-
竹子作为一种特殊的森林资源,可为人类提供大量非木材产品,又因其速生特性,已成为替代塑料的首选材料[1−4]。毛竹Phyllostachys edulis是中国最具特色的笋材两用竹种,具有极强的生长能力和更新能力,以及较高的经济、生态和文化价值[5−7]。与多数双子叶植物不同,毛竹茎中存在居间分生组织[8]。居间分生组织和顶端分生组织一样都拥有很强的分裂能力,毛竹依赖于细胞的快速分裂分化能力,快速拔节长高[9−10]。
WUSCHEL-related homeobox (WOX)基因家族在植物形态建成的全过程中具有重要调控作用,参与植物根系顶端分生组织、维管发育、胚胎发生与发育以及不定器官的发生与发育,同时还参与植物对逆境(如干旱、盐和低温等)的响应[11−12]。WOX4基因是该基因家族中参与植物生长发育调控的关键基因之一,在拟南芥Arabidopsis thaliana和杨树Populus等双子叶植物中已有了广泛的研究,而在单子叶植物中的研究相对较少。AtWOX4在维持拟南芥形成层细胞中起着重要作用,AtWOX4突变体植株矮小,维管组织中积累未分化的细胞,并且木质部、韧皮部和纤维细胞的分化减少[13]。在多种杨树物种中,WOX4a/b RNAi和双突变体均表现出次生生长显著减弱,维管形成层明显减少,毛白杨Populus tomentosa中过表达PtoWOX4a的植株也表现出髓部发育受抑制,但木质部宽度增加[14]。这一结果表明拟南芥和杨树之间存在保守的CLE/TDIF-TDR/PXY-WOX4调控途径。研究发现:山新杨Populus davidiana × P. bolleana中WOX4基因的过表达降低了活性氧(ROS)的清除能力,加剧了盐胁迫诱导的细胞损伤和死亡,并通过抑制PdbDREB2C的表达负调控山新杨的耐盐性[15]。棉花Gossypium hirsutum中的GhWOX4_A01在胚性愈伤组织中的表达水平高于普通愈伤组织,而沉默植株(pCLCrVA: GhWOX4_A01)积累了更多的活性氧,且对干旱处理更为敏感,表明GhWOX4_A01参与调控组织再生和非生物胁迫[16]。OsWOX4是水稻Oryza sativa中WOX4的同源基因,其不仅调控维管系统和叶片发育,还参与了顶端分生组织的维持,通过超表达水稻FCP1抑制OsWOX4的表达,会导致水稻分生组织发育缺陷、叶片生长畸形和维管系统发育紊乱,形成异常的顶端分生组织(SAM)[17−18]。毛竹WOX4家族同源基因WOX4s (PheWOX4a、PheWOX4b、PheWOX4c)均在毛竹笋期表达活跃,其中PheWOX4c在不同高度笋的顶端组织中,例如鞭梢、冬笋和不同发育阶段幼竹的顶端组织中表达更加明显,推测PheWOX4c的功能可能与水稻OsWOX4相近,都参与顶端分生组织活性的维护[19]。还有研究发现PheIAA15与PheWOX4c相互作用,可能通过拮抗机制调控PheCLE25的表达,区别于其他植物的生长素响应模式,共同调控维管组织的分化[20]。
启动子(promoter)作为基因表达的启动引擎,主要位于DNA序列的5′上游,这一特殊位置的DNA序列被称为启动子区域。它能够与RNA聚合酶结合,激活或抑制后续的转录过程,通常包含多个关键的顺式作用元件,在基因表达中发挥着关键的调节功能[21]。已有研究发现在竹亚科 Bambusoideae中WOX启动子的顺式作用元件分布特征十分类似,且在不同竹类WOX的直系同源基因的同一启动子区域存在特殊顺式元件,可能在竹子的生长发育过程中发挥重要调控作用[19]。然而毛竹PheWOX4c基因启动子区域存在的调控元件以及不同环境因子对该基因的影响,仍有待进一步研究。
基于此,本研究拟克隆PheWOX4c上游启动子序列,分析启动子上的顺式作用元件,分析PheWOX4c启动子全长和5′端缺失启动子的活性,探究各顺式作用元件对不同处理的应答模式,以期阐明其生物学功能和表达调控机制。
-
选取干燥新鲜的毛竹种子,冲洗干净在水中浸泡2~3 d后,均匀铺撒在育苗托盘中,在培养箱中遮光培养1~2周后,将发芽生根的毛竹种子栽至土培盆中,日常光照培养至三叶一心,提取毛竹总DNA。
将种植培养土加水吸收至湿润松软的状态,将本氏烟草Nicotiana tabacum种子均匀少量撒播在土壤表面,使用聚乙烯膜覆盖花盆顶部并用皮筋固定,膜上扎出少许透气孔洞。处理好的烟草放置在透气温暖湿润的环境中,待烟草萌发去除覆膜。2~3周后,将长出2~3片叶的烟草单株移栽,在温度25 ℃,光照强度10 klx,光照时间16 h,黑暗时间8 h的培养箱中培养,用于侵染实验。
-
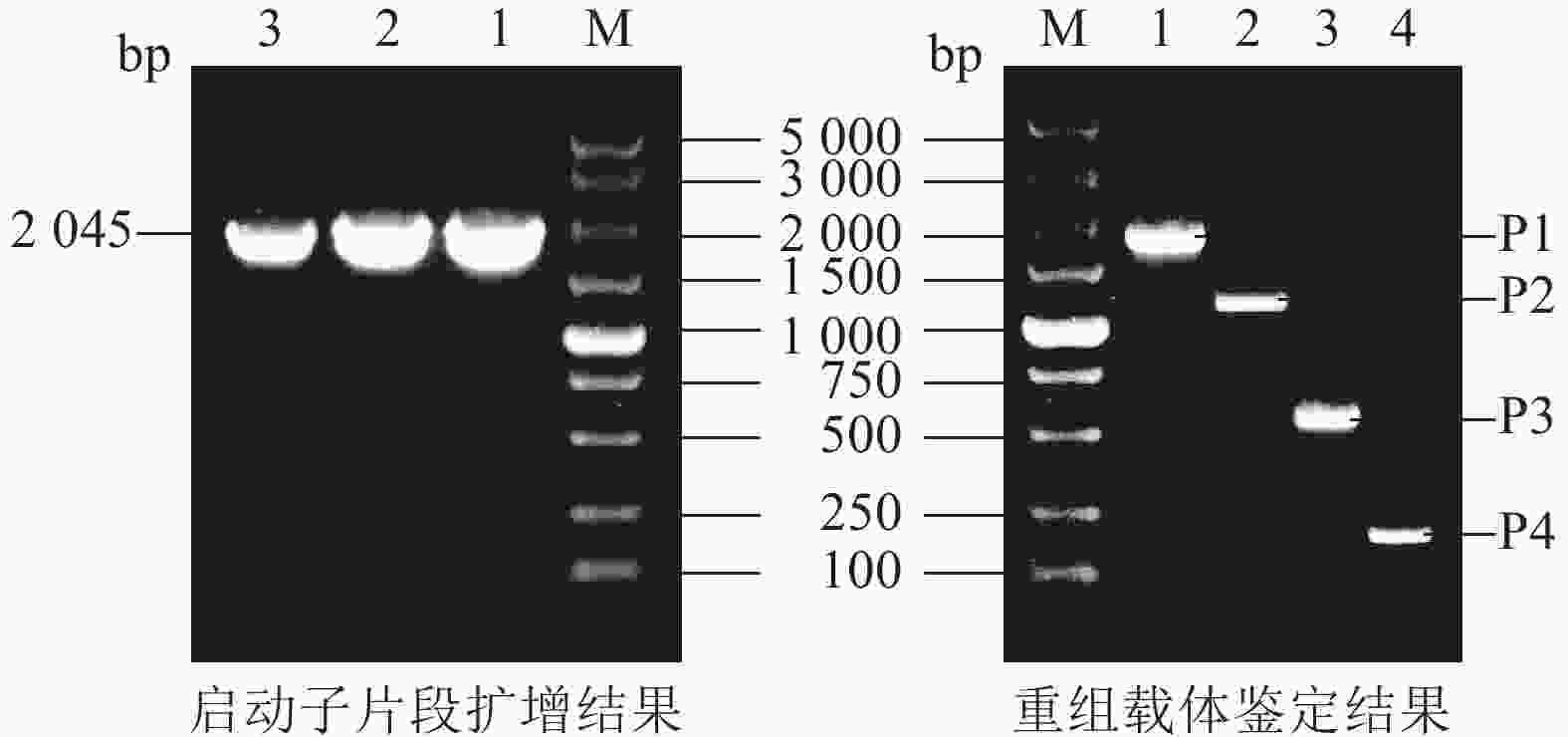
在毛竹全基因组数据库中,检索得到PheWOX4c基因转录起始位点(A)上游2 045 bp的启动子序列。根据植物DNA提取试剂盒说明书,将毛竹叶片放入液氮中研磨,提取毛竹总DNA,以此为模板设计引物进行扩增。选择Hind Ⅲ和XbaⅠ作为PBI121-GUS载体构建的双酶切位点,通过胶回收纯化扩增片段和酶切过的线性化载体,再使用同源重组试剂盒获得重组质粒后转化大肠埃希菌Escherichia coli,选择菌检结果阳性的单菌落进行测序并与已知序列比对。
-
根据Plant CARE在线网站提供的预测结果,通过生物信息学方法分析克隆得到的PheWOX4c基因启动子序列,并预测了其可能包含的顺式作用元件。
-
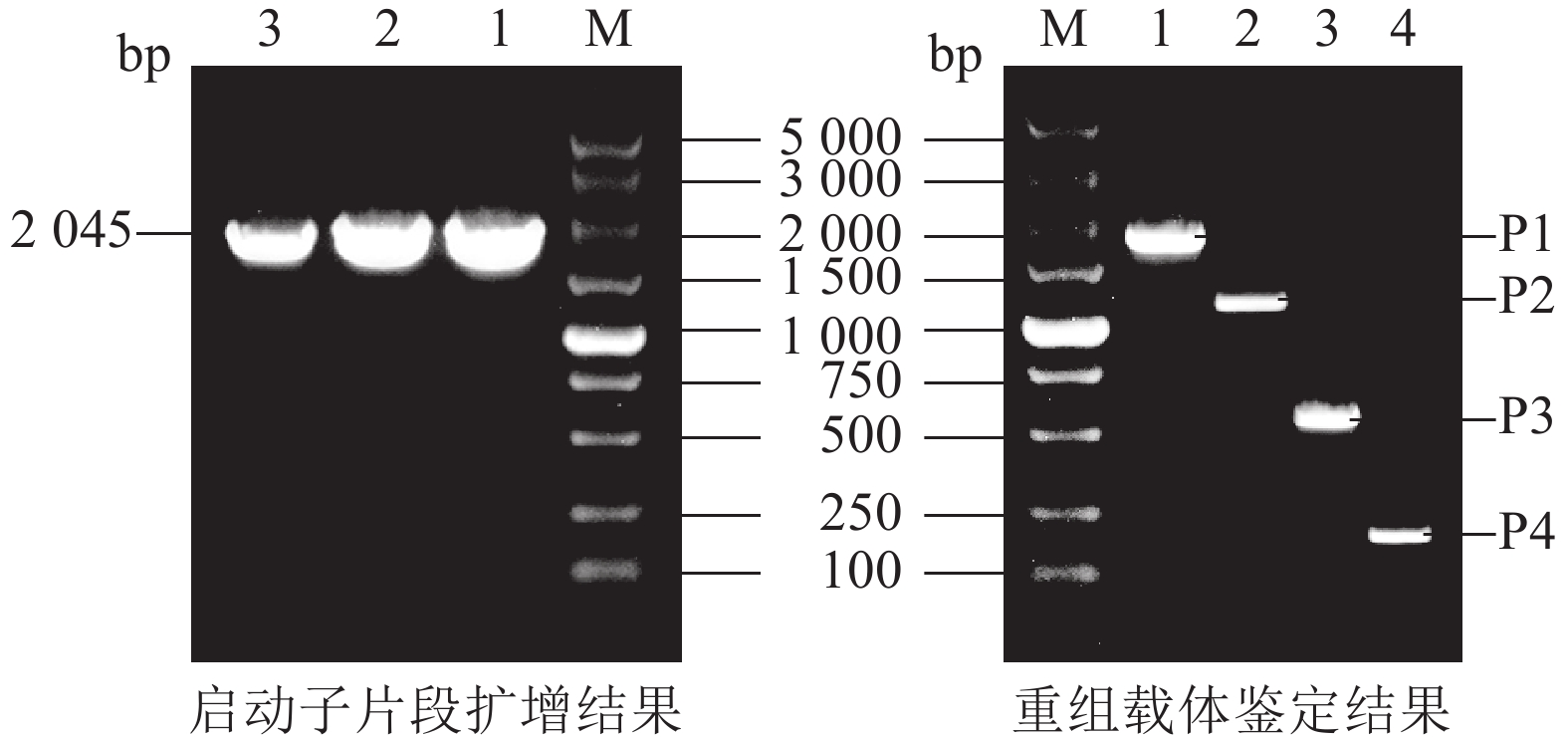
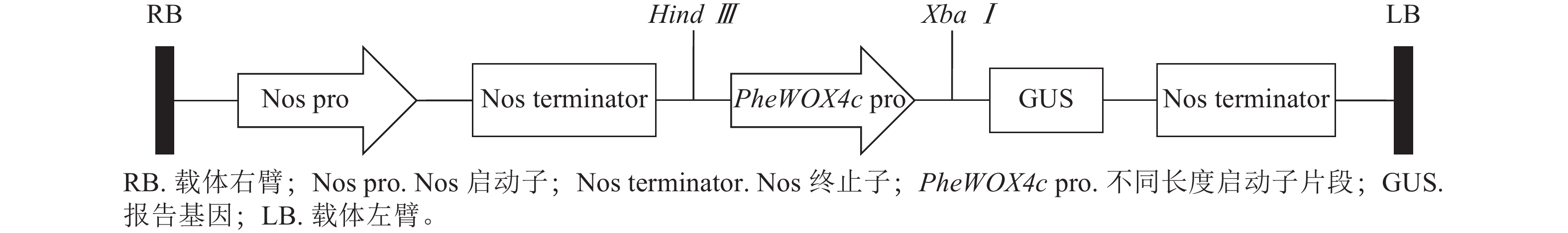
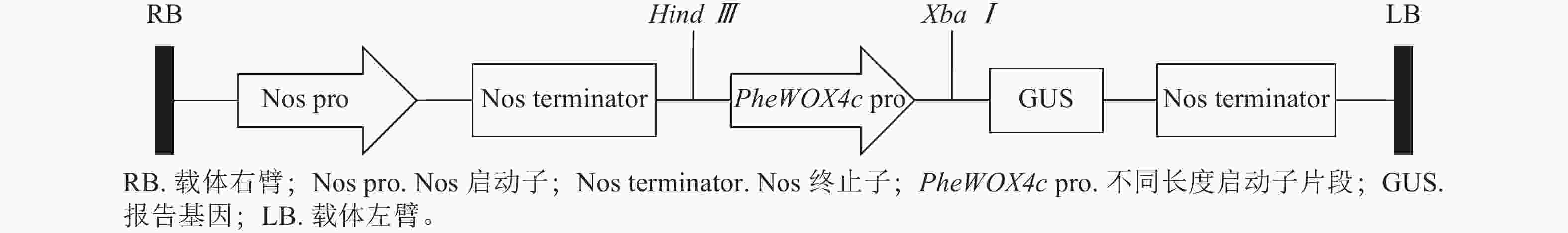
根据PheWOX4c基因2 045 bp启动子序列上顺式作用元件预测的功能及定位,设计不同长度5′端缺失克隆引物(表1),选择植物表达载体PBI121,报告基因为GUS,构建全长及各不同长度启动子片段的5个植物表达载体(图1)。对PBI121载体上Hind Ⅲ和XbaⅠ等2个酶切位点进行酶切后胶回收线性载体,与分离并回收得到的不同长度启动子片段进行同源重组,将重组质粒转化大肠埃希菌Escherichia coli,对菌检阳性的菌株进行测序,获得与目标序列比对一致的 P1~P4重组表达载体:PBI121-P1∷GUS、PBI121-P2∷GUS、PBI121-P3∷GUS、PBI121-P4∷GUS。
引物名称 引物序列(5′→3′) 引物功能 Promoter-F GCTTGTAGTTTAATTCAAAGTTTGTATGG 启动子全长克隆 P1-F CCGGGAGGCCTCTTTTAGC 缺失片段1克隆 P2-F TGCTTTGCATCTGGGTCCAT 缺失片段2克隆 P3-F GCGCCAATTTTCAGAGCACA 缺失片段3克隆 P4-F TCAAAACCCGGCCTCACCA 缺失片段4克隆 Promoter-R CGGCAGAGCTAGCAAGCACT 启动子全长及缺失片段克隆 Table 1. Primers used in this experiment
-
将构建成功的PheWOX4c启动子全长载体及P1~P4重组表达载体转化农杆菌Agrobacterium GV3101,28 ℃振荡培养至菌液初步混浊后,每30 min测量1次吸光度D(600),D(600)为0.5时进行离心,用配置好的烟草侵染液重悬收集到的菌体,重悬液D(600)保持不变。在避光环境中,室温静置至少3 h。选择生长出第4或5轮叶片的本氏烟草进行注射,每处理4个重复,其中阳性对照为PBI121-GUS空载体,阴性对照为去除GUS基因前CaMV35S启动子的PBI121表达载体,分析PheWOX4c基因启动子在烟草中驱动基因表达的特征。
-
注射农杆菌72 h后采集各处理烟草叶片,使用圆形打孔器(直径为1 cm)打孔,染色液的配置参考GUS染色试剂盒说明书,将各叶盘放置于10 mL离心管中,加入没过样品的GUS染色液,真空抽滤30 min后,37 ℃摇床振荡培养12 h。将染色反应后的叶盘取出,去除多余染色液,使用体积分数为75%的乙醇进行脱色,完全脱色后用清水润洗,置于体视镜下观察并拍照记录。
-
配置浓度均为100 μmol·L−1的生长素(IAA)、脱落酸(ABA)、茉莉酸甲酯(MeJA)和水杨酸(SA),分别均匀喷洒在注射农杆菌48 h后的烟草叶片上,置于室温;另选择一组烟草喷洒蒸馏水,置于4 ℃进行低温处理;以室温喷洒蒸馏水的烟草作为对照组。每处理4个重复,采集处理24 h后的烟草叶片并分组标记。使用Bradford法[21]蛋白提取试剂盒提取烟草叶片总蛋白,测量各样品595 nm波长处的吸光度,根据标准样品数值绘制的牛血清蛋白(BSA)标准曲线,计算各启动子片段在不同处理下烟草中瞬时表达的总蛋白浓度。根据GUS基因定量检测试剂盒所述方法,将酶促反应终止液提前置于37 ℃温浴,取150 μL蛋白提取物,加入150 μL 4-MUG底物液,37 ℃温浴反应,分别在10、20、30 min时,从反应液中取100 μL加入900 μL终止液中,避光放至测量结束。使用激发波长365 nm、发射波长455 nm测量各样品,3次荧光值测定后,形成酶促反应的速率折线,选择斜率较大的时间段作为最终计算数值,计算标准时间内单位质量的总蛋白水解底物产生的4-MUG的物质的量(nmol·g−1·min−1)。
-
将毛竹全基因组DNA作为模板,利用PCR技术,克隆了PheWOX4c启动子序列,全长为2 045 bp (图2)。测序结果显示扩增出的条带与预期目的序列一致,表明PheWOX4c启动子片段已被成功克隆。
-
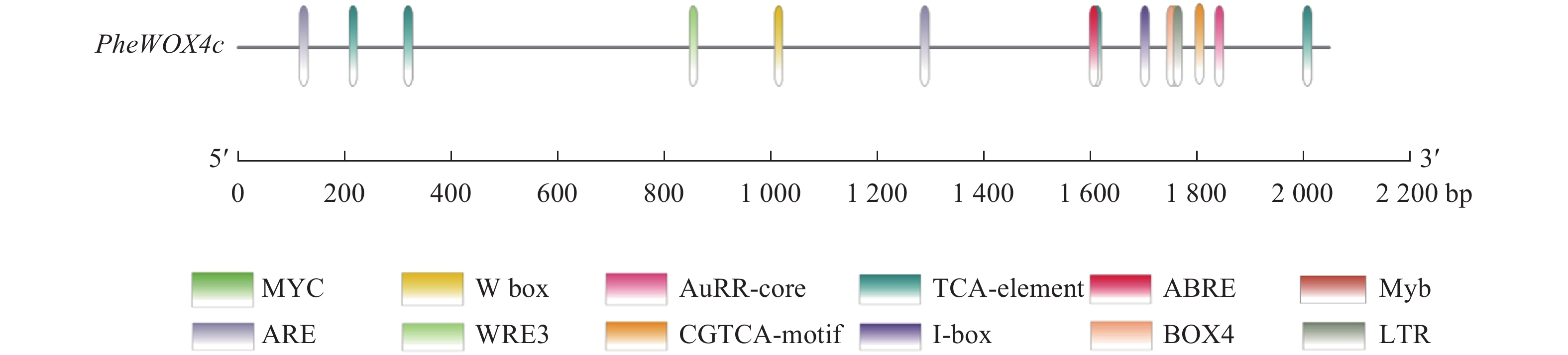
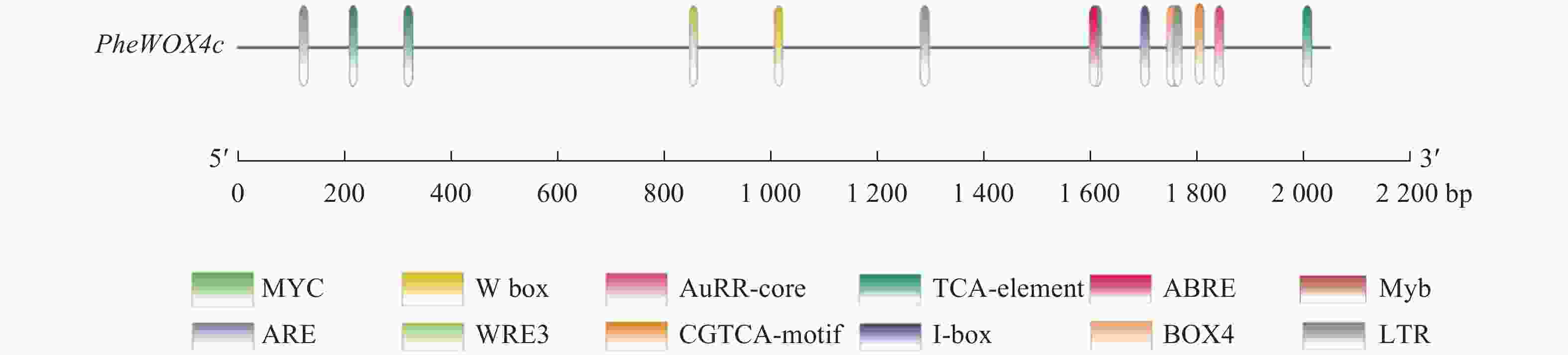
使用Plant CARE在线网站分析启动子序列,发现该启动子上含有ABA响应元件(ABRE)、SA响应元件(TCA-element)、MeJA响应元件(TGACG-motif)和IAA响应元件(AuxRR-core)等植物激素应答元件,低温响应元件(LTR)、转录因子MYB结合位点Myb和bHLH结合位点MYC、厌氧诱导响应元件(ARE)等胁迫应答元件,Box 4、I-box等光响应元件(图3和表2)。此外,还存在种子特异性元件(RY-element)和愈伤调节响应元件(WRE3)。以上表明PheWOX4c基因很可能受到多种植物激素的调节并能对各种逆境做出响应,还可能参与植物生长发育的许多重要过程。
顺式作用元件 序列(5′→3′) 功能 数量 位置/bp TGACG-motif TGACG MeJA响应元件 4 −2 014、−1 619、−326等 I-box CGATAAGGCG 光响应元件 1 −1 817 Box 4 ATTAAT 光响应元件 1 −1 759 ABRE CGTACGTGCA/AACCCGG ABA响应元件 2 −1 301、−132 LTR CCGAAA 低温响应元件 1 −1 710 TCA-element CCATCTTTTT SA响应元件 1 −1 854 AuxRR-core GGTCCAT IAA响应元件 1 −864 ARE AAACCA 厌氧诱导型元件 1 −1 772 WRE3 CCACCT 愈伤调节响应元件 2 −120、−61 RY-element CATGCATG 种子特异性元件 2 −1 768、−1 025 Myb TAACTG MYB转录因子结合位点 1 −516 MYC CATGTG/CATTTG bHLH转录因子结合位点 3 −1 206、−1 144、−230 TATA-box ATATAT/TATA/TATAA/TATATA等 转录起始−30 bp处启动子核心元件 32 −1 783、−1 655、−987等 CAAT-box CAAAT/CAAT/CCAAT 启动子和增强子区域顺式作用元件 58 −1 805、−1 423、−770等 Table 2. Partial cis-acting elements in PheWOX4c gene promoter predicted by Plant CARE
-
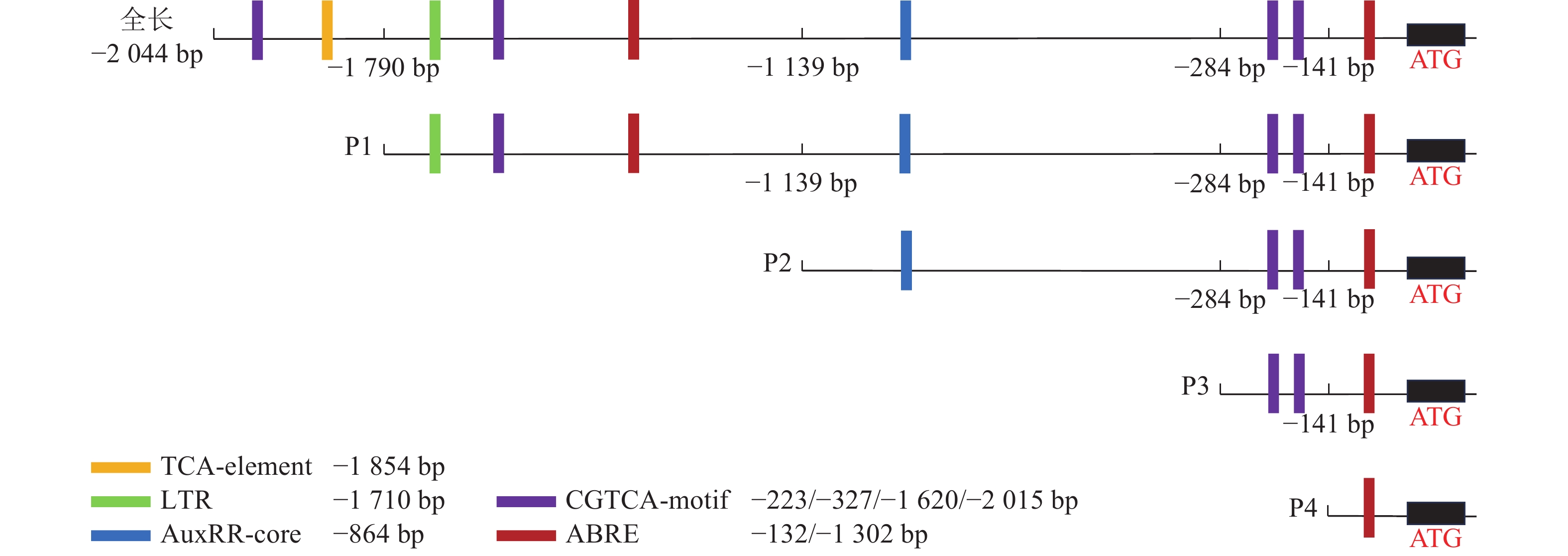
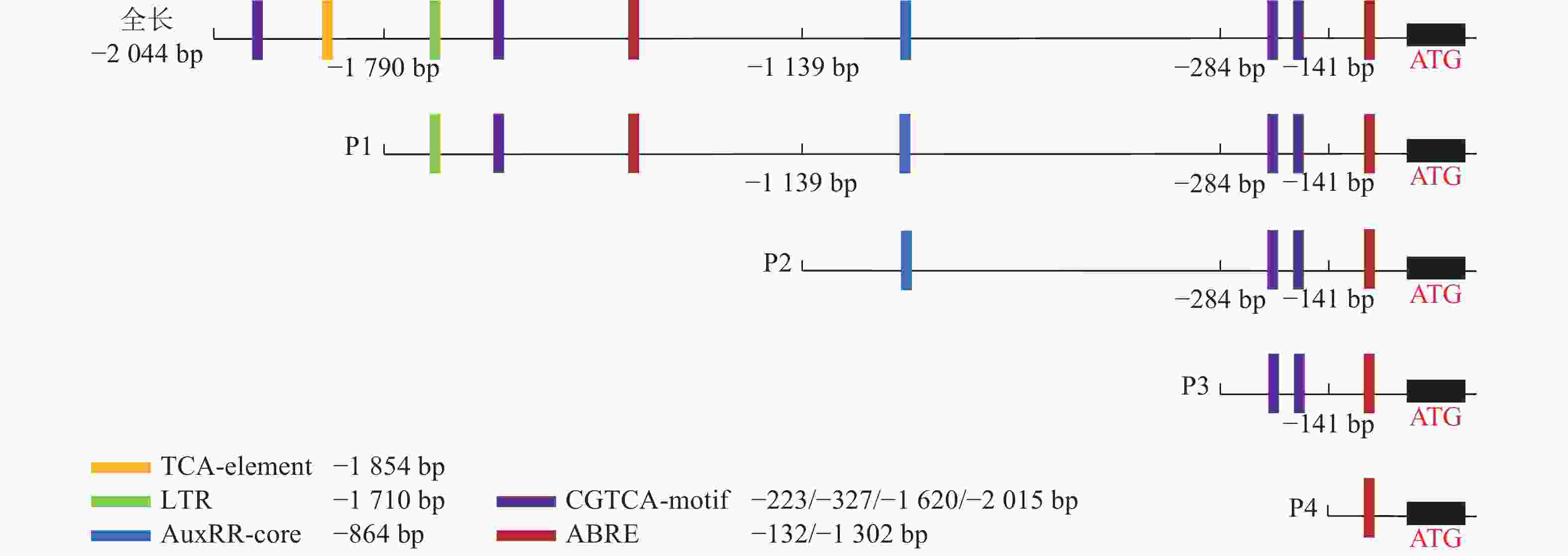
为了探究毛竹PheWOX4c启动子中预测出的激素相关顺式作用元件能否调控基因的表达,根据Plant CARE预测的相关顺式作用元件功能及分布区域,以PheWOX4c启动子序列全长为模板,克隆获得长度为1 745、1 140、507、137 bp的4个5′端缺失片段(图2),分别命名为P1、P2、P3、P4。与全长启动子相比,P1~P4片段分别缺失了SA响应元件TCA-element、LTR响应元件TGA-element、IAA响应元件AuxRR-core、MeJA响应元件CGTCA-motif,最终短片段仅含有ABA响应元件(图4)。
-
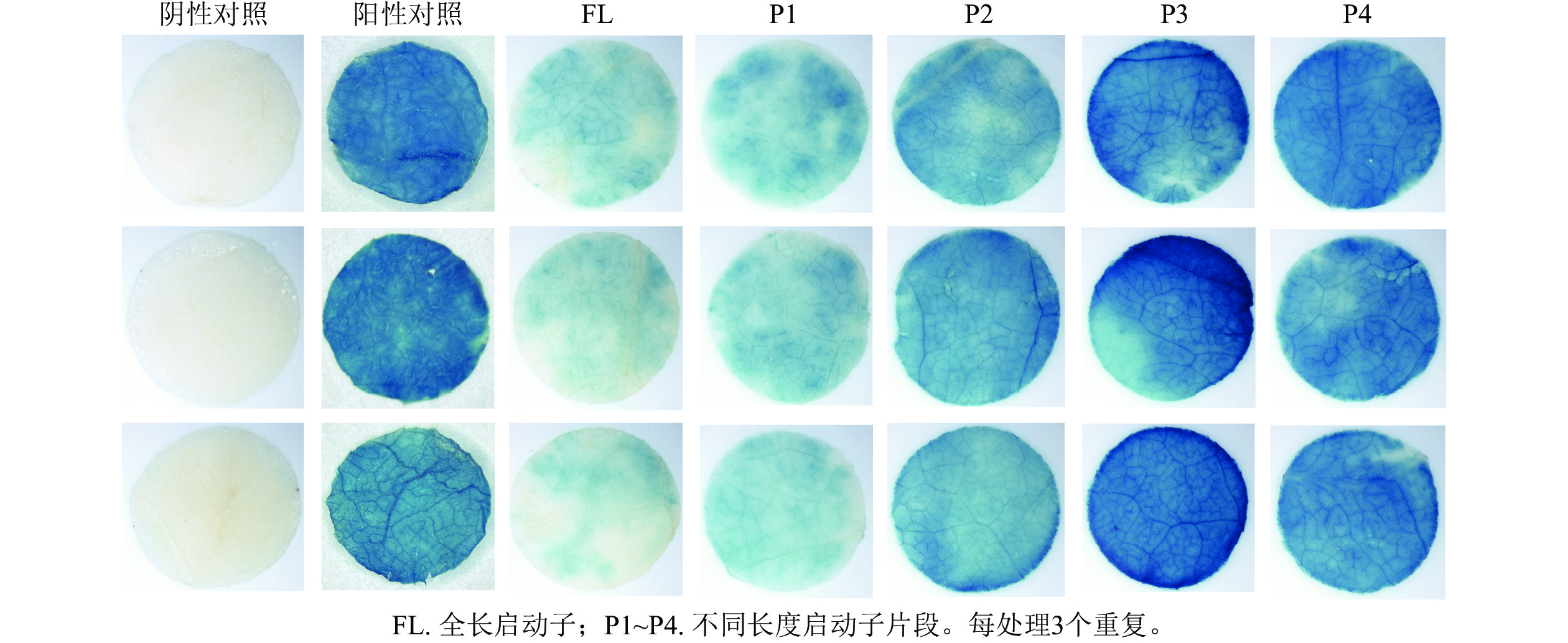
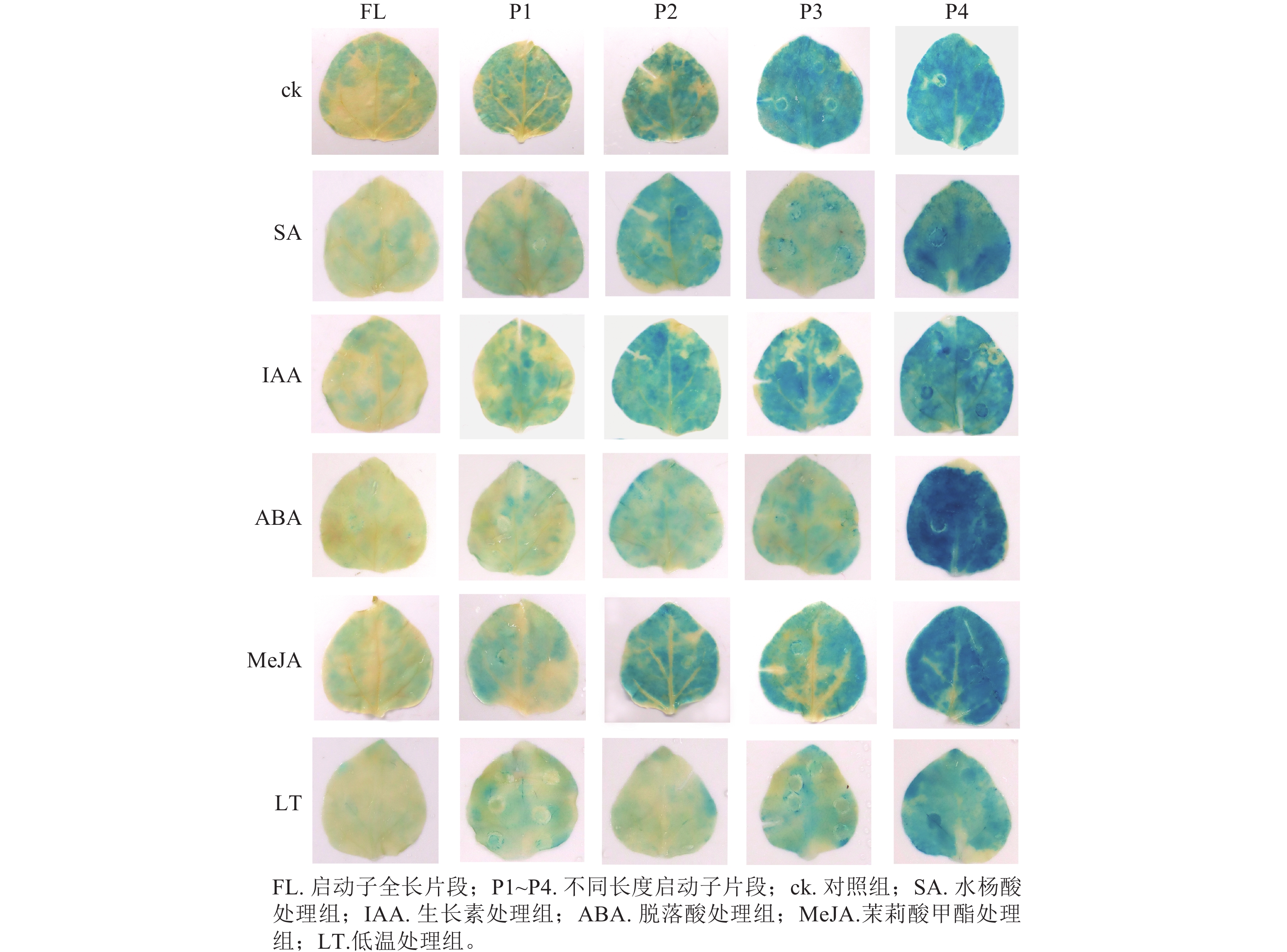
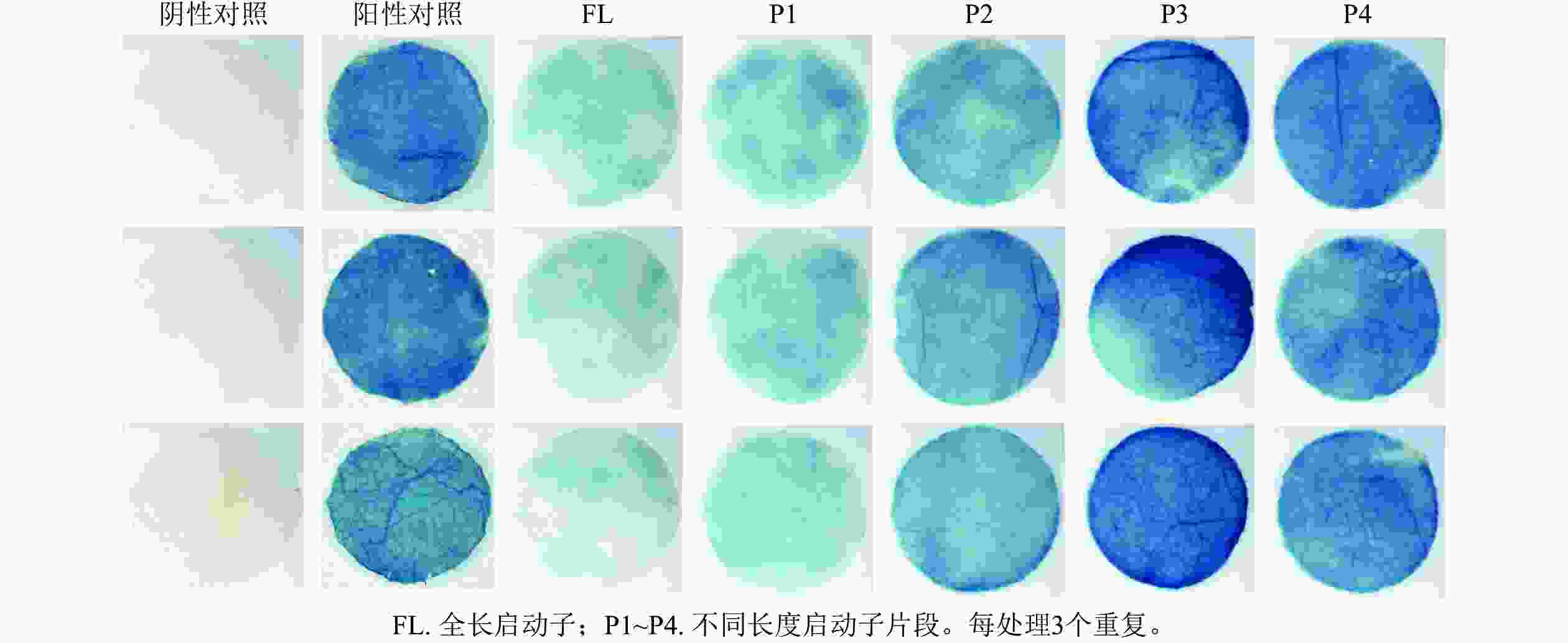
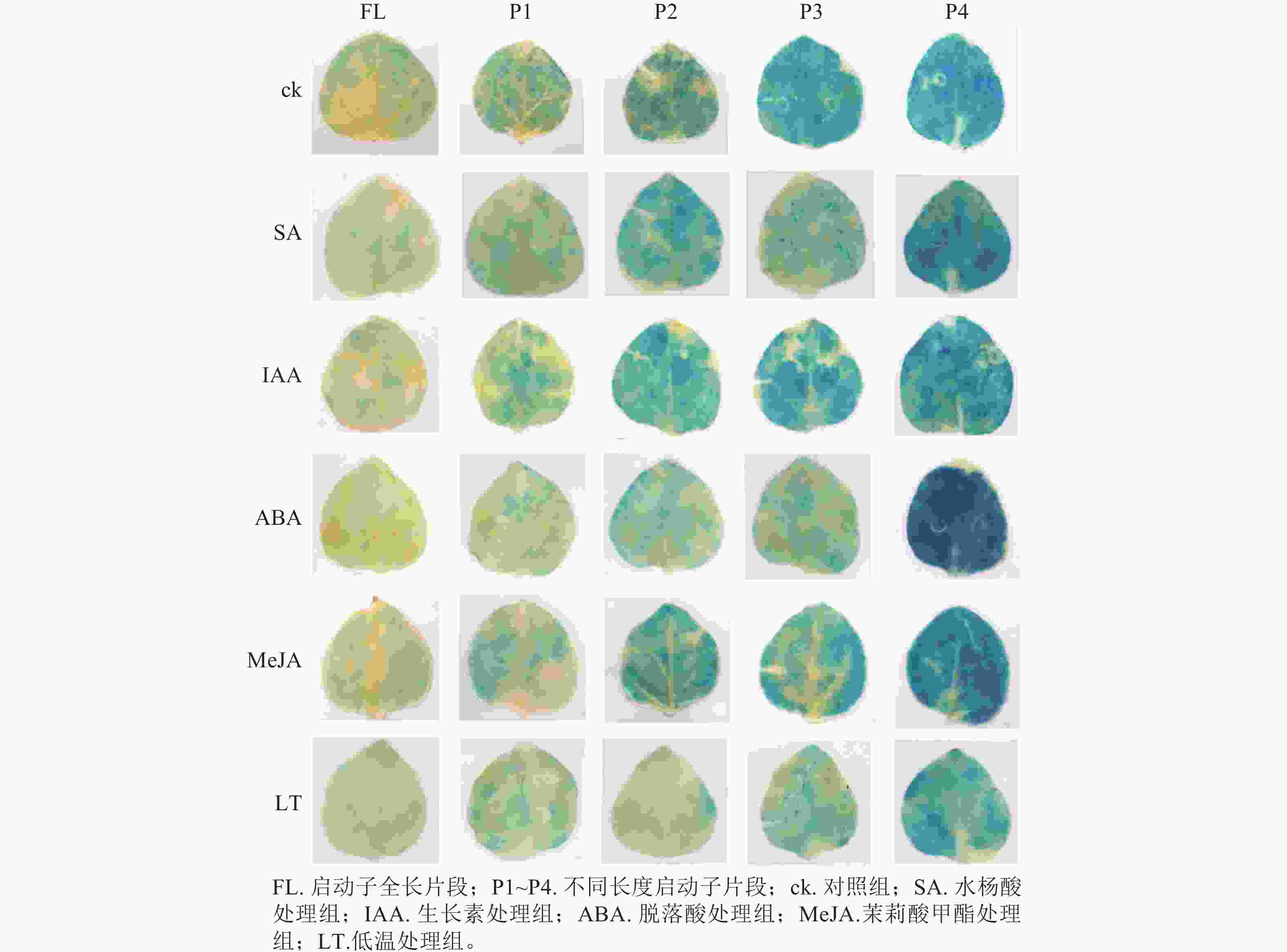
PheWOX4c启动子全长载体及P1~P4载体注射烟草叶片72 h后,对取下的叶片进行 GUS染色并分析不同长度启动子片段的活性,不带有CaMV35S启动子的阴性对照PBI121载体注射烟草叶片无法染上颜色,说明染色处理无背景污染;阳性对照野生型PBI121空载体注射的烟草叶片经过染色变蓝,说明可以正常表达GUS基因;注射启动子全长及P1~P4片段的烟草叶片经GUS染色后均出现明显蓝色,表明PheWOX4c基因启动子全长和4个不同长度启动子缺失片段都具有活性,从染色深浅可以看出启动子活性呈现先升高后降低的趋势(图5)。
-
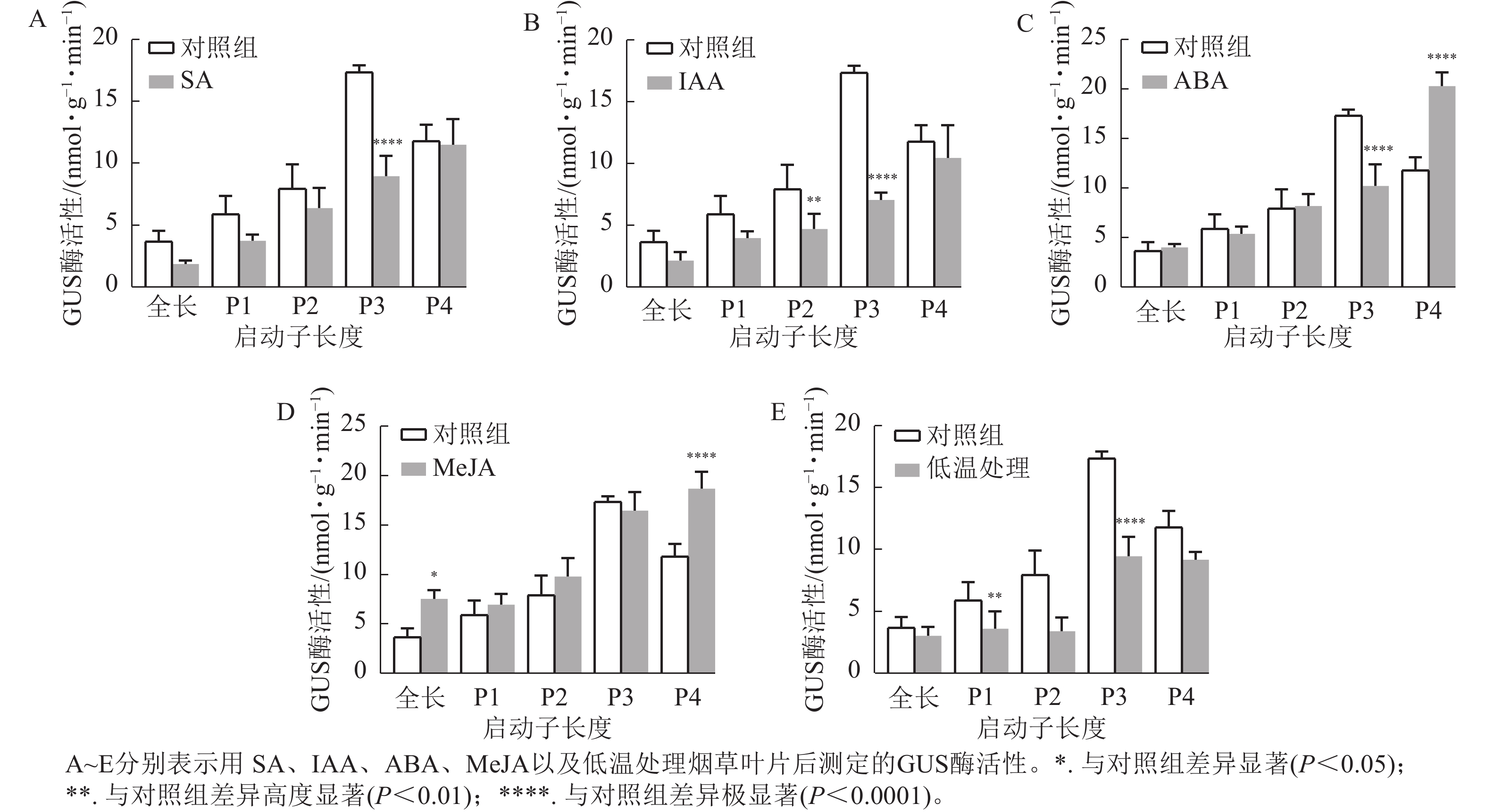
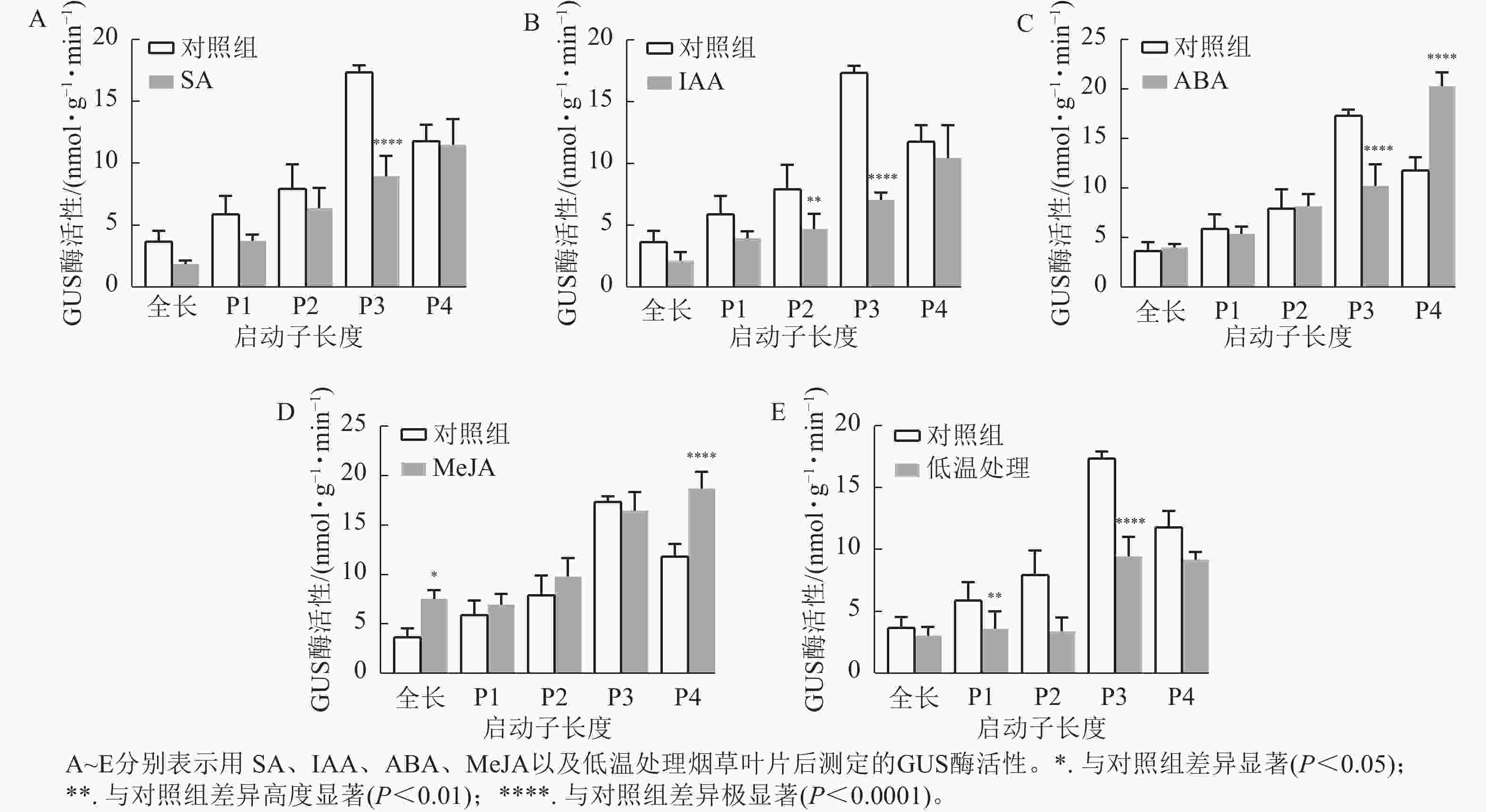
通过测定GUS酶活性和观察GUS染色结果分析各响应元件功能。ABA、SA、IAA以及4 ℃低温处理均不同程度抑制了PheWOX4c基因启动子驱动GUS基因表达的活性。由图6A~B可知:SA和IAA处理下P3缺失片段GUS酶活性比其他片段显著下降(P<
0.0001 ),结合图7中P3片段颜色相较于对照组明显变浅,推测在启动子−507~−130 bp中存在SA的负调控响应元件。由图6C可知:ABA处理下,P3片段GUS酶活性抑制作用显著(P<0.0001 ),而P4片段在ABA处理下的染色结果明显深于对照组(图7),酶活性呈现显著促进作用(P<0.0001 ),推测在启动子−507~−130 bp存在负调控的ABA响应元件,在−137~0 bp存在ABA应答增强元件。由图6D可知:MeJA处理下,启动子全长和P4片段GUS酶活性提高,结合染色结果(图7),推测启动子−2 045~−1 745 bp和−137~0 bp存在正调控的MeJA响应元件。由图6E可知:在低温处理下,启动子全长与各长度片段GUS酶活性均有不同程度下降,其中P3片段抑制作用最显著(P<0.0001 ),与图7的叶片染色结果相吻合。 -
WOX家族基因在植物的生长发育中扮演着重要角色,同时在植物对干旱、盐和低温等逆境的响应中也发挥着重要功能[22−24]。启动子是基因序列中非常重要的一部分,能引导RNA聚合酶以一种精确而有序的方式进行转录。这种特性使得启动子能够控制基因的起始、延伸以及终止,进而影响基因的时空表达[25−26]。因此,对PheWOX4c基因启动子进行结构与功能研究,可以更全面地了解其背后的调控机理。通过克隆并分析PheWOX4c基因的启动子序列,发现PheWOX4c启动子在具有基本顺式作用元件TATA-box、CAAT-box的同时,还存在多种植物激素调控元件、低温响应元件、光响应元件以及种子特异性反应元件等,这表明PheWOX4c基因可能与种子萌发等生长发育过程相关,并且光照、激素以及各种胁迫也可能影响该基因的表达。根据毛竹PheWOX4c启动子序列上含有的植物激素应答元件以及低温响应元件,设计不同长度启动子片段植物表达载体,对瞬时转化启动子全长以及P1、P2、P3和P4片段长度的烟草叶片喷施SA、ABA、IAA和MeJA 等4种激素以及低温处理,GUS染色与酶活性测定结果显示不同处理对各启动子片段的活性有不同影响。
已有研究分析了不同生长期的毛竹笋茎秆上、中以及下部内源激素含量,发现IAA含量与笋的生长发育密切相关。IAA通过促进茎秆细胞的分裂来调控竹子的高生长[27−29],而在木本植物中IAA诱导的MP/ARF5会抑制WOX4基因活性,并限制干细胞数量[30]。双分子荧光互补实验验证了PheIAA15和PheWOX4c能够直接互作,推测毛竹具有独特的IAA响应模式[20]。本研究中IAA处理明显抑制PheWOX4c启动子活性,推测PheWOX4c基因表达会受到IAA影响,从而参与调控毛竹生长发育的重要过程。研究表明ABA通过调控细胞壁发育以及木质素合成相关功能基因的上游调控因子,促进毛竹单节间(SCW)增厚[31],而SCW增厚相关的基因在毛竹快速生长阶段节间中的表达水平远高于快速生长阶段,这表明细胞壁的快速木质化对竹子茎秆快速生长尤为重要[32]。PheWOX4c启动子序列存在2个ABA应答元件ABRE,注射P4片段的烟草叶片在ABA处理下启动子活性明显增强,推测PheWOX4c启动子序列上受ABA诱导的顺式作用元件与毛竹竹笋快速生长密切相关。
此外,植物中WOX基因的表达也受逆境胁迫的影响,包括构树Broussonetia papyrifera、茶树Camellia sinensis、小黑杨Populus nigra和麻风树Jatropha curcas在内的木本植物中的大多数WOX基因在不同程度上响应干旱、盐和低温等各种非生物胁迫刺激[15, 33]。如黄瓜Cucumis sativus叶片中CsWOX4基因表达量会在受低温胁迫后下降[34];乌拉尔图小麦Triticum urartu在低温胁迫下TuWOX4基因表达量显著降低[35]。PheWOX4c启动子上存在低温响应元件,GUS酶活性和染色结果证实低温抑制了PheWOX4c启动子活性。植物激素也是植物体应对生物胁迫重要的防御信号分子[34],如SA主要通过受体蛋白NPR调控相关的bHLH及MYB类转录因子[36],影响下游相关抗逆基因及次生代谢物合成途径基因的表达,提高植物抗逆性[37]。MeJA主要通过靶蛋白茉莉酸ZIM结构域(JAZ)的泛素化降解释放MYC2、MYC3、MYC4等转录因子活性,进而激活JA信号通路相关基因的表达[38]。PheWOX4c启动子存在转录因子MYB结合位点Myb和bHLH结合位点MYC。在本研究中,发现SA和MeJA处理对启动子活性有抑制作用,而P4片段启动子活性在MeJA处理下显著升高,推测该位置具有响应MeJA的正调控元件,PheWOX4c基因表达可能受MeJA和SA的调控,进而影响植物对逆境胁迫的应答。
综上,PheWOX4c基因启动子上不同顺式元件驱动基因表达效率的不同,反映出激素和低温调控PheWOX4c基因表达是一个相对复杂的过程。
-
本研究克隆了毛竹PheWOX4c基因启动子,并通过生物信息学分析预测了启动子序列上可能存在的顺式作用元件,通过对瞬时转化后的烟草进行激素处理,发现PheWOX4c基因启动子可能存在SA、IAA和低温负调控元件,存在MeJA和ABA正调控元件。
Cloning and regulatory element analysis of PheWOX4c gene promoter in Phyllostachys edulis
doi: 10.11833/j.issn.2095-0756.20250198
- Received Date: 2025-03-13
- Accepted Date: 2025-06-23
- Rev Recd Date: 2025-06-20
- Available Online: 2026-01-27
- Publish Date: 2026-02-20
-
Key words:
- Phyllostachys edulis /
- WOX4 gene /
- promoter activity /
- cis-acting element /
- hormone
Abstract:
| Citation: | SHEN Zhuting, REN Zheng, ZHOU Mingbing. Cloning and regulatory element analysis of PheWOX4c gene promoter in Phyllostachys edulis[J]. Journal of Zhejiang A&F University, 2026, 43(1): 66−75 doi: 10.11833/j.issn.2095-0756.20250198 |









 DownLoad:
DownLoad: