-
甘油脂包括甘油磷脂和甘油糖脂,是细胞膜及信号分子的重要组成部分,参与广泛的生理生化过程,在植物生长发育过程中发挥重要作用[1−5]。在高等植物中,甘油脂的合成涉及2条途径,即在质体外进行真核合成途径和质体中进行原核合成途径[6−7]。在原核合成途径中,由ATS1基因编码的3-磷酸甘油酰基转移酶(glycerol-3-phosphate acyltransferases,GPAT)催化甘油脂生物合成途径的第一步酰化反应,该反应被认为是关键的限速步骤[8−10]。
有关质体中ATS1基因的克隆与功能已有较多研究[11−13]。随着现代分子生物学的发展,人们已从南瓜Cucurbita moschata、红花Carthamus tinctorius、向日葵Helianthus annuus和油菜Brassica napus等植物中分离鉴定了多个与拟南芥Arabidopsis thaliana ATS1同源的基因[14−17]。这些ATS1基因表现出多种生理功能,在植物生长发育和抗逆性中发挥着重要作用。如YAN 等[18]研究发现:在烟草Nicotiana tabacum中异源表达甜椒Caspsicum frutescens质体ATS1基因可增强转基因烟草对高温胁迫的耐受性。KANG等[17]报道甘蓝型油菜BnATS1的过表达增加了细胞膜中多不饱和脂肪酸的积累,从而促进了甘蓝型油菜在低温条件下的生长。另有研究表明:ATS1在植物高盐和低磷等非生物胁迫中具有重要作用[19−20]。
然而,ATS1在植物正常生长发育中的功能并不完全清楚。KUNST等[21]利用EMS诱变创制了多个拟南芥ats1突变体,尽管这些突变体叶片的质体中脂肪酸组分发生了急剧变化,但是ATS1基因上的点突变并未对种子发育产生明显影响。相反,在高于28 ℃的温度条件下,突变体的生长速度比野生型略快。与上述ats1表型不一致的是,ATS1基因的T-DNA插入纯合突变体呈现败育现象, 并且发现运用RNAi干扰技术下调ATS1基因的表达会导致植株变小、胚胎发育受阻、种子结实率下降[9]。目前,尚不清楚造成这种不一致性的真正原因,但一种可能原因是,转基因植株中的T-DNA可能会干扰其插入位点或上下游基因的功能,从而对表型产生某种影响。
为了进一步明确ATS1在拟南芥正常生长发育中的功能,本研究利用现代基因编辑技术,采用优化后的CRISPR/Cas9基因编辑载体对ATS1基因进行定点编辑,创建功能丧失型突变体,并分析 ATS1基因功能的丧失对拟南芥生长发育的影响,有助于进一步了解高等植物中甘油脂原核合成途径在植物生长发育过程中的作用。
-
野生型拟南芥为哥伦比亚生态型(Col-0),购自美国索尔克生物研究所(Salk Institute for Biological Studies),编号为SALK_063776。
-
参照 WANG 等[22]和朱丽颖等[23]的方法进行CRISPR/Cas9靶序列的设计和目的基因载体的构建。运用CRISPR在线设计软件筛选目标基因的靶序列。并对选择的靶序列进行分析,最终从拟南芥ATS1基因中分别选取了GC含量较高、基因特异性较强的2个关键片段ATS1 target sequence 1 (5′- CGAAGAGTCGACGAAGCGAG-3′)和 ATS1 target sequence 2 (5′-TAGTCATTCCCGTACTTTCT-3′)作为靶序列。之后,以1 mg·L−1的pCBC-DT1T2 为模板进行四接头引物(5′-GGAAGAGTCGACGAAGCGA-3′,5′-AGAAAGTACGGGAATGACT-3′,5′-GGAAGAGTCGTCGACGAAGCGAG-3′和 5′-AGAAAGTACGGGAATGACTC-3′) PCR 扩增并纯化回收PCR产物。 同时用BsaI酶切回收的PCR产物和骨架载体pHEE401,经T4连接酶连接,获得具有2个靶序列的CRISPR/Cas9基因编辑载体。
-
所用植物材料为拟南芥Col-0,植物生长的昼夜温度为22 ℃/18 ℃,湿度为40%,光照/黑暗时长分别为14 h/10 h。参考李丹丹等[24]的方法使用农杆菌Agrobacterium tumefaciens转化法将基因编辑载体转化至拟南芥。
-
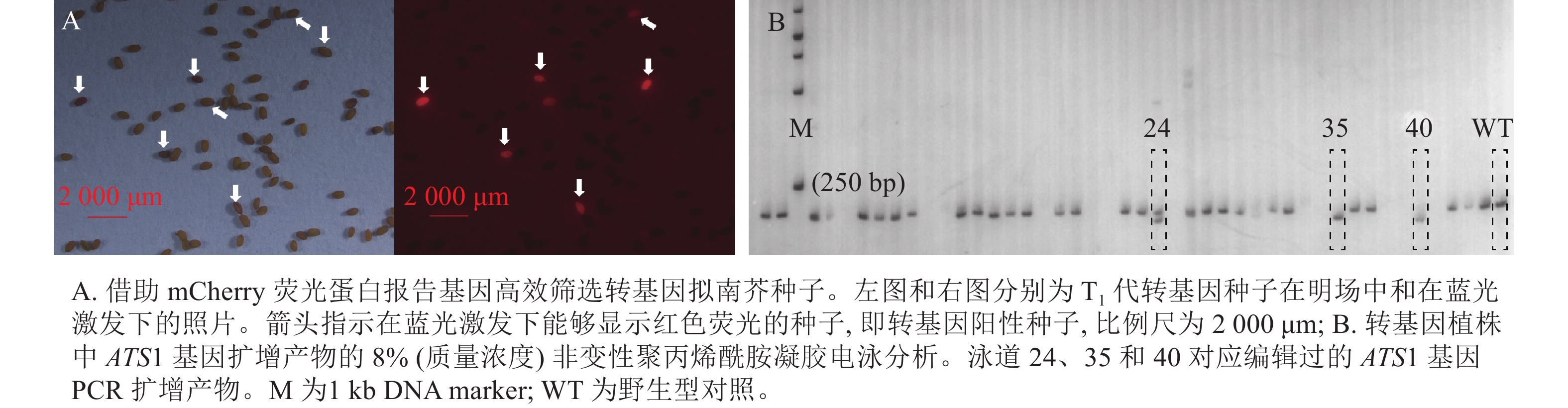
以种子专一表达的At2S3基因启动子驱动荧光蛋白报告基因 mCherry 的表达,将这一筛选标记克隆至CRISPR/Cas9编辑载体中[23]。由于mCherry荧光蛋白在蓝光激发下会发出红光,因此可将获得的成熟转基因拟南芥T1代种子置于荧光显微镜下,筛选蓝色激发光下发出红光的种子,即为转基因阳性种子。
种植筛选获得的T1代转基因阳性种子,30 d后,提取植株叶片的DNA,作为模板进行PCR扩增。根据拟南芥参考基因组,分别在靶序列上下游约100 bp设计PCR引物(ATS1-FP:5′-TCACCAAACACGCTTTAATGAC-3′和ATS1-RP:5′-AGACATGGCTCTCACACTAACG-3′)。将PCR产物经质量浓度为8%的非变性聚丙烯酰胺凝胶电泳(PAGE),筛选出与对照电泳条带不同的株系,即为发生了基因编辑的株系。将这些株系的PCR产物进行测序验证,并收获T2代种子。
每个株系挑选16粒不含红色荧光的T2代种子进行种植。1个月后提取叶片基因组DNA进行PCR扩增。综合PCR产物的PAGE和测序结果,挑选靶序列发生纯合突变的植株,即获得了不含转基因的ATS1基因突变株系。将这些株系重新编号,并收获T3代种子,进行扩繁,用于后续实验。
-
脂质提取步骤参考徐雪珍等[25]的方法。取播种4周的拟南芥叶片至研钵中,加入液氮充分研磨成粉末,称取100 mg样本转入12 mL离心管。经过6 mL氯仿-甲醇-甲酸溶液(体积比为10∶10∶1)和 2 mL氯仿-甲醇-水溶液(体积比为5∶5∶1)提取液的2次抽提,并合并2次上清液,加入3 mL含0.2 mol·L−1磷酸和1.0 mol·L−1氯化钾的混合溶液,提取下层氯仿相。 萃取液用氮气吹干,加入200 μL氯仿溶解萃取物,再加入2 mL 体积分数为1%硫酸-甲醇溶液,80 ℃加热2 h,对油脂的脂键进行充分的水解。之后置于冰上,加入2 mL 正己烷及1 mL质量浓度为 0.9%的生理盐水,对脂肪酸甲酯进行萃取,取上层相转至新的12 mL离心管中,萃取2次,合并萃取液,萃取液通过氮吹法浓缩至100 μL。最后,利用气相色谱仪分析叶片脂肪酸组分。每个株系设置3 个生物学重复。
-
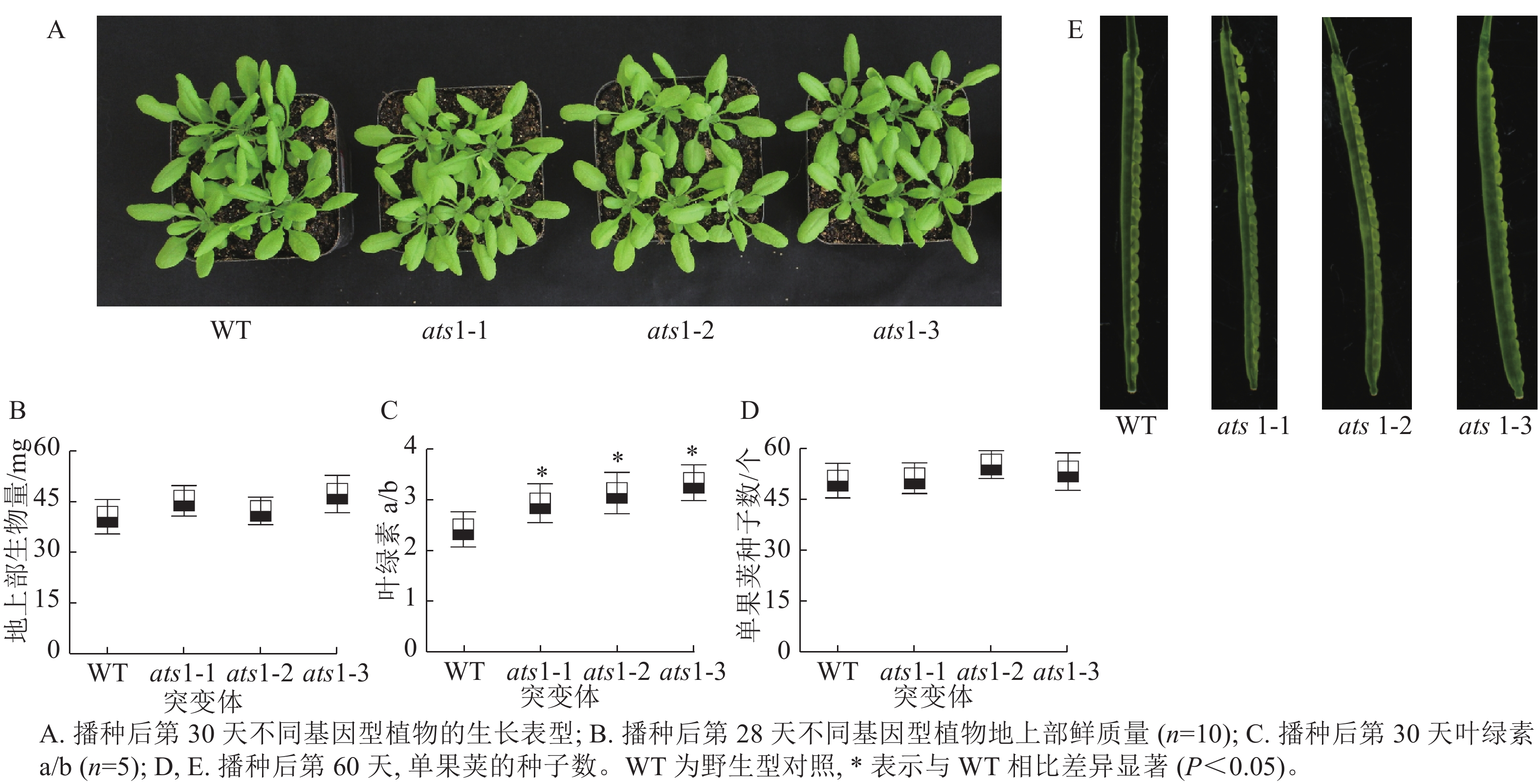
取播种后30 d的植株整个地上部,放入12 mL玻璃管中,加入3 mL 体积分数为80%的丙酮溶液, 4 ℃下避光保存 14 h后,测定叶绿素。 每个株系设置5个生物学重复。
-
选取播种后28 d的拟南芥植株整个地上部分,称取鲜质量。 每个株系设置 10 个生物学重复。
-
选取播种后60 d的植株果荚,测量每个果荚的种子数量,并通过体视显微镜进行拍照。 每个株系设置5 个生物学重复。
-
数据以平均值±标准差表示,并通过GraphPad Prism 6 软件进行统计分析。通过t检验或单因素方差分析进行组间差异比较,显著性水平为0.05。
-
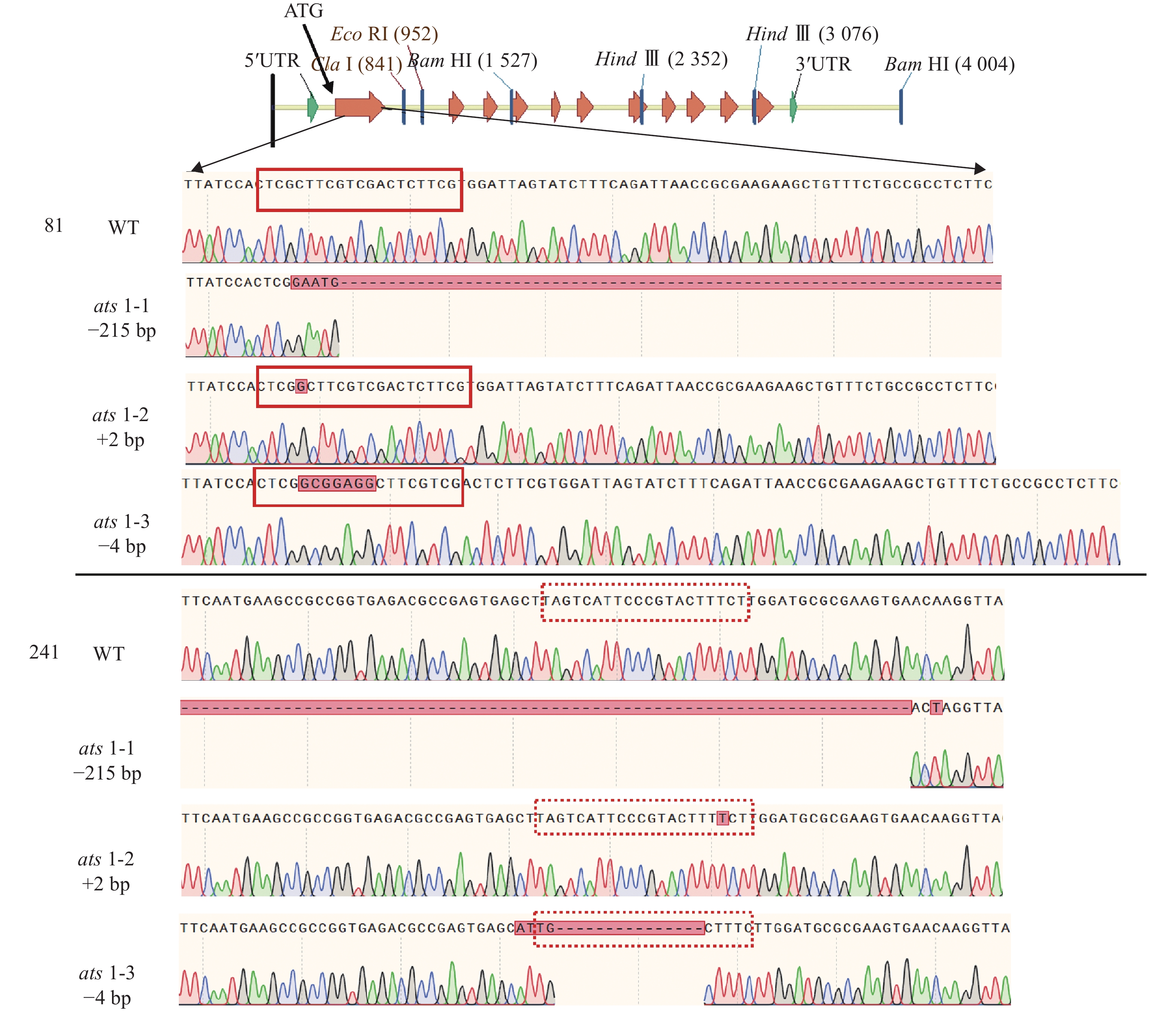
运用农杆菌介导法将含ATS1靶序列的CRISPR/Cas9基因编辑载体(含mCherry报告基因)转到拟南芥中,并筛选带荧光的T1代转基因种子(图1A)。随后,采用聚丙烯酰胺凝胶电泳法鉴定转基因阳性植株中 ATS1 基因编辑产物的PCR扩增片段特性(图1B)。经连续多代筛选,从不同转基因株系的后代分离群体中获得 3个纯合且稳定遗传的突变体,分别命名为ats1-1、ats1-2、ats1-3。同时,对这些突变体的自交后代进行连续多代的PCR检测与荧光观察,获得不含任何外源T-DNA插入片段的突变体,这些突变体中既不含Cas9基因,也不带荧光蛋白报告基因(mCherry)。
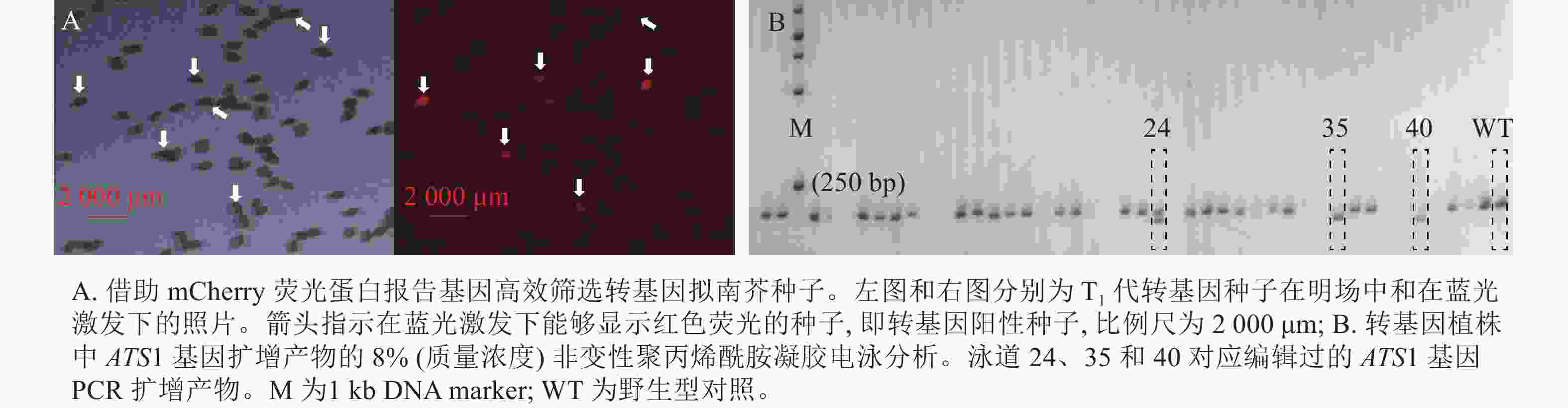
Figure 1. Screening of transgenic plants carrying the mCherry fluorescent protein and those with CRISPR/Cas9-edited ATS1 gene product
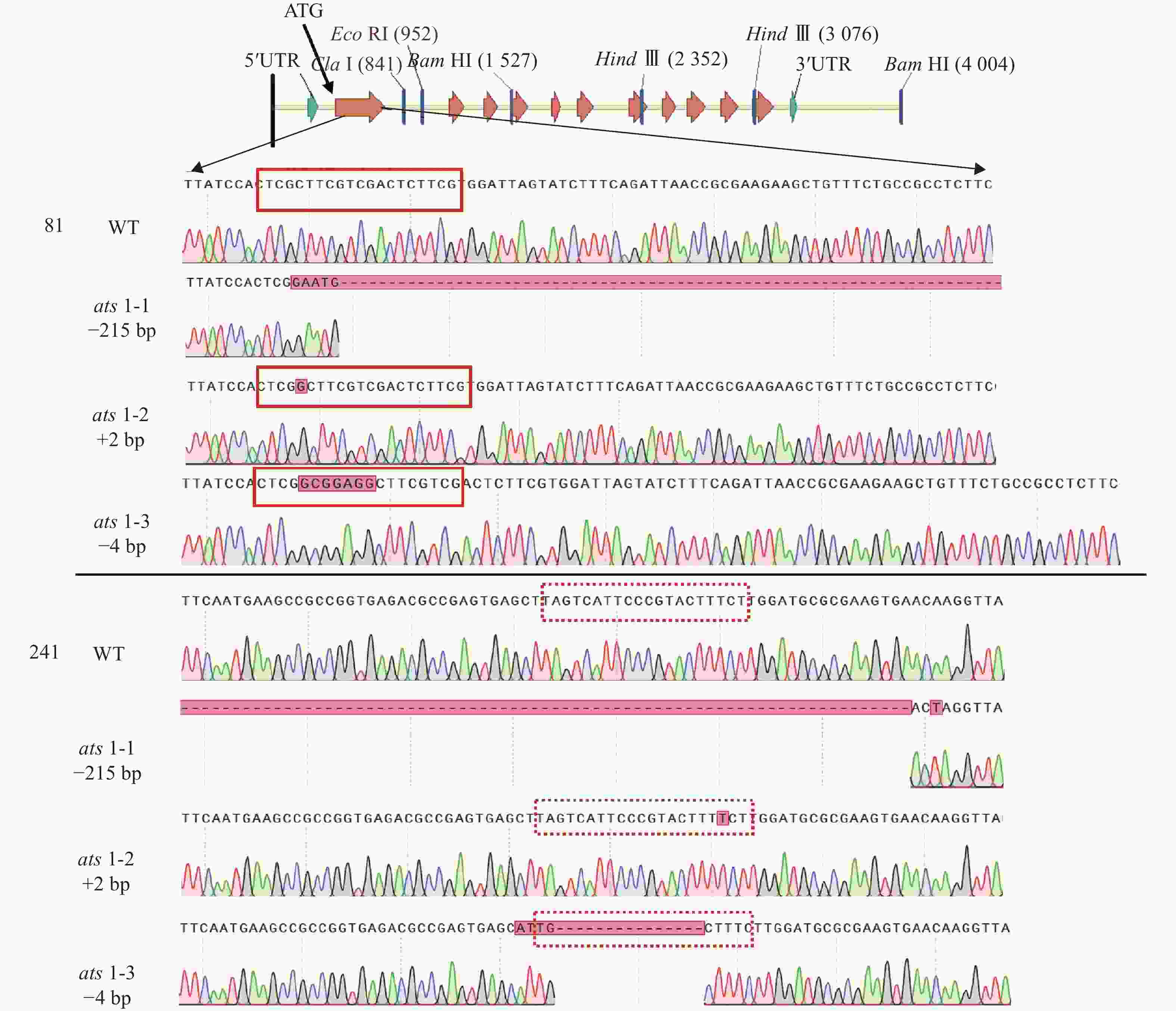
进而,对这些突变体的靶位点附近序列进行测序分析,结果显示这些突变体的突变位点均位于第 1 个外显子上(图2)。在ats1-1突变体中,ATS1基因的92~314 bp (相对起始密码子ATG的位置)处发生215 bp 碱基缺失和8 bp 碱基替换。在ats1-2中,ATS1基因在2个位置发生1 bp 碱基插入,分别位于91 和293 bp 处。在ats1-3中,ATS1基因的91~92 bp之间存在7 bp 碱基插入,而在275~289 bp 间发生11 bp 碱基缺失和4 bp 碱基替换(表1)。上述这些突变大多位于 PAM 序列(NGG)的切割位点附近,其特点是ATS1基因的第1个外显子的碱基数呈非3的倍数的插入或缺失,从而导致移码突变或翻译提前终止,且由之产生的蛋白不含酰基转移酶保守结构域。这些结果表明,上述3个ATS1基因突变体均属功能丧失型突变体。这些突变体可成为ATS1基因功能研究的理想遗传材料。
突变体 突变位点 ats1-1 92~314 bp:215 bp缺失;8 bp替换 ats1-2 91~92 bp:插入1 bp;293~294 bp:插入1 bp ats1-3 91~92 bp:7 bp插入;275~289 bp:11 bp缺失,
4 bp替换Table 1. Designation of different ats1 mutants and the sequences of corresponding mutational sites
-
ATS1是甘油脂原核合成途径中参与第一步酰化反应的关键酶,过去的研究表明,ATS1基因突变会改变膜脂组分及脂肪酸组分,特别是质体中的C16:3含量急剧下降[6]。对生长 4周的拟南芥植株叶片进行脂肪酸组分分析显示:与野生型相比,3个突变体(ats1-1、ats1-2和ats1-3)中不饱和脂肪酸C16:3的含量急剧下降,而不饱和脂肪酸C18:3的含量显著增加(表2),这与过去基于EMS诱变产生的ATS1突变体的脂肪酸组分变化完全一致[6]。因为质体外的甘油脂不含C16:3,其通常存在于质体中的单半乳糖基二酰基甘油 (monogalactosyldiacylglycerol,MGDG)骨架的sn-2位置[6, 21],因此,ats1-1、ats1-2和ats1-3中C16:3的大幅降低,印证了这些突变体中参与甘油脂原核合成途径中第一步酰化反应的ATS1基因的功能丧失。
脂肪酸 脂肪酸组分含量/% C16:0 C16:1 C16:3 C18:0 C18:1 C18:2 C18:3 WT 14.91±0.73 a 7.35±0.53 a 11.56±0.38 a 6.17±1.55 a 4.37±0.59 b 14.89±1.30 b 38.50±3.04 b ats1-1 11.91±0.65 b 5.55±0.69 b 0.70±0.15 b 3.89±0.87 a 8.65±0.75 a 18.61±0.54 a 49.14±2.24 a ats1-2 11.20±0.18 b 5.93±0.89 ab 0.65±0.15 b 4.67±0.32 a 8.89±1.06 a 18.67±0.98 a 48.31±1.68 a ats1-3 12.29±0.81 b 6.00±0.93 ab 0.57±0.18 b 6.02±1.62 a 9.08±1.02 a 18.28±0.88 a 46.04±1.45 a 说明:WT为野生型对照,n=3,不同小写字母表示不同株系间显著差异(P<0.05)。 Table 2. Leaf fatty acid composition of ats1 mutants and wild-type A. thaliana
-
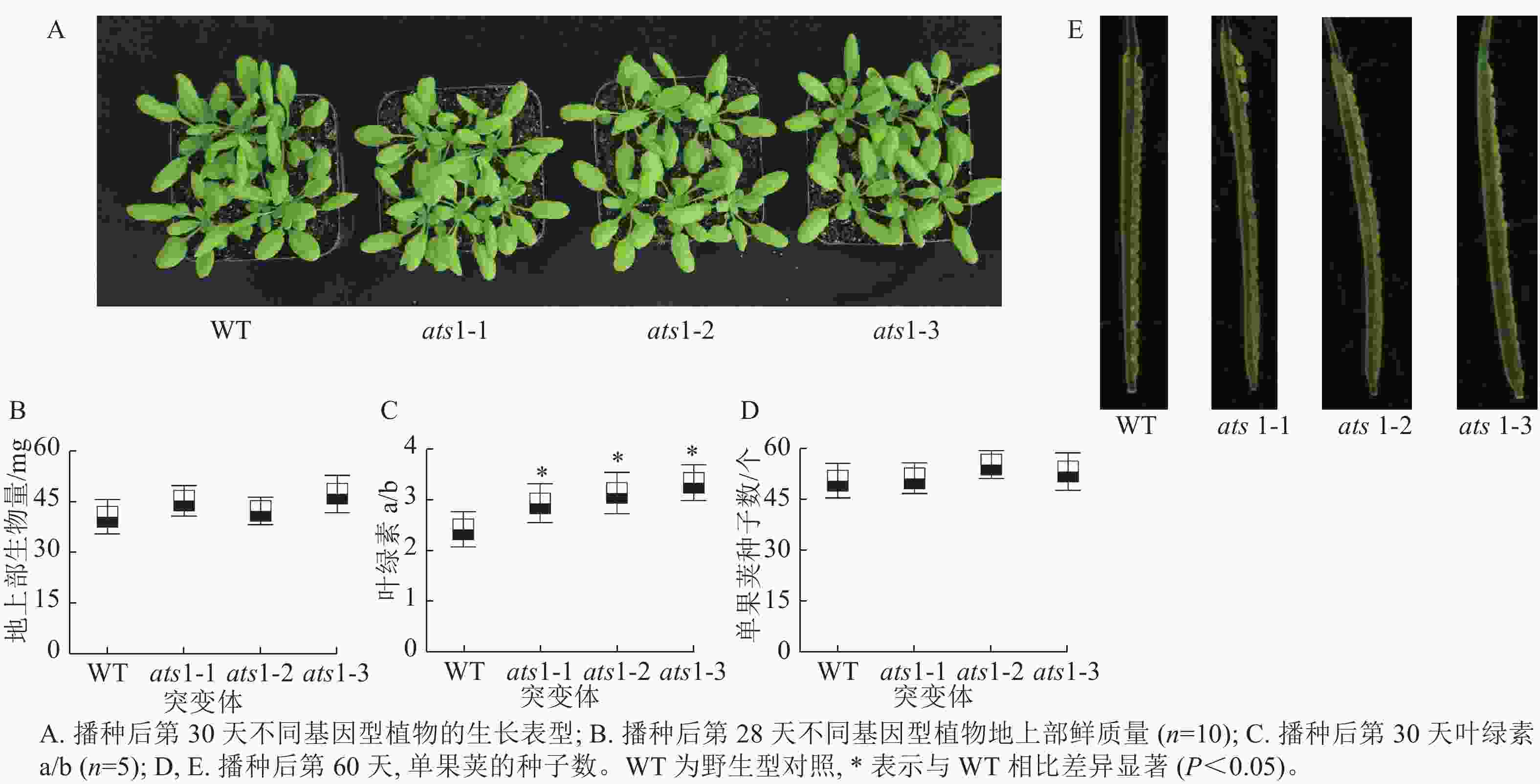
如图3 A所示:在营养生长期,与野生型相比,突变体(ats1-1、ats1-2和ats1-3)有时会出现叶片略微变黄的现象,但植株叶片发育与野生型相比无明显差异。对植株地上部生物量检测结果显示:与野生型相比,突变体植株地上部生物量无显著差异(图3 B)。对植株叶片叶绿素检测结果显示:与野生型相比,突变体植株叶绿素a/b约上升29.5%(图3 C)。拟南芥果荚生长分析显示:与野生型一样,突变体株系的种子发育正常,无败育现象出现(图3 D和E),这一结果不支持XU等[9]的研究结果。本研究结果表明在正常生长条件下ATS1 基因的功能丧失对拟南芥种子发育并不产生可见影响。
-
之前,研究者利用EMS诱变获得的ats1突变体和T-DNA插入突变体,对ATS1基因的功能进行了大量研究,然而基于不同突变体的研究得出的结论不一致[6, 9]。这可能存在2个原因,一是,EMS诱变产生的点突变可能不会使基因产物完全丧失活性,因而在某些特定条件下,突变体的表型变得不明显;二是,T-DNA插入虽然可以导致目标基因的功能完全丧失[9],但T-DNA插入可能会干扰插入位点附近基因的表达,从而对突变体的表型产生额外的影响。为了排除上述因素对ATS1基因功能研究产生的干扰,本研究运用现代基因编辑技术创制了不含外源T-DNA插入片段的ATS1功能丧失型突变体。
对其中的3个突变体(ats1-1、ats1-2和ats1-3)进行了分子与生化鉴定,发现这些突变体在ATS1第1个外显子上发生了插入、缺失、替换等几种不同类型的突变,这些突变导致非3的倍数的碱基插入或缺失,使阅读框发生移码及翻译提前终止,最终使得ATS1基因丧失功能。与此一致,脂肪酸组分分析显示:所有突变体的叶片中不饱和脂肪酸C16:3 (来源于叶绿体中的甘油糖脂)的含量大幅降低,而C18:3的含量显著升高。这一结果与基于EMS诱变产生的ats1突变体的分析结果相吻合[6]。总之,分子与生化鉴定的结果表明本研究获得的突变体为ATS1功能丧失型突变体。
-
目前,对ATS1基因在植物生长发育中的作用存在某些争议。由EMS诱变产生的ats1突变体呈正常的种子发育过程[21],而当用RNAi干扰技术下调ATS1基因的表达,拟南芥的种子发育异常,结实率下降[9]。为了完善人们对ATS1基因功能的认知,本研究利用不含外源DNA插入片段的多个ATS1功能丧失型突变体分析其在正常生长发育过程中的作用。表型分析显示:在正常生长条件下,这些突变体植株生长良好,除了其叶片有时会略显黄色,种子生长发育正常、无败育现象,这一表型与源于EMS诱变的ats1突变体分析结果一致[21],因此有充足理由推断拟南芥ATS1并非种子发育所必需的。
ATS1对种子发育的非必需性,需要重新评估甘油脂原核合成途径对植物正常生长发育的贡献,并调查植物细胞的质体中是否存在其他酰基转移酶参与甘油脂合成的第1步酰化反应。另外,期望本研究获得的功能丧失型突变体,能够更好地剖析植物细胞中真核合成途径与原核合成途径产生的不同甘油脂分子之间的交换机制。
Loss-of-function mutations in ATS1 reveal its dispensable role in normal seed development of Arabidopsis thaliana
doi: 10.11833/j.issn.2095-0756.20220738
- Received Date: 2022-11-29
- Accepted Date: 2023-03-06
- Rev Recd Date: 2023-03-01
- Available Online: 2023-07-13
- Publish Date: 2023-08-20
-
Key words:
- Arabidopsis thaliana /
- ATS1 /
- gene editing /
- ats1 mutant
Abstract:
| Citation: | KE Xingxing, LIU Yakun, XU Xuezhen, et al. Loss-of-function mutations in ATS1 reveal its dispensable role in normal seed development of Arabidopsis thaliana[J]. Journal of Zhejiang A&F University, 2023, 40(4): 707-713. DOI: 10.11833/j.issn.2095-0756.20220738 |









 DownLoad:
DownLoad: