-
无机磷酸盐(inorganic phosphates, Pi)的消耗是影响全球植物生长的严重问题[1]。所有生物体的生命都依赖于脂质双层细胞膜的存在,该膜将细胞内部与环境分开,单乳糖基和二乳糖基二酰基甘油(分别为MGDG和DGDG)构成植物叶绿体中膜脂质的大部分[2]。膜脂质重塑是植物用来在Pi耗尽条件下生存的防御机制[3]。在Pi饥饿期间,磷脂被降解为Pi提供其他必需的生物过程,而质体中的半乳糖脂合成通过单半乳糖基二酰基甘油合酶3(MGD3)的转录激活上调[4]。在拟南芥Arabidopsis thaliana中,已经鉴定出3种功能性MGD合成酶(MGD1,MGD2和MGD3),并根据其氨基酸身份将其分为A型(MGD1)和B型(MGD2和MGD3)酶[5−6]。A型和B型酶在特异性、亚细胞定位和基因表达谱方面存在一些差异,A型酶定位于质体的内膜,而B型酶定位于质体外膜。在这三者中,MGD1是叶绿体中最丰富的亚型。MGD1的表达在光合组织中广泛存在,而MGD2和MGD3的表达在根等非光合组织中特异性检测到,在绿色组织中很少检测到[7]。2种类型的MGD酶对于植物在低磷酸盐条件下的生存至关重要,但尚未报道对MGD基因分歧的详细分析。这些基因对于植物适应磷素缺乏症可能是必不可少的。
棉花Gossypium是一种油籽和纤维作物,在全球70多个国家种植,在全球经济中发挥着重要作用[8-10]。然而,棉花的生长和生产力受到非生物和生物胁迫[11]。磷是核酸的主要组成部分,在植物生长发育和代谢过程中极为重要,是细胞分裂和根系生长不可缺少的,低磷胁迫不利于作物生长发育[12-16]。因此,研究低磷胁迫的分子适应机制和加强抗逆性对棉花生产至关重要。张学昕等[17]通过田间试验对不同施磷处理下的棉花各时期取样进行养分和产量分析表明:当其他条件一定时,随着施磷量的递增,棉株的单株铃数以及单铃质量也会随之增加,从而实现增产的目标。王刚等[18]研究发现:棉株地上和地下两部分的干物质都随有效磷含量的增加而增加,根冠比则随其增加而降低。通过有效磷含量影响糖类物质的合成转化以及棉株叶绿素含量,进而作用于整个棉株的生长。刘耘华等[19]研究表明:施用有机肥能够增加土壤有效磷含量,除了有机肥中有一部分磷素之外,还可以促进有效磷的转化。磷素与有机肥混合施用对土壤磷库、棉花产量和磷素吸收量都会产生积极影响。
本研究以前期研究筛选出的磷高效棉花品种陆地棉‘新陆早19’Gossypium hirsutum‘Xinluzao 19’为材料,克隆了GhMGD3基因,采用生物信息学工具分析其编码蛋白理化性质,并对其进行表达特性分析,旨在为深入解析棉花GhMGD3基因的生物学功能提供科学参考,并为培育磷高效利用的棉花新品种提供基因资源。
-
陆地棉‘新陆早19’种植在河南科技大学农学院大田,不施加磷肥。选取长势一致的植株,采集根、茎、叶、花,用于分析GhMGD3基因在植株4个组织中的表达状况。
将‘新陆早19’种子播种于含干净湿润细沙的塑料盆内,28 ℃恒温培养箱,光照14 h/黑暗10 h,培养至棉苗三叶期,选择长势良好且一致的幼苗移至水培盆中,用1/2 Hoagland 营养液培养1周后分为2个处理水平,即适磷处理(SP,1.00 mmol·L−1)和低磷处理(LP,0.01 mmol·L−1),磷源采用KH2PO4,为保证K+的浓度一致,以1.0 mmol·L−1为标准,低磷营养液中以KCl补齐K+,其他营养成分含有2 mmol·L−1 Ca(NO3)2·4H2O、2.5 mmol·L−1 KNO3、0.5 mmol·L−1 NH4NO3、1.4 mmol·L−1 MgSO4·7H2O、1.0×10−3 mmol·L−1ZnSO4·7H2O、1.0×10−3 mmol·L−1 MnSO4·H2O、0.1×10−3 mmol·L−1 CuSO4·5H2O、0.01 mmol·L−1 H3BO3、0.05×10−4 mmol·L−1 (NH4)6MO7O24·4H2O、0.1 mmol·L−1 EDTA-FeNa[20]。分别处理0、4、12、24、72 h后,选取3株生长一致的棉花植株,并混合取其根部组织,然后迅速置于液氮中,−80 ℃保存备用。每日调节营养液pH 6.5,3 d更换1次营养液。
-
根据前期陆地棉根部低磷胁迫基因表达谱芯片差异表达基因序列进行分析,在美国国家生物信息中心(NCBI)网站的EST数据库中检索差异表达基因序列,得到该基因的相似序列,将所得相似性序列(覆盖率>50%,相似度>90%)使用DNASTAR的Seqman进行拼接得到重叠群,将所得序列继续检索与拼接直至没有新的相似序列出现,所得即为结果序列conting,利用ORFfinder在线平台查找开放阅读框,进行目标基因GhMGD3的克隆与分析。根据ORFfinder在线平台查找的开放阅读框,使用Primer 5.0设计引物,由生工生物工程有限公司合成,见表1。
引物名称 引物序列(5′→3′) 上游引物 GGGTTCTTTGTCTTTCTTG 下游引物 ACCTGGATTTGGGACTCTT M13F TGTAAAACGACGGCCAGT M13R CAGGAAACAGCTATGACC GhACTIN-F ATCCTCCGTCTTGACCTTG GhACTIN-R TGTCCGTCAGGCAACTCAT GhMGD3-F CTTGTTTTCTTTTCTTCTTGGTGGA GhMGD3-R TGATAATAATAGCCGTTGTTGTTGA Table 1. Primers used in the study
-
取液氮速冻整株植株后在研钵中迅速研磨成粉末转移至离心管,利用十六烷基三甲基溴化铵(CTAB)法提取基因组DNA,向其中添加200 μL的TE缓冲液溶解后置于−20 ℃保存备用。
-
以基因组DNA为模板设计PCR扩增反应体系(20 μL),冰上操作:2×M5 蓝色染料高保真Taq酶10.0 μL,上游引物(10 μmol·L−1) 0.5 μL,下游引物(10 μmol·L−1) 0.5 μL,模板 DNA 0.5 μL,无核酸酶ddH2O 8.5 μL。反应程序为:95 ℃ 3 min;94 ℃ 25 s,53 ℃ 25 s,72 ℃ 40 s,32个循环;72 ℃ 5 min;4 ℃低温保存。120 V,25 min,质量浓度为1%琼脂糖凝胶电泳检测。将目的DNA片段进行胶回收纯化后连接pTOPO-T载体,具体体系为(10 μL):M5 HiPer pTOPO-TA Vector 1.0 μL,10×增强剂 1.0 μL,纯化后的PCR产物 3.9 μL,灭菌水 4.1 μL。取5.0 μL连接产物转化50.0 μL大肠埃希菌Escherichia coli DH5ɑ 感受态细胞,将菌液涂布在含氨苄青霉素的LB固体培养基上,37 ℃培养过夜。随机挑取单菌落,扩大培养4 h。以所得菌液作模板进行菌液PCR反应,体系如下(20 μL):2×M5 蓝色染料高保真Taq酶10.0 μL,通用引物M13F 0.5 μL,通用引物M13R 0.5 μL,模板DNA 0.5 μL,无核酸酶ddH2O 8.5 μL。反应程序与以基因组DNA为模板PCR扩增反应体系相同。检测符合后送至生工生物工程有限公司测序。
-
取液氮速冻整株‘新陆早19’植株后在研钵中迅速充分研磨成粉末,后续步骤依照RNAprep Pure多糖多酚植物总RNA提取试剂盒说明书提取RNA,150 V,15 min,质量浓度为1%琼脂糖凝胶电泳检测。参考HiScript® cDNA一链合成试剂盒说明书完成cDNA一链的合成,−20 ℃保存备用。
-
以‘新陆早19’的cDNA为模板设计PCR扩增反应体系(20 μL),冰上操作:2×M5蓝色染料高保真Taq酶10.0 μL,上游引物(10 μmol·L−1) 0.5 μL,下游引物(10 μmol·L−1) 0.5 μL,cDNA 1.0 μL,无核酸酶 ddH2O 8.0 μL。PCR反应程序为:95 ℃ 3 min;94 ℃ 25 s,56 ℃ 25 s,72 ℃ 35 s,35个循环;72 ℃ 5 min;4 ℃低温保存。120 V,25 min,质量浓度为1%琼脂糖凝胶电泳检测。将目的DNA片段进行胶回收纯化后连接pTOPO-T载体,体系(5 μL)如下:M5 HiPer pTOPO-TA Vector 0.5 μL,10×增强剂 0.5 μL,纯化后的PCR产物1.5 μL,灭菌水2.5 μL。取5 μL连接产物转化50 μL大肠埃希菌DH5ɑ感受态细胞,培养后随机挑取单菌落扩大培养后进行菌液PCR反应,电泳检测符合后送至生工生物工程有限公司进行测序。
-
使用在线平台及软件对基因进行生物信息学分析,预测分析基因的结构、性质等。
-
利用半定量RT-PCR技术,分析GhMGD3基因在根、茎、叶、花中的表达状况。以各组织的cDNA为模板,GhACTIN作内参基因,所用引物见表1。设计PCR扩增反应体系(20 μL),冰上操作:2×M5蓝色染料高保真Taq酶10.0 μL,GhMGD3-F(10 μmol·L−1) 0.5 μL,GhMGD3-R(10 μmol·L−1) 0.5 μL,cDNA 1.0 μL,无核酸酶ddH2O 8.0 μL。PCR反应程序为:95 ℃ 3 min;94 ℃ 25 s,60 ℃ 25 s,72 ℃ 10 s,35个循环;72 ℃ 5 min;4 ℃低温保存。120 V,25 min,质量浓度为1%琼脂糖凝胶电泳检测。
-
采用实时荧光定量PCR (RT-qPCR)技术检测GhMGD3基因在不同组织中及低磷胁迫处理下的表达模式。以GhACTIN作为内参基因,所用引物见表1。采用SYBR® Green Pro Taq HS预混型qPCR试剂盒进行荧光定量,使用的荧光定量PCR仪器型号是CFX96,数据分析采用
$2^{{-\Delta \Delta C}_{t}}$ 法,用Origin 9.0对数据进行统计分析并作图。 -
利用前期低磷胁迫差异表达序列为探针,共检索发现并下载了8条相似序列,利用DNASTAR将所得序列全部进行拼接,得到了一个新的conting重叠群,序列长度为1 276 bp。
-
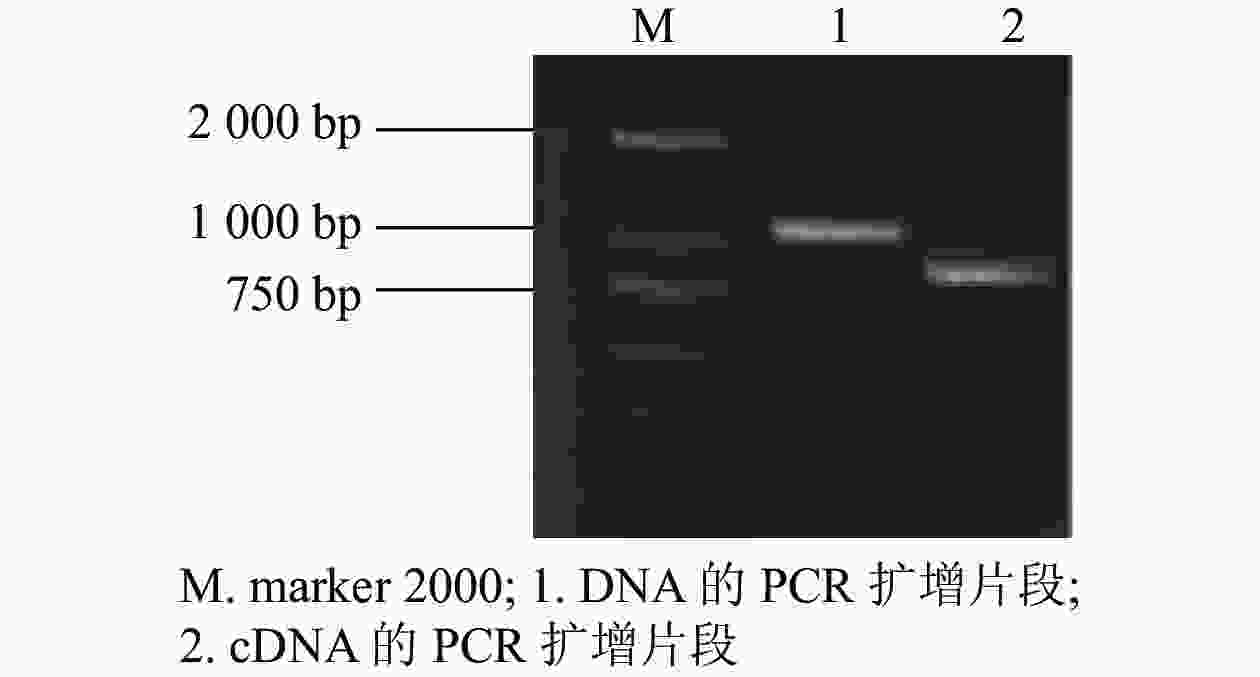
根据GhMGD3基因的编码序列(coding sequence,CDS)设计特异性引物,以植株的DNA和cDNA为模板,克隆得到GhMGD3基因的CDS (图1),其条带符合目的条带大小。通过无缝克隆连接到pTOPO-T载体上,挑选阳性克隆测序。获得DNA序列1 040 bp左右(泳道1)与conting序列的一致性较高,为93.14%。获得cDNA序列780 bp左右(泳道2)与conting序列比对一致性为99.43%。开放阅读框为长度为681 bp,共编码226个氨基酸。该基因GhMGD3是MGD家族成员。
-
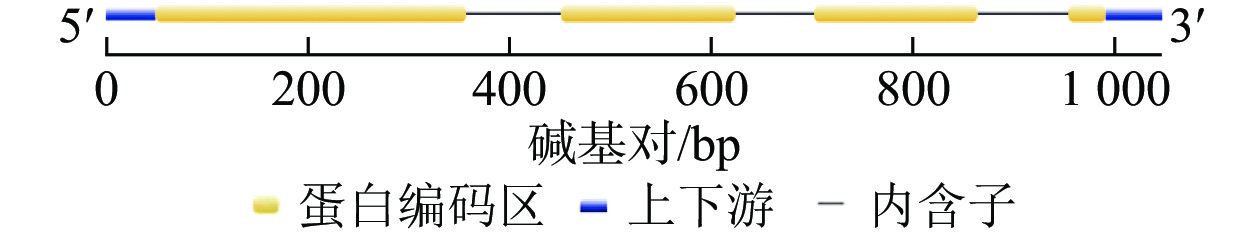
将DNA测序结果序列与cDNA测序结果的最大开放阅读框序列导入Gene Structure Display Server 2.0对该基因组序列与编码序列进行分析,表明该基因编码序列包含5′与3′端非编码序列,含有3个内含子,4个外显子(图2)。
-
将基因编码蛋白导入ExPASy-ProtParam tool在线软件,分析可知GhMGD3共编码了226个氨基酸,编码蛋白的分子式是C1201H1832N328O333S13,脂肪族氨基酸指数是76.28,分子量是26 610.54 Da,等电点为8.74,属于碱性蛋白。其中缬氨酸的质量分数占比最大,为8.4%,共19个。在该蛋白中带正电荷的氨基酸有30个,另外有26个带负电荷。氨基酸组分会直接影响蛋白的亲疏水性,总亲水性平均系数为−0.471,不稳定系数是37.83,是亲水性稳定蛋白。ProtScale analysis在线软件显示此蛋白中精氨酸亲水性最强,亲水指数为−4.500,异亮氨酸疏水性最强,亲水指数为+4.500。
-
在NPS@:SOPMA secondary structure prediction网站预测该蛋白二级结构,其中占比最大的无规则卷曲占39.38%,包含89个氨基酸,其次是α-螺旋,占比为34.96%,包含79个氨基酸;此外此蛋白还含有延伸链(extended strand)和β-转角(beta turn),其中延伸链占比相比较多,占比21.24%,有48个氨基酸,β-转角仅有10个氨基酸,占比4.42%,是最少的。表明此蛋白的二级结构由α-螺旋和无规则卷曲占据主体地位所组成。由于无规则卷曲占比较多,预测其结构比较复杂。运用TMHMM-2.0分析该基因所编码蛋白不含有跨膜结构域,属于非跨膜蛋白,226个氨基酸并未形成跨膜螺旋区。通过SignalP-5.0进行信号肽分析可知:此基因编码蛋白有信号肽的概率是0.001 6,其他的可能性则高达0.998 4。226位氨基酸中并未出现典型的信号肽趋势,即该蛋白不存在信号肽。在线网站内输入基因编码序列进行基因编码蛋白的亚细胞定位预测,预测定位在叶绿体,该蛋白可能在叶绿体发挥作用。
-
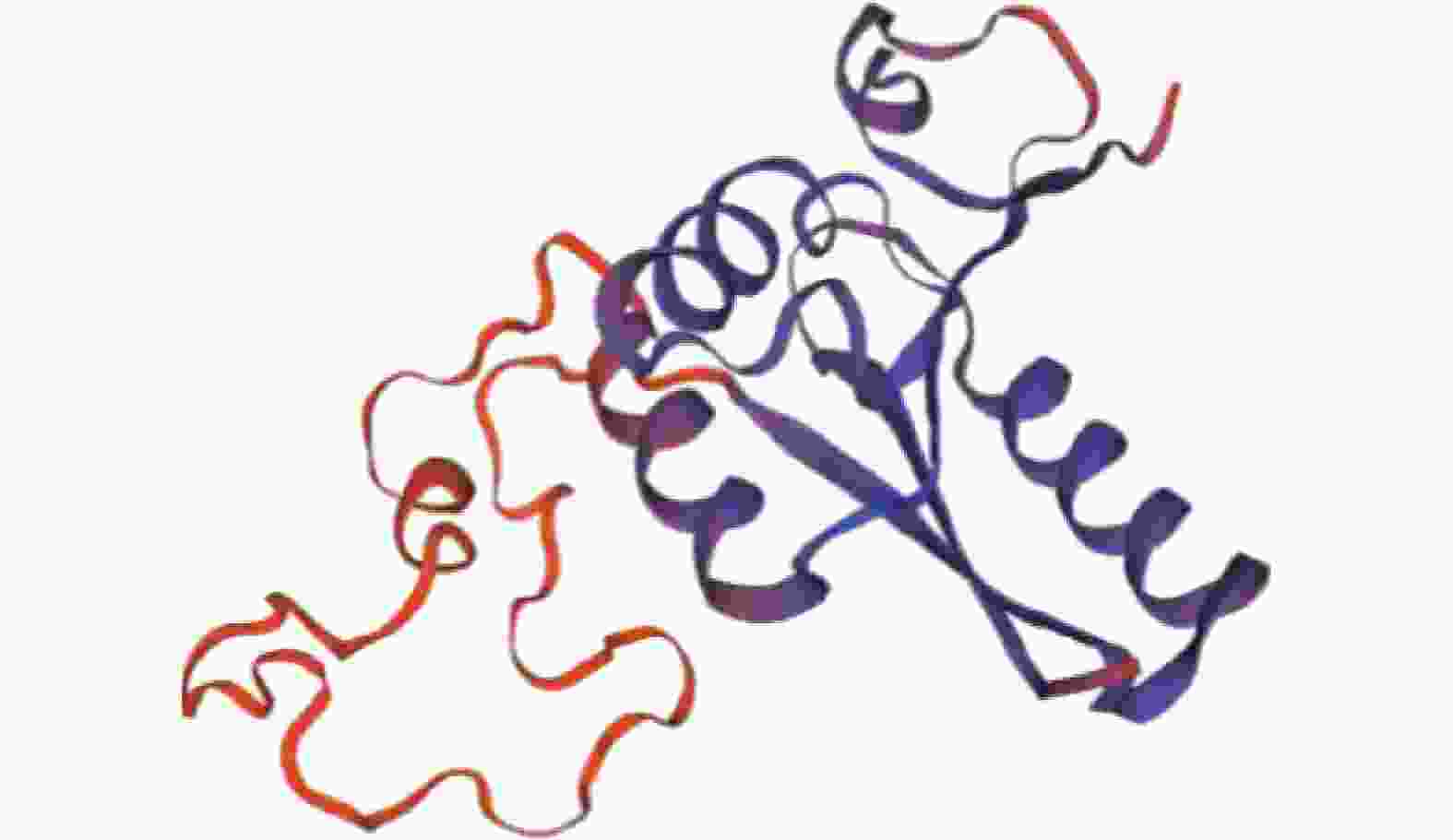
通过SWISS-MODEL Interactive Workspace进行同源建模,预测蛋白三级结构。如图3所示:QMEAN值为0.60,GMQE值为0.12,预测其结构与单半乳糖基二酰基甘油合酶相似,与拟南芥半乳糖脂合酶MGD1的结构类似。蛋白的三级结构与二级结构分析结果一致,均以α螺旋和无规则卷曲为主体。
将获得的基因编码序列提交到STRING 11.0在线网站预测该基因可能的互作网络,分析结果显示:此基因编码蛋白能够预测到蛋白的相互作用网络,因此预测该基因可能是通过多个蛋白互作进行调控。糖基化对蛋白稳定及功用均会产生极其重要的影响。将获得的基因编码序列提交到NetNGlyc 1.0在线软件可知:该蛋白不存在N-糖基化位点。磷酸化位点与蛋白的功能、结构都有很大的关系,通过NetPhos 3.1在线网站分析:此蛋白中占比最大的是丝氨酸和苏氨酸磷酸化位点,而酪氨酸占比较少。它除了可以调控蛋白的活性以外,在细胞的信号转导中也占据了重要的位置。
-
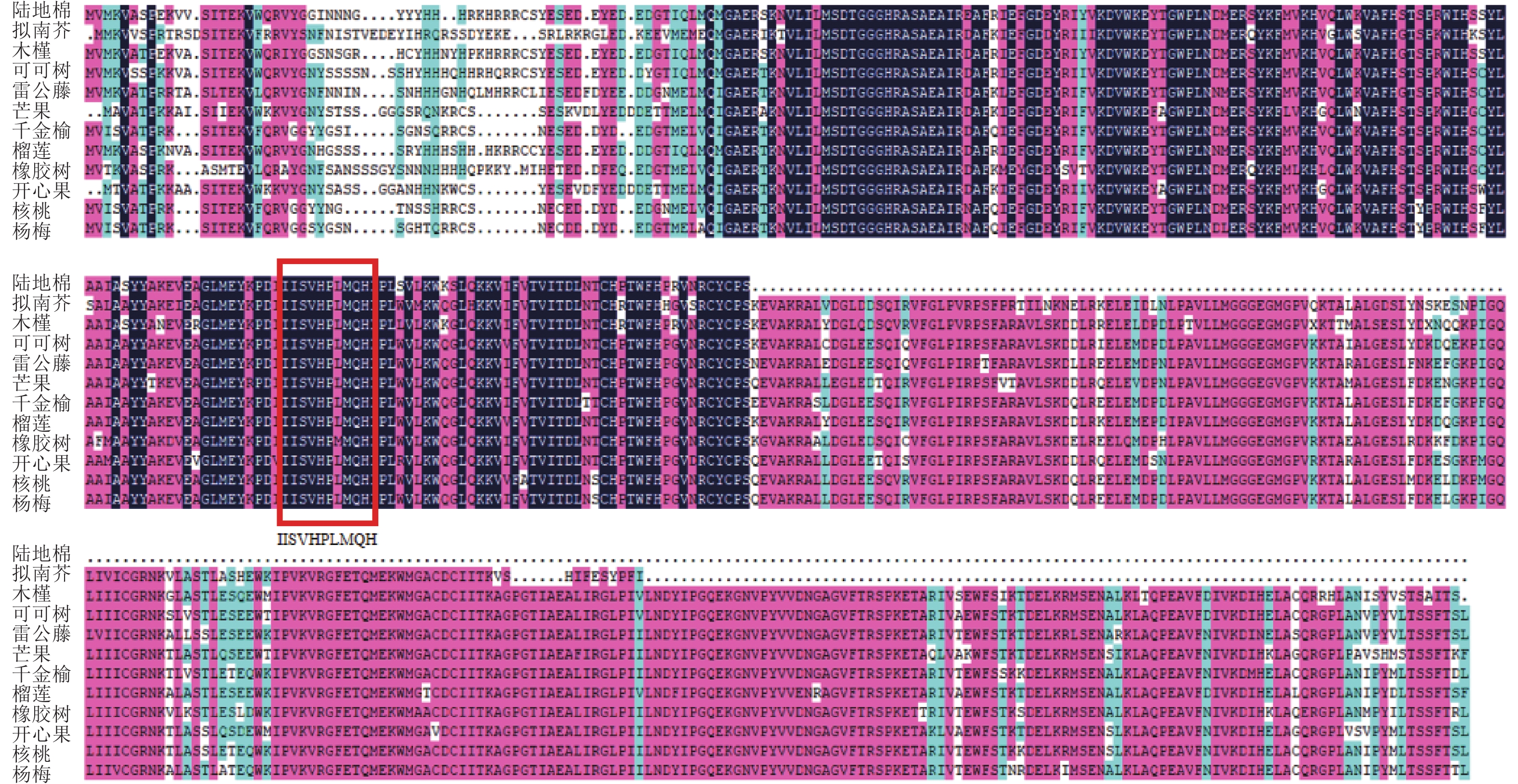
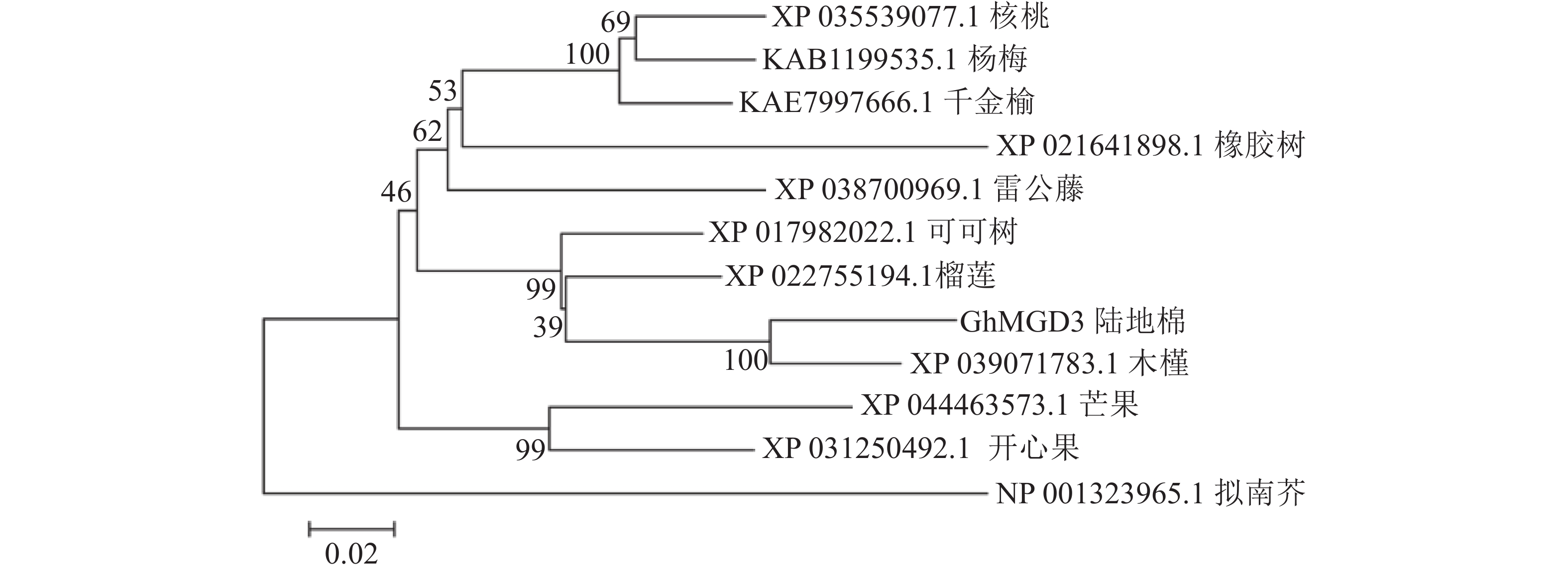
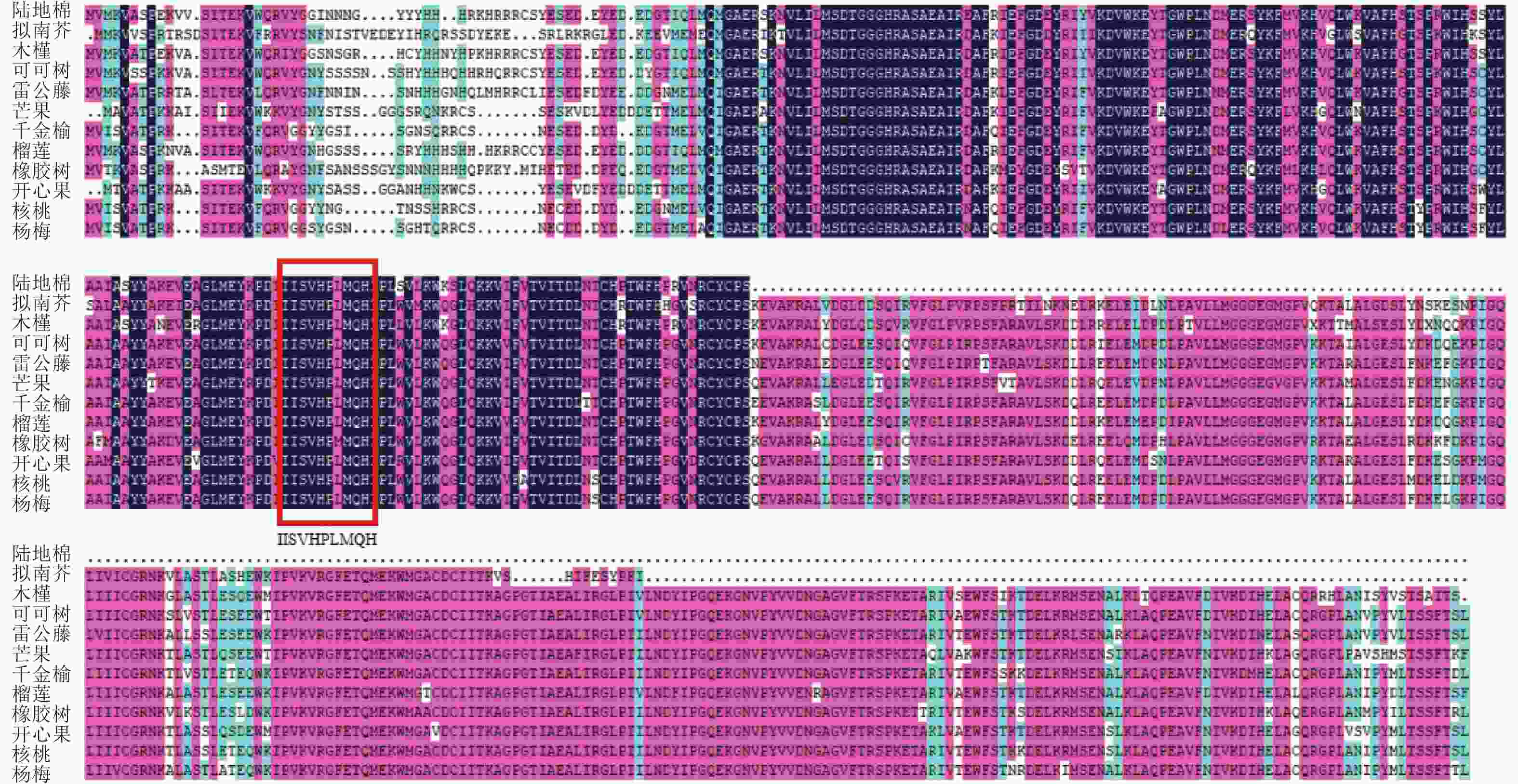
使用NCBI的保守结构域数据库(CDD-search)查找此氨基酸序列的保守结构域,结果显示:该基因所编码蛋白匹配所属的超家族是PLN02605,是一类包含多个结构域的模型。经与其他物种MGD家族同源蛋白进行多序列比对(图4),该基因存在1个MGD家族特有的保守结构域(图4红框所示),说明该基因属于陆地棉MGD家族成员。在NCBI数据库利用BLASTp检索氨基酸序列的同源序列,下载了11条不同物种的蛋白序列:雷公藤Tripterygium wilfordii、拟南芥、可可树Theobroma cacao、芒果Mangifera indica、千金榆Carpinus fangiana、木槿Hibiscus syriacus、榴莲Durio zibethinues、核桃Juglans regia、橡胶树Hevea brasiliensis、开心果Pistacia vera、杨梅Morella rubra。通过软件MEGA5构建系统进化树(图5),棉花蛋白序列与木槿亲缘关系最近,其次是榴莲、可可树,与其余物种的亲缘关系较远。
-
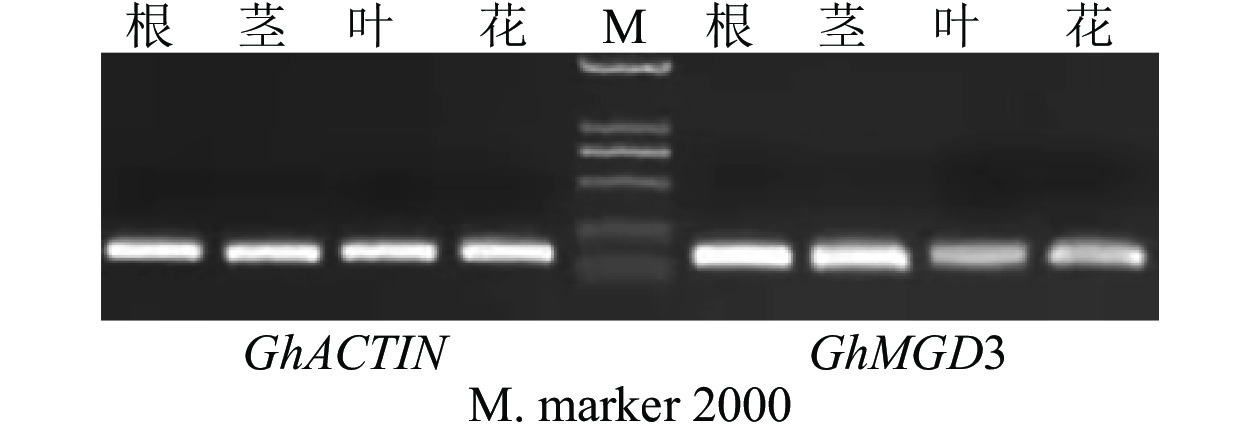
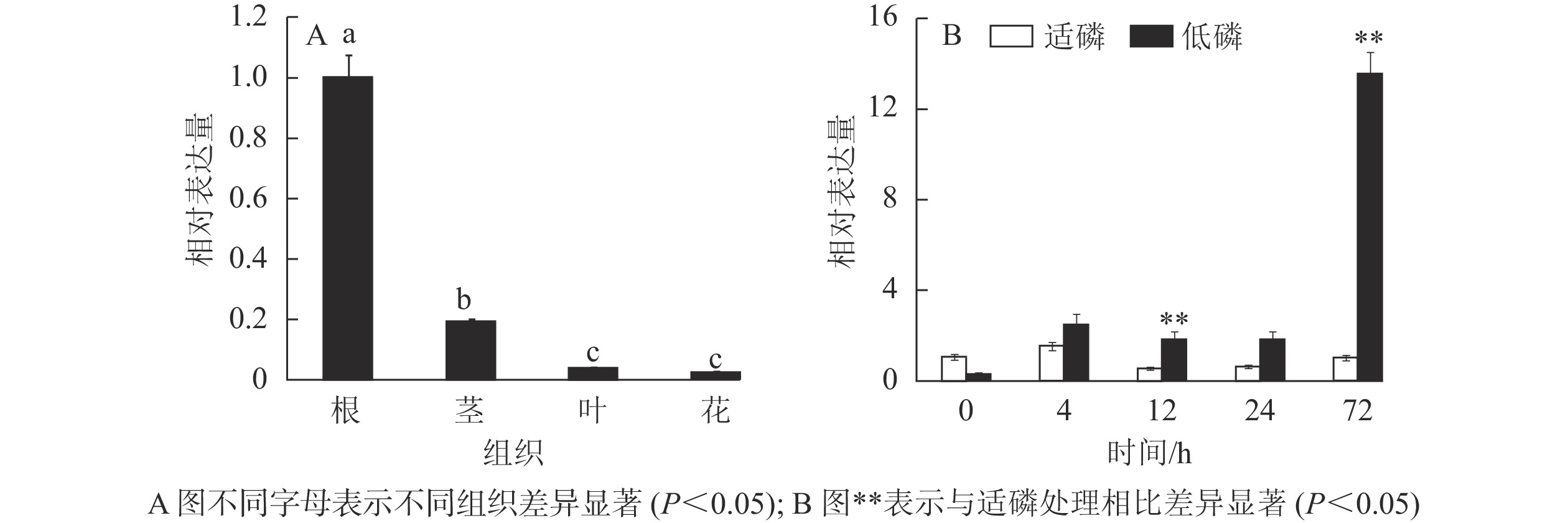
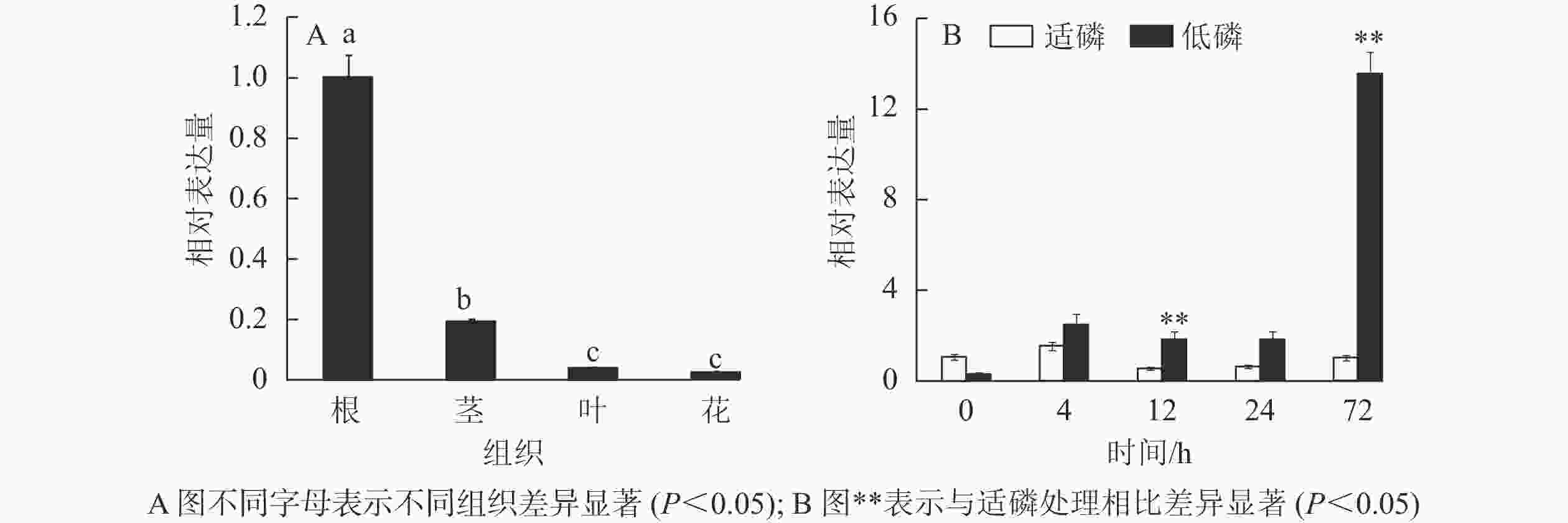
以棉花的GhACTIN为内参基因,通过半定量RT-PCR技术对GhMGD3基因在‘新陆早19’的4个组织中的表达情况进行分析。从图6可知:该基因高表达于根,微量表达于叶和花,中量表达于茎。利用qPCR检测GhMGD3基因在陆地棉‘新陆早19’植株的根、茎、叶、花等4个组织中的表达结果(图7A)同半定量RT-PCR一致。由图7B可以看出:在低磷胁迫下4~24 h,表达量逐渐下调,但是与适磷处理相比,其表达量都相对较高,且在低磷处理72 h时表达量达到最高,其中12和72 h时低磷处理比适磷处理显著表达(P<0.05),说明该基因在受到低磷胁迫的刺激后,能够对低磷胁迫作出应答反应。
-
大部分脂质由非磷且不带电荷的半乳糖甘油脂类单半乳糖基二酰基甘油(MGDG)和二半乳糖基二酰基甘油(DGDG)组成,它们占总脂质的80%。单半乳糖基二酰基甘油是质体的生物活动以及光合活性所需的主要和必需的植物脂质,这需要富含半乳糖脂的环境。MGDG也是二半乳糖基二酰基甘油(DGDG)的关键前体,它在磷酸盐胁迫下转移到质体膜外。因此,MGDG的合成对植物至关重要。大部分MGDG是由位于质体膜上的MGDG合酶合成的[21]。尽管植物组织中磷含量之间的平衡可能对植物生长很重要,但细节仍然未知。
拟南芥mgd3和mgd2mgd3突变体在磷胁迫下植物会出现严重的生长迟缓和DGDG含量降低,清楚地表明MGD3介导的MGDG合成在磷稀缺时对生存具有重要作用[22]。史中惠等[23]将OsMGD基因转入烟草Nicotiana tobacum植株得到了超表达OsMGD基因烟草植株,通过低磷胁迫下各生理生化指标变化情况的分析表明:该基因可以提高磷脂与半乳糖脂的含量,进一步增强植株耐低磷的特性。MGDG仅存在于质体中,但DGDG可以存在于非质体膜中,在特定条件下,例如磷胁迫下,它可以替代磷脂[24]。半乳糖脂被认为在种子萌发过程中原层体向类囊体膜的光依赖性转化以及响应光变化的高度堆叠的颗粒膜和光合机制的动态组织中发挥重要作用[25]。MGDG和DGDG还可以稳定叶绿体中的光系统蛋白复合物[26-27]。在磷胁迫的条件下,植物激活体内相关的机制,以应对细胞对环境条件变化的需求。
本研究所得基因序列与拟南芥中的MGD家族中的单半乳糖基二酰基甘油合酶同源性较高,从陆地棉‘新陆早19’中克隆得到了1个MGD3类似基因,并将其命名为GhMGD3,该基因属于MGD超家族成员。该基因存在3个内含子,GhMGD3蛋白相对分子质量为26 610.54 Da,等电点为8.74,是碱性蛋白,具备稳定性与亲水性。此蛋白的二级结构由α-螺旋和无规则卷曲占据主体地位所组成,通过同源建模可知该基因三级结构预测其结构与拟南芥半乳糖脂合酶MGD1结构相似。该基因编码蛋白不存在信号肽和跨膜结构域。该蛋白不存在潜在的N-糖基化位点,但含有多个酸化位点。亚细胞定位结果显示该基因编码蛋白在叶绿体中,预测其能够通过蛋白质的相互作用参与调控。基因表达分析结果表明:该基因在棉花根中的表达量是最大的,中量表达于茎,微量表达于叶和花,在低磷胁迫72 h时其相对表达量达到最高值。GhMGD3基因可能在根系发挥作用,在棉花的磷高效利用信号调控过程中具有重要的作用。
Cloning and expression of phosphorus efficient gene GhMGD3 in Gossypium hirsutum
doi: 10.11833/j.issn.2095-0756.20220145
- Received Date: 2022-01-25
- Accepted Date: 2022-08-28
- Rev Recd Date: 2022-07-14
- Available Online: 2022-11-21
- Publish Date: 2022-12-20
-
Key words:
- Gossypium hirsutum /
- low phosphorus stress /
- gene cloning /
- expression analysis
Abstract:
| Citation: | MENG Chaomin, GENG Feifei, QING Guixia, et al. Cloning and expression of phosphorus efficient gene GhMGD3 in Gossypium hirsutum[J]. Journal of Zhejiang A&F University, 2022, 39(6): 1203-1211. DOI: 10.11833/j.issn.2095-0756.20220145 |














 DownLoad:
DownLoad: