土壤,起到补充绿地改善不良景观的作用。山麦冬属Liriope植物只有8种,中国栽培6种,其中包含3个特有种,但山麦冬属植物分布广泛,除极寒地区及高海拔地区外,中国各省均有分布,其地理分布受人为栽培引种因素影响很大,没有特定的地理分布规律[1]。山麦冬成熟时果实表皮由绿转黑,9月结果后观果时期可长达整个冬季,且其花葶较长多矗立于叶子的上方,易于观察,具有很高的园林应用价值。目前,针对山麦冬成熟过程中呈色物质及调控基因尚未报道,但花青素合成途径在植物中是保守的,合成途径中上游合成基因是决定植物组织能否积累花青素的关键[2],而下游修饰基因的表达常与花青素的积累一致,是加深果色花色的关键基因[3-5]。此外,花青素的积累还受转录因子的调控,其中以MYB转录因子与bHLH转录因子最为常见[6]。
用于基因表达定量分析的方法比较多,其中实时荧光定量PCR(RT-qPCR)由于定量准确、成本低且高通量,被广泛应用于基因表达水平研究。但其结果常受RNA质量、反转录效率、引物特异性、初始样品量及扩增效率等因素的影响[7-8],需要引入1个或多个表达稳定的内参基因(reference genes, RGs)来评估目的基因的相对表达[9]。在植物学研究中,曾以肌动蛋白(actin,ACT)[10-12]、组蛋白(histone)[11]、蛋白磷酸酶(protein phosphatase,PP2A)[13]、甘油醛-3-磷酸-脱氢酶(glyceraldehyde-3-phosphate dehydrogenase,GAPDH)[12]、泛素结合酶(ubiquitin conjugating enzyme, UBC)[14-15]以及18S核糖体RNA(18S ribosomal RNA,18S)[16]等基因作为内参基因。但是常见的内参基因也并非适用于任何研究,且目前还未见山麦冬内参基因的报道。鉴于此,本研究基于山麦冬转录组数据,对山麦冬果实发育中稳定表达的内参基因进行研究,为提高果色转变关键基因RT-qPCR分析的准确性提供科学依据。
-
在浙江农林大学资源圃,选取生长环境相同,且植株生长状况良好、长势整齐的山麦冬,随机均匀采集15~20株山麦冬植株的各一簇花葶的上、中、下部分果实,基于山麦冬果实生长特性,采集山麦冬幼果期(2020年9月)及成熟期(2020年11月) 2个时期样品,果实从花葶中取下后立即存于−80 ℃冰箱备用。设置3次生物学重复。
-
使用天根离心柱型RNA试剂盒(天根生物科技有限公司)从每个时期样本中提取总RNA。采用质量分数为1%的琼脂糖凝胶电泳检测RNA的完整性。总RNA的纯度和质量浓度采用NanoDrop ONE微量核酸蛋白浓度测定仪(Therm,美国)测定。总RNA样本质量浓度均高于4×10−5 ng·L−1以上,总RNA纯度[D(260)/D(280)]为1.9~2.1。cDNA的合成使用PrimerScript™ RT Master Mix cDNA (Perfect Real Time)反转录试剂盒,所有样本总RNA加入量按照3×10−5 ng·L−1稀释至同一质量浓度,cDNA置于−20 ℃冰箱保存。
-
基于已获得的山麦冬转录组数据及京都基因与基因组百科全书(KEGG)注释,筛选了多条通路的基因作为内参基因参考库,包括参与山麦冬果实运输和分解代谢的基因(SLC36等),参与代谢过程的基因(PP2C、MGL、PDP、G6PD等),参与信号传导与转运的基因(AUX、GPR107、CNNM等),参与细胞过程的基因(CFL等),参与植物免疫的基因(Trx等),参与遗传信息处理的基因(UGT、PP2A、EF1-α等)共1 648个,参考前人对内参基因的筛选阈值稍作修改后[11-13],以每千个碱基转录每百万映射读取的片段(FPKM)高于5的基因(低表达的难以检测)、变异系数<0.1、变化倍数<0.2为筛选条件,得到前15个候选内参基因(表1)。
基因名 基因注释 变异
系数变化
倍数基因名 基因注释 变异
系数变化
倍数SLC36 solute carrier family 36 0.003 0.001 CFL cofilin 0.061 0.178 PP2C protein phosphatase 2C 0.007 0.019 UGT UDP-glucose: glycoprotein glucosyltransferase 0.064 0.184 Trx-1 thioredoxin 0.037 0.107 PP2A protein phosphatase 2A 0.064 0.185 MGL monoacylglycerol Lipase 0.043 0.123 EF1-α elongation factor 1-alpha 0.067 0.193 AUX auxin influx carrier 0.050 0.144 G6PD-1 glucose-6-phosphate dehydrogenase 0.068 0.197 GPR107 G protein-coupled receptor 107 0.056 0.161 G6PD-2 glucose-6-phosphate dehydrogenase 0.045 0.130 PDP pyruvate dehydrogenase phosphatase 0.058 0.169 Trx-2 thioredoxin 1 0.065 0.186 CNNM cation transport mediators 0.061 0.177 Table 1. 15 candidate reference genes of L. spicata
根据转录组获得的核酸序列信息,利用primer 5软件设计引物,并交由杭州有康生物技术有限公司合成(表2)。利用TB Green染料(Takara)预反应,体积20 μL,并使用LightCycler® 480 Ⅱ型荧光定量PCR仪(罗氏,瑞士)进行RT-qPCR。反应程序:95 ℃预变性5 min;95 ℃变性10 s;60 ℃退火延伸30 s,40个循环。实验设置3次生物学重复。扩增效率(cDNA稀释浓度梯度为5−1、5−2、5−3、5−4、5−5)计算公式为E=[10(−1/K)–1]×100%,其中:E为扩增效率,K为斜率。15个候选内参基因的扩增效率为91.7%~108.0%(表2)。
基因名 正向引物序列(5′→3′) 反向引物序列(5′→3′) 产物长度/bp 扩增效率/% 相关系数 SLC36 GTAAGTTTCGCCGAGTGCTT ACTGCAGTAGCAGACCAGTT 148 91.7 0.982 PP2C TGGGCCATGATGTTCCAGAT AGTACACGCAGTCTTCACCT 77 94.8 0.999 Trx-1 TTGTTGGCACCCACAAGTTT CATTCGTGCCACTCCAACAT 72 102.0 0.999 MGL AATGCCTTCACTGGAACAGC GCCGCCAAGTGAGTAAACAA 138 101.0 0.994 AUX TGCAGAGAAACCACCCTTCT CCGAATCCAAATCCGACCAC 99 91.7 0.949 GPR107 ACAGGTGATTGCGAACATCG CTTCGACGTCTCCTTCAACG 166 105.0 0.906 PDP GACGGAGGTCGGTTGGATTT CTGCACATGCATCATCACGA 124 96.2 0.976 CNNM GCTGCACTAACTCCAGCTTC GGCACAACTGTGGTCAACAT 86 96.8 0.999 CFL CGAGGAGAACTGCCAGAAGA GTTGGATCGGTCGCTTGTAG 153 107.0 0.992 UGT TGGAAGCATCCTCACTTGACT TGTCTTCAAATTAGGGTTAGCGA 83 93.5 0.994 PP2A GAGTCGGAGAGGTCGAAGAG GCGGAGCAATTCCTACCATC 121 99.2 0.975 EF1-α CAAGCGTCCCACTGACAAG CCAGGCTTGAGGATACCAGT 111 101.0 0.998 G6PD-1 GATGCAACAGGCCAGAAGAG AGTGCAAACAGTGCAGGAAA 104 97.9 0.996 G6PD-2 ATAACGTTGCCCTCTCCACA ATCCAACTGCAATCCAAGCC 107 108.0 0.999 Trx-2 GTGGTGCACCGTCAGTAAAC CGCTGTGGTTGATGTCTCTG 113 96.0 0.992 Table 2. Primer sequences and amplicon characteristics of 15 candidate reference genes
-
通过4种方法分析内参基因的稳定性:ΔCt值法[17]、geNorm[18]、NormFinder[19]和BestKeeper[20]。利用Excel 2010计算4种方法对候选内参基因几何平均数的排名,综合筛选最适的内参基因。同时根据前期转录组数据筛选了10种目的基因,涵盖花青素合成通路上下游基因以及调控基因。这10种基因在转录组数据加权共表达分析中属于中枢基因,表达量高、与花青素相关性强,且在果实成熟过程中显著上调。目的基因包括C4H、CHS、MT、UFGT、MYB、bHLH,上述基因引物序列及扩增子特征见表3,最后利用SPSS 19.0与Graphpad Prism 8.0分析及作图。
基因名 正向引物序列(5′→3′) 反向引物序列(5′→3′) 产物长度/bp 扩增效率/% 相关系数 C4H TCTTTGATCACGGCTTGCAG ATGAGATCGACACCGTCCTC 88 109.0 0.992 CHS-1 TGCATTGCACCAGTAGTAGC GCCCTCCTGATCTCCTCAAC 122 104.0 0.995 CHS-2 TTGTTGGCACCCACAAGTTT CATTCGTGCCACTCCAACAT 82 91.7 0.997 MT CCACCGAGAGCAAGAACAAC GGGTACACACTGGTCTCCAA 112 96.2 0.999 UFGT-1 AGCAAGGTGTTGAAGGAGGA AAATTCCGAACCGAGCTTCC 110 91.7 0.935 UFGT-2 CGACGGATCCCATTCGACTA CGCCGCTCCTCCTATTAAC 57 92.9 0.996 MYB-1 GCAAGATCAGGTCCTCCTCA CAAAGTACGTGGCGAAGGAG 162 107.0 0.975 MYB-2 ATGGGAAGATGGTGGCCTTT GAAGGGTGCACAGCTTCAG 70 91.7 0.986 MYB-3 CGAGGAGAACTGCCAGAAGA GGTGCTTGTTGAGAGAGCTG 172 105.0 0.996 bHLH TGCTTAGCAATGGCAACAGG GGCTGCTGACCAGAAGATTG 123 101.0 0.998 Table 3. Primer sequences and amplicon characteristics of 10 target genes
-
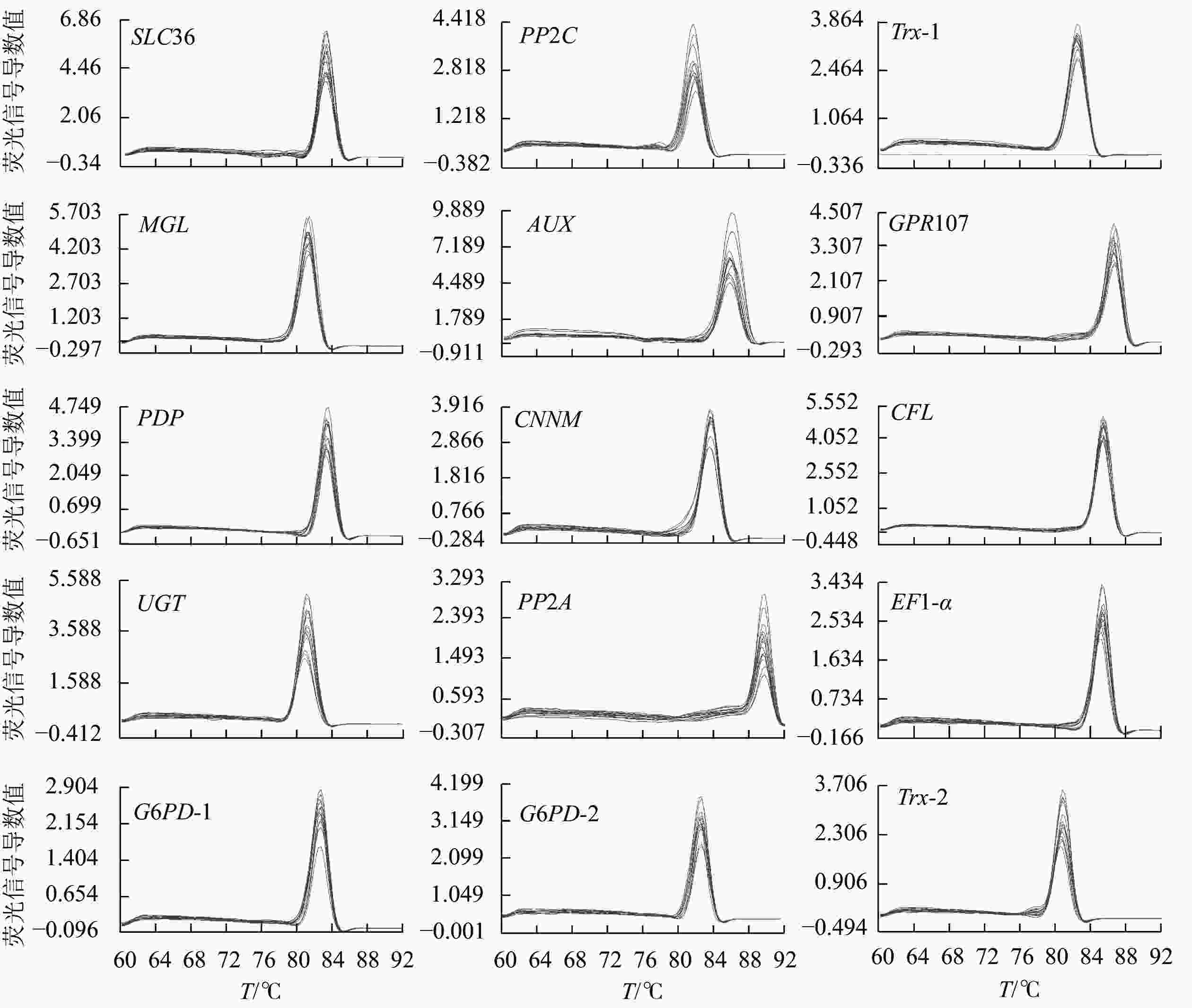
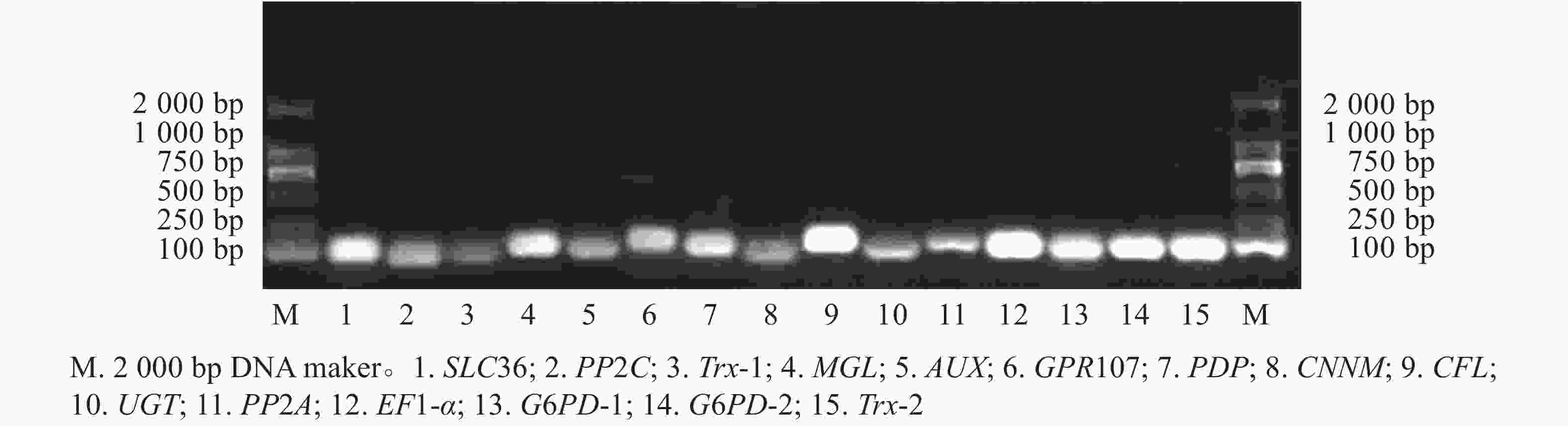
15个候选内参基因的溶解曲线均为单一峰(图1),琼脂糖凝胶电泳检测后出现与预期大小一致的单一条带(图2)。该结果表明引物具有良好的特异性。
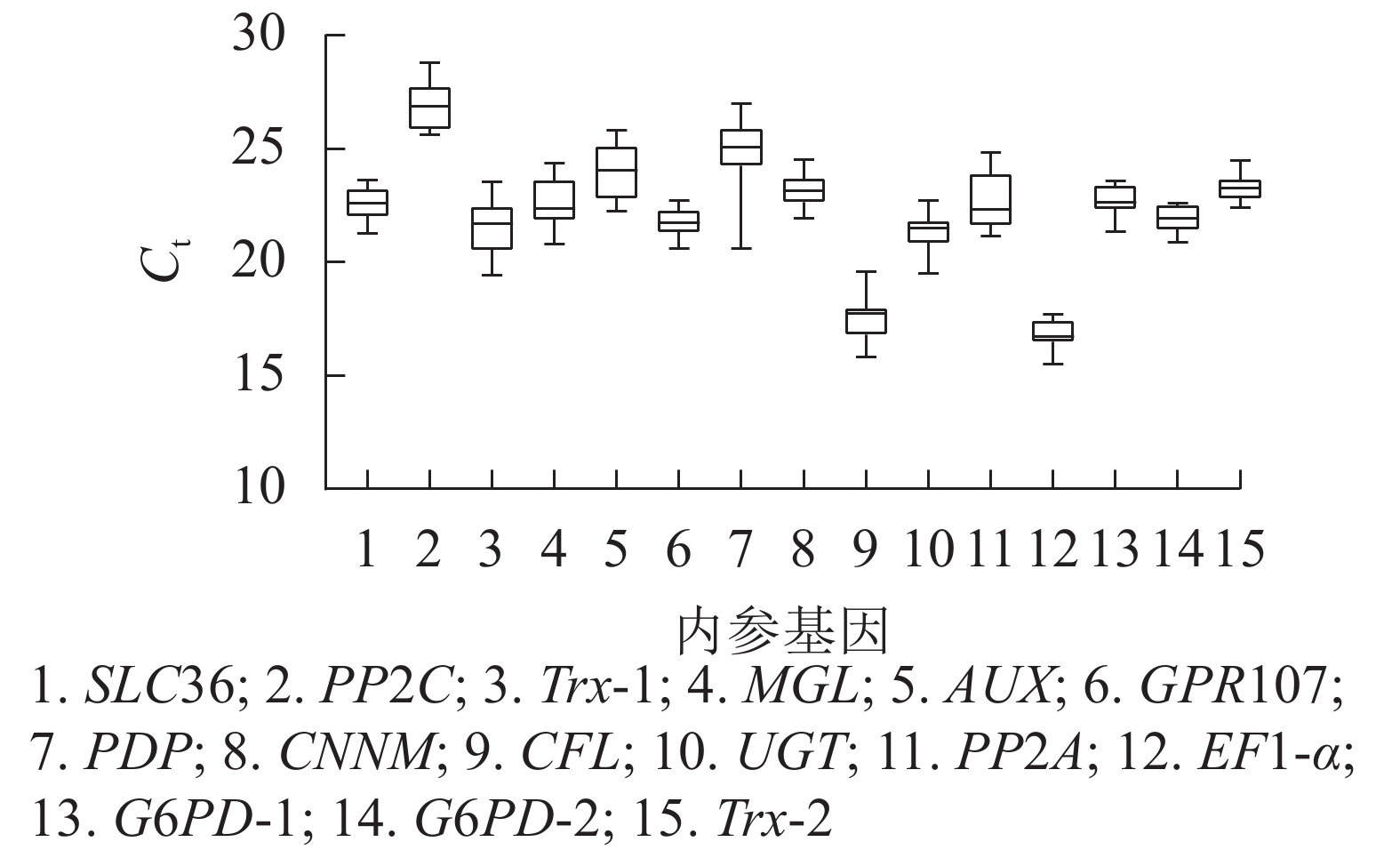
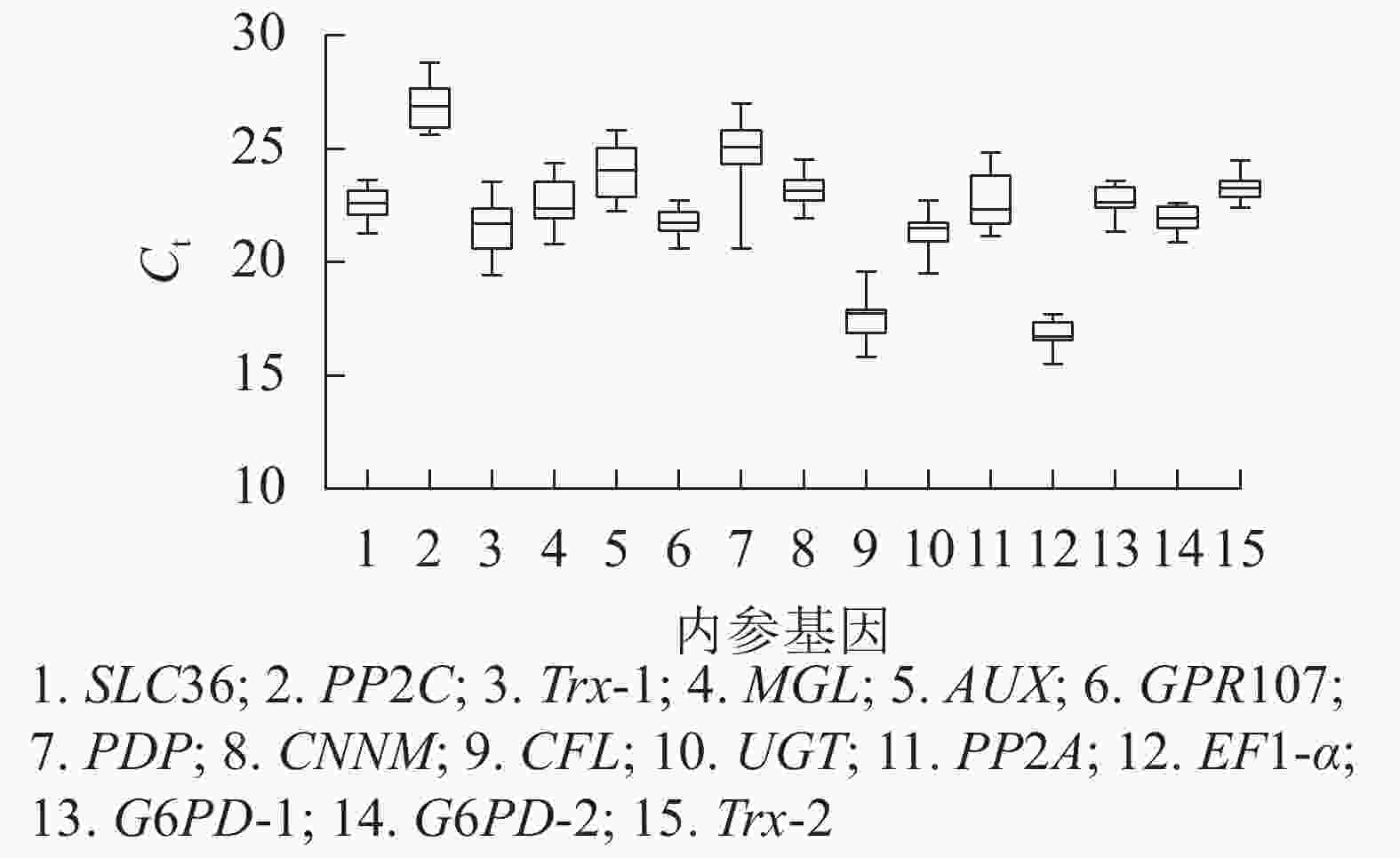
根据原始循环阈值(Ct)分布发现:所有候选内参基因的Ct为15.53~28.81,Ct越高,基因的表达量越低,反之表达量越高。本研究中,EF1-α基因表达量最高,PP2C基因表达量最低,其余基因表达量介于两者之间。此外,由箱线图(图3)跨度可初步判定内参基因的稳定性。PP2C、Trx-1、AUX、PP2A、PDP基因的Ct跨度广,不稳定,而GPR107、CNNM、EF1-α、G6PD-2、Trx-2基因最为稳定,其中GPR107、CNNM、G6PD-2基因的Ct中位数与平均数接近,即上述基因相对表达量离散程度低,表达更稳定。然而对原始Ct分析内参基因稳定性的不足,还需引入其他的方法。
-
利用ΔCt法、geNorm、NormFider和BestKeeper对15个候选内参基因的稳定性进行分析(表4)。
内参基因 ΔCt geNrom NormFinder Beatkeeper 标准差 基因平均表达值 基因稳定值 标准差 变异系数 相关系数 SLC36 2.632 0.854 0.173 0.569 2.523 0.671 PP2C 2.321 0.927 0.416 0.828 3.070 0.824 Trx-1 2.663 1.130 0.510 0.852 3.964 0.832 MGL 2.673 1.007 0.493 0.885 3.918 0.918 AUX 2.652 1.094 0.598 1.063 4.430 0.882 GPR107 2.617 0.817 0.167 0.489 2.253 0.728 PDP 2.737 1.390 0.831 0.642 2.571 0.462 CNNM 2.615 0.847 0.157 0.468 2.015 0.721 CFL 2.274 1.094 0.346 0.532 3.038 0.781 UGT 2.613 0.923 0.237 0.517 2.418 0.651 PP2A 2.693 1.054 0.568 1.057 4.671 0.511 EF1-α 2.127 0.895 0.286 0.393 2.347 0.687 G6PD-1 2.763 1.204 0.692 0.469 2.065 0.009 G6PD-2 2.636 0.880 0.334 0.290 1.323 0.750 Trx-2 2.663 0.989 0.465 0.417 1.790 0.487 Table 4. Expression stability of 15 candidate reference genes evaluated by 4 methods
ΔCt法是在原始Ct值的基础上,计算每个基因所有样本与其他基因的Ct值之差,并计算其标准差。一般平均标准差越低,基因稳定性越高。该方法中,EF1-α、PP2C、CFL、CNNM是山麦冬果实发育阶段最稳定的内参基因;PDP、G6PD-1、PP2A是最不稳定的内参基因。
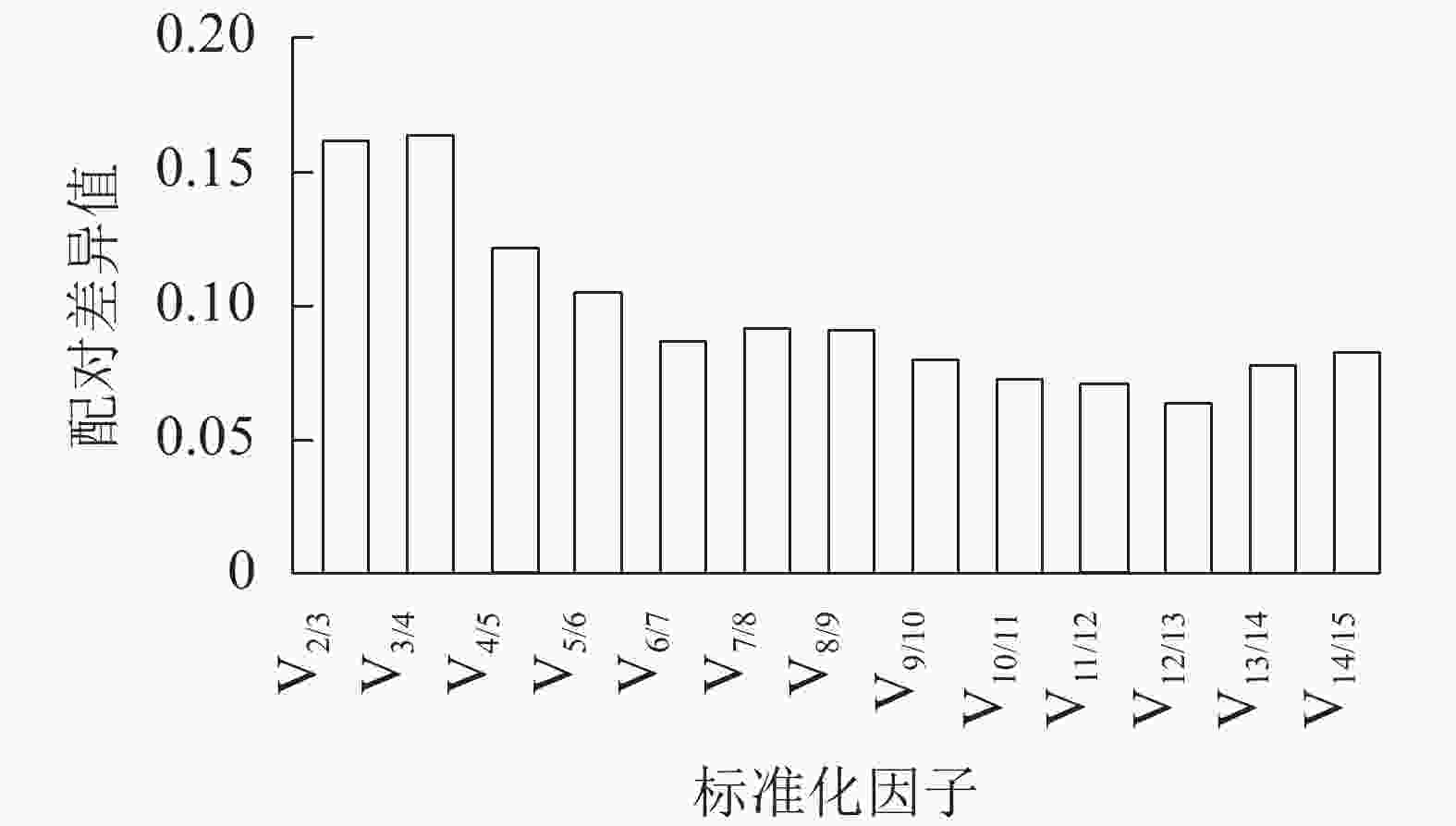
geNorm软件通过平均表达值来描述候选内参基因的稳定性,同时还能计算归一化因子之间的两两变异(Vn/n+1,其中n为可使RT-qPCR结果准确的最少基因数目)。该方法中,所有基因的平均表达值都在1.5以下(稳定内参基因的临界值),即该方法判定下的所有基因都可作为内参基因,其中GPR107(0.817)与CNNM(0.847)基因的平均表达值最低,说明最稳定。同时PDP、G6PD-1基因的平均表达值最高,分别为1.390、1.204,最不稳定,这与ΔCt法判定结果一致。此外,利用geNorm计算2个归一化基因的Vn/n+1,确定适合量化果实生长过程的最优内参基因数目。geNorm首先计算2个最稳定的候选内参基因的归一化因子值,然后将剩余候选内参基因按其表达稳定性下降的顺序依次相加。如果基因之间的Vn/n+1大于或等于0.15,则进行RT-qPCR分析时应该再添加1个基因才能达到可靠的结果,一旦Vn/n+1低于0.15,就不需要添加额外的基因[21]。由图4可见:从V4/5开始Vn/n+1小于0.15,即需要使用4个内参基因才能得到可靠的RT-qPCR结果。
NormFinder软件可分析候选内参基因的两两变异性,其中稳定值越小,候选内参基因越稳定。CNNM与GPR107基因的稳定值最小,分别为0.157、0.167,即CNNM与GPR107基因最稳定,这与geNorm分析结果一致;此外,对最差的内参基因评价也与上述2种方法一致:PDP、G6PD-1、AUX是量化果实发育阶段最不适合的内参基因。
Bestkeeper与geNorm、NormFinder软件不同,需导入原始Ct值平均数,计算候选内参基因在所有样品中的标准差、变异系数、相关系数。一般地,稳定的内参基因拥有低的标准差、变异系数及高的相关系数。在Bestkeeper评价中,与geNorm、NormFinder分析结果一致,CNNM与PDP基因分别还是最稳定与最不稳定的内参基因。除此之外,还发现G6PD-2为该方法中最稳定的内参基因,其标准差与变异系数最低,分别为0.290、1.323,相关系数为0.750。
最后通过几何平均数对这4种方法的分析结果进行综合性排序(表5)。根据表5的排名与geNorm推荐的内参基因数目,筛选CNNM、GPR107、EF1-α、G6PD-2作为标准化山麦冬果实RT-qPCR的最优内参组合,PDP为最差内参基因,通过4种算法得出的结果也与最初候选内参基因原始Ct值分布箱线图分析结果一致。
基因名 几何平均数 排名 基因名 几何平均数 排名 CNNM 2.340 1 PP2C 6.557 9 GPR107 2.913 2 MGL 8.572 10 EF1-α 3.162 3 AUX 10.602 11 G6PD-2 3.722 4 Trx-1 11.199 12 SLC36 5.350 5 G6PD-1 11.977 13 UGT 5.826 6 PP2A 12.368 14 CFL 5.925 7 PDP 14.491 15 Trx-2 6.160 8 Table 5. Comprehensive ranking of reference genes for normalization
-
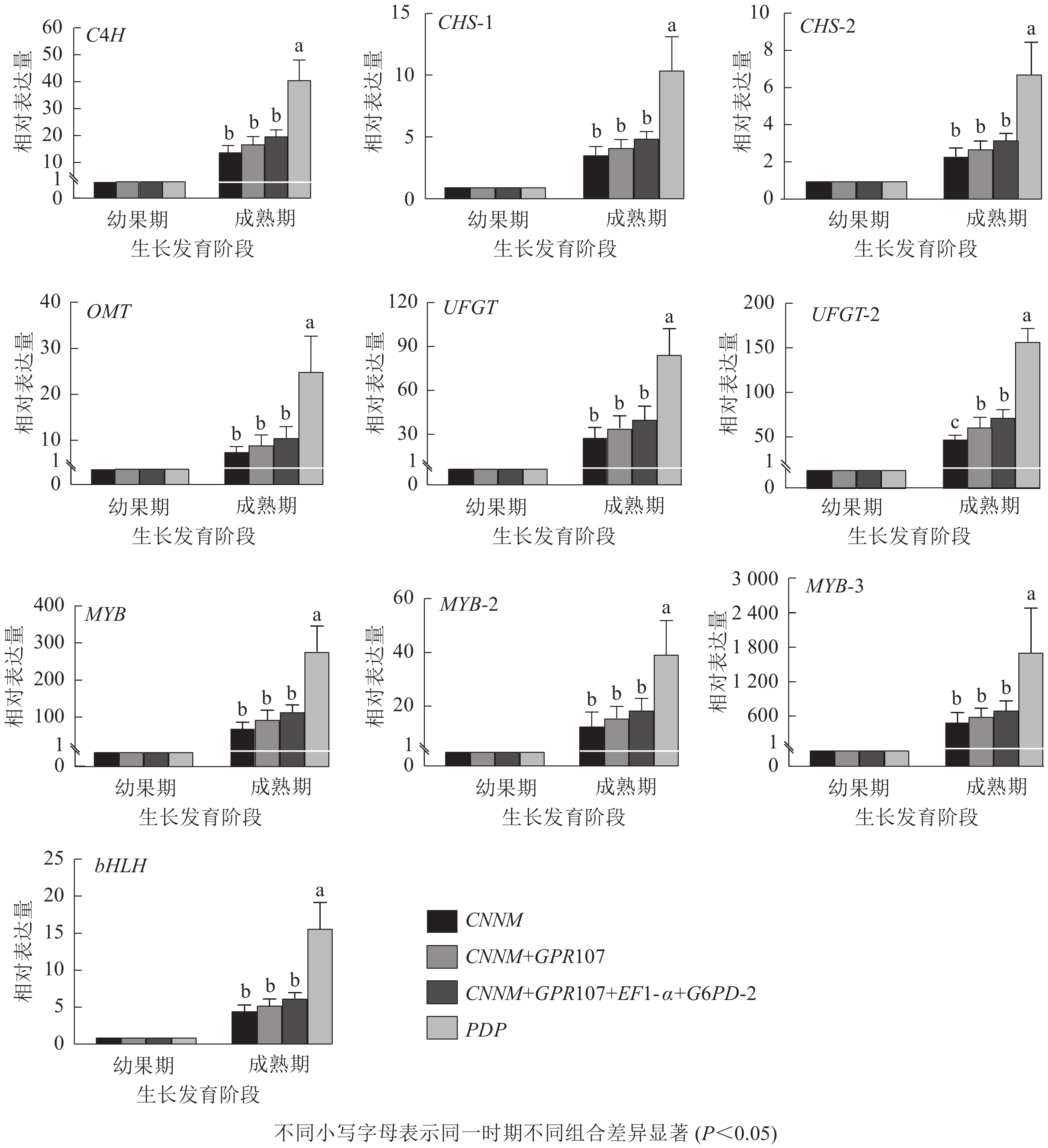
为验证内参基因的有效性,选择10种花青素合成结构基因与调控基因作为目的基因。用单一内参基因:最优内参(CNNM)、最差内参(PDP),及2种内参组合:排名前2位的内参基因(CNNM、GPR107)和排名前4位的内参基因(CNNM、GPR107、EF1-α、G6PD-2)进行归一化。从图5可见:在山麦冬果实花青素合成过程中,使用4种内参方式归一化时,所有的目的基因都上调表达,但变化倍数稍有不同。在山麦冬果实成熟期,使用PDP基因作为内参时,所有目的基因相对表达量均显著高于其他3类,特别是对转录因子bHLH基因的量化时产生严重偏差,使用PDP基因与CNNM+GPR107+EF1-α+G6PD-2基因组合作为内参,bHLH基因的相对表达量分别为6.28与15.70,两者差异高达2.5倍。然而,当使用最优内参基因CNNM进行标准化时,除UFGT基因外,CNNM、GPR107、EF1-α、G6PD-2内参组合无显著差异,使用CNNM基因标准化时,UFGT相较幼果期上调表达50.71倍,使用4种内参组合时,UFGT上调72.49倍。此外,本研究还分析了候选内参排名前2位的基因(CNNM、GPR107)作为目的基因的表达量,发现选用2种内参基因与geNorm软件推荐使用4种内参基因,在10个目的基因中均无显著差异。
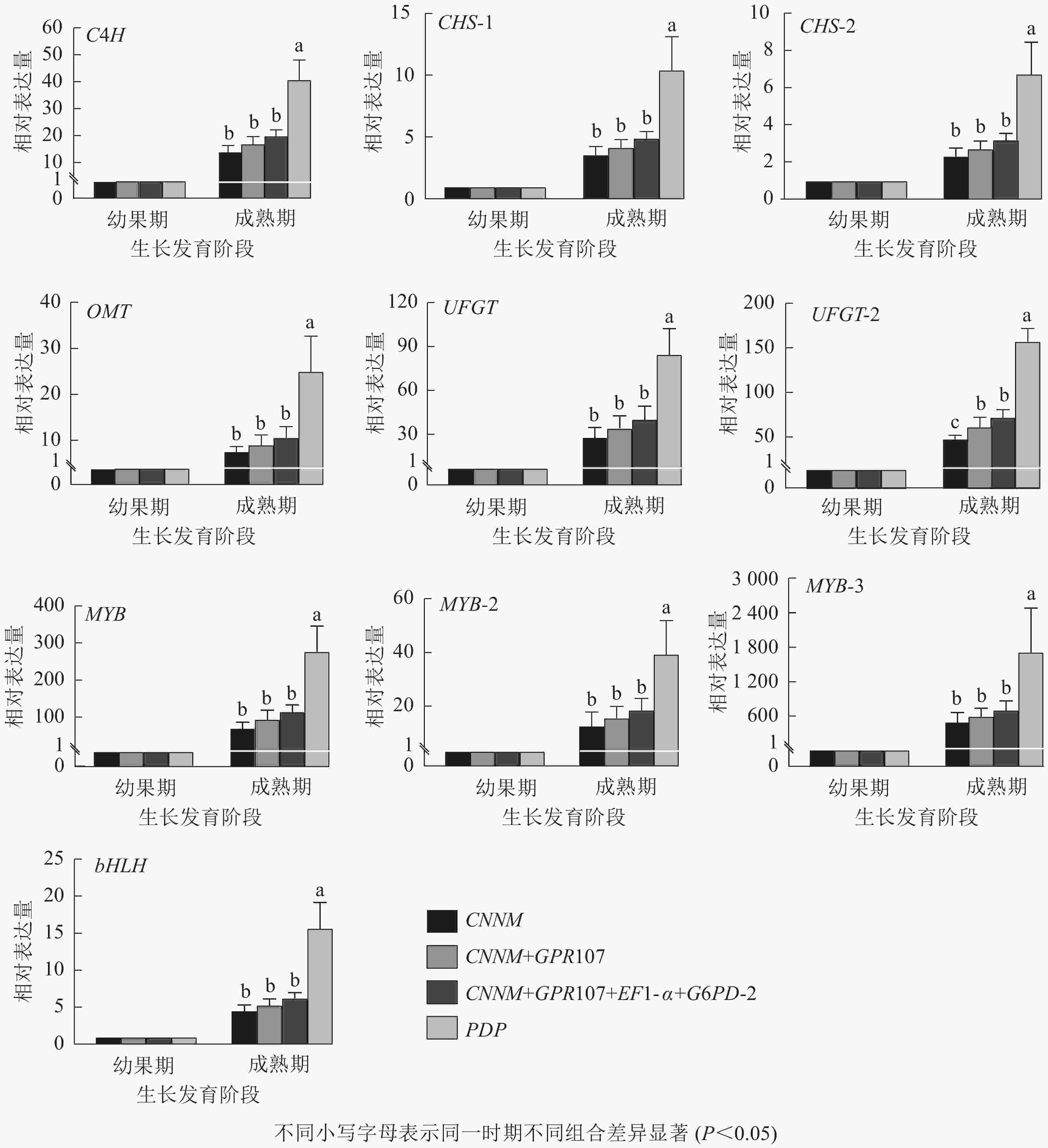
Figure 5. Relative expression levels of ten target genes after normalized by different reference genes
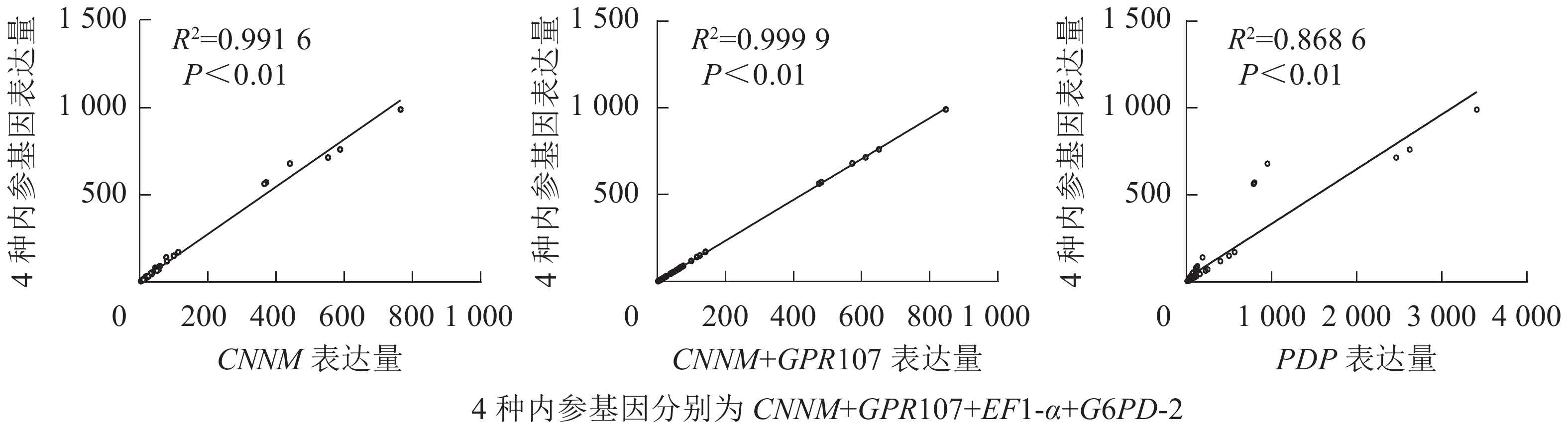
从图6可见:利用最差内参PDP得到的目的基因表达量与4种内参基因组合得到的目的基因表达量相关系数为0.868 6 (P<0.01),当使用最优基因CNNM作为内参时,与4种内参组合相关系数可达0.991 6 (P<0.01)。对2种内参组合与geNorm推荐的4个数目内参组合比较发现:通过这2种方法标准化得到的目的基因相关性可达0.999 9 (P<0.01),即仅使用CNNM、GPR107基因作为双内参也可达到geNorm软件推荐的4个内参数目组合的效果。
-
山麦冬作为一种优良的地被园林植物及药用植物,研究多集中于提高栽培技术及块茎产量,而针对园林观赏应用的研究较少。在本研究之前没有山麦冬内参的研究报道,作为沿阶草族植物,其近源种也仅有麦冬Ophiopogon japonicus抗逆性研究中曾以微管蛋白基因(tubulin)[22]及Actin [23]作为参考基因。但这2类基因在前期转录组筛选中由于变异系数及变化倍数在候选内参中就已经被排除。本研究根据几何平均数的综合排名,推荐使用内参基因CNNM、GPR107、EF1-α、G6PD-2作为研究山麦冬花青素合成的最优内参组合。EF1-α、G6PD-2属于常见的内参基因,在植物生长发育、抗逆反应、代谢合成中已被广泛应用[24-25]。基于前期转录组数据,新型内参基因CNNM、GPR107也可作为RT-qPCR分析的内参基因,CNNM编码过渡金属转运蛋白,可参与多种金属吸收、排除及区分化[26],GPR107编码G蛋白偶联受体107,广泛存在于细胞表面的膜蛋白,可参与植物体多种细胞信号转导及调控机制保守[27]。上述2种基因在山麦冬果实中表达稳定,其相对表达量平均值与中位数相近,离散程度低,且表达量适中,符合内参基因的标准。在观赏植物中,由于新型内参基因稳定性强于传统内参基因,常被选用标准化目的基因的表达。例如,在异型花柱连翘Forsythia suspensa中,转录组中变化微小的未知基因是研究花开放最适合的内参基因[28];太行花Taihangia rupestris花器官有复杂的性别决定机制,鉴定两性花与雄性花的内参基因是编码铁硫簇组装蛋白、3-巯基丙酮酸硫转移酶与跨膜蛋白50的新型内参基因[11]。SmDnaJ基因在旱柳Salix matsudana各种非生物胁迫下表达最为稳定[29]。bHLH在观赏百合Lilium oriental×Trumpet hybrid体胚诱导、体胚发育中表达最稳定[30],但bHLH是植物颜色育种中的重要靶基因,并不适合作为本研究的内参基因候选,这也证实了不同目标性状需采用不同的内参基因,没有一种内参基因是普适的。
花青素合成路径在植物中是保守的,其中MYB转录因子与bHLH转录因子可形成二元复合体,激活花青素合成酶基因[31-32]。大量研究表明:MYB、bHLH转录因子基因与花青素合成酶基因在紫色系植物组织发育过程中协同上调[3, 33-34]。为验证内参基因的结果,挑选了10个在山麦冬花青素合成调控网络的中枢基因(相关性强且表达量高)作为验证,其中包括转录因子与结构基因(C4H、CHS、MT、UFGT、MYB、bHLH),这10种基因在4种归一化方法下表达模式均显著上调,但趋势稍有不同,选用较差内参PDP标准化结果偏差最大,在山麦冬成熟黑果中所有基因都显著高于其他基因。尽管最优内参基因CNNM对目的基因的归一化可以达到与4种内参组合很高的相关系数,但对UFGT基因的量化存在显著差异,而UFGT基因作为花青素合成通路的下游修饰,对花青素积累至关重要,特别是在山麦冬这类组织颜色深即富含花青素的类型[2, 35],例如在葡萄Vitis vinifera果皮[36]、玫瑰Rosa rugosa [37]、紫皮石刁柏Asparagus officinalis[33]中UFGT都被验证为关键基因,因此仅选用单一基因作为研究山麦冬果皮花青素积累的内参是不合适的,继而在CNNM基因基础上又引入GPR107来规避单内参基因的误差,该内参组合与geNorm推荐的内参组合相关系数最高,在10种目的基因的验证结果中与4种内参组合均无显著差异,且选用双内参组合比4种内参组合可操作性强,因此判定使用CNNM、GPR107作为双内参即可得到可靠的RT-qPCR结果。双内参组合联合使用可以减少实验因素对基因表达的影响,且结果更为准确。暴露于UV-B辐射下的番茄Lycopersicon esculentum幼苗不同组织都应选用特定的内参组合,例如叶中选用肌动蛋白基因与微管蛋白基因,而根中选用微管蛋白与UV-B抗性位点基因更加适合[38];UBQ和EF1-α基因由于表达稳定,可作为内参基因用于鹅掌草Anemone flaccida各器官的不同发育阶段[39]。
-
本研究基于转录组数据筛选了15个候选内参基因,分析其在山麦冬果实不同时期的表达稳定性。经过10种目的基因验证后,表明以CNNM、GPR107基因作为组合是山麦冬果实花青素生物合成研究的最佳内参基因,而常用的内参基因却并不适用于本研究,这为筛选新型内参基因提供了新思路。
Selection and validation of reference genes for anthocyanin biosynthesis in Liriope spicata fruits
doi: 10.11833/j.issn.2095-0756.20210332
- Received Date: 2021-04-29
- Accepted Date: 2021-12-22
- Rev Recd Date: 2021-11-30
- Available Online: 2022-01-27
- Publish Date: 2022-03-25
-
Key words:
- Liriope spicata /
- quantitative real-time PCR (RT-qPCR) /
- anthocyanin biosynthesis /
- reference gene
Abstract:
| Citation: | GAN Sichen, SHI Yue, LIANG Lijun. Selection and validation of reference genes for anthocyanin biosynthesis in Liriope spicata fruits[J]. Journal of Zhejiang A&F University, 2022, 39(2): 307-317. DOI: 10.11833/j.issn.2095-0756.20210332 |











 DownLoad:
DownLoad: